Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (1): 225-240.DOI: 10.12211/2096-8280.2022-051
• Research Article • Previous Articles
Mutator-driven continuous genome evolution of Saccharomyces cerevisiae
PAN Yingjia1,2, XIA Siyang1, DONG Chang2, CAI Jin1, LIAN Jiazhang1,2
- 1.Key Laboratory of Biomass Chemical Engineering of Ministry of Education,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310027,Zhejiang,China
2.ZJU-Hangzhou Global Scientific and Technological Innovation Center,Zhejiang University,Hangzhou 310000,Zhejiang,China
-
Received:2022-09-21Revised:2022-12-14Online:2023-03-07Published:2023-02-28 -
Contact:CAI Jin, LIAN Jiazhang
基因增变器驱动的酿酒酵母基因组连续进化
潘颖佳1,2, 夏思杨1, 董昌2, 蔡谨1, 连佳长1,2
- 1.浙江大学化学工程与生物工程学院,浙江 杭州 310027
2.浙江大学杭州国际科创中心,浙江 杭州 310000
-
通讯作者:蔡谨,连佳长 -
作者简介:潘颖佳 (1992—),女,博士研究生。研究方向为合成生物学。E-mail:12228048@zju.edu.cn夏思杨 (1996—),女,硕士研究生。研究方向为基因组进化。E-mail:21828174@zju.edu.cn蔡谨 (1960—),男,博士,副教授。研究方向为工业微生物学。E-mail:caij@zju.edu.cn连佳长 (1984—),男,博士,研究员。研究方向为合成生物学。E-mail:jzlian@zju.edu.cn -
基金资助:国家自然科学基金(21808199);国家重点研发计划(2018YFA0901800);浙江省杰出青年基金(LR20B060003);中央高校基本科研业务费专项资金(226-2022-00214)
CLC Number:
Cite this article
PAN Yingjia, XIA Siyang, DONG Chang, CAI Jin, LIAN Jiazhang. Mutator-driven continuous genome evolution of Saccharomyces cerevisiae[J]. Synthetic Biology Journal, 2023, 4(1): 225-240.
潘颖佳, 夏思杨, 董昌, 蔡谨, 连佳长. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-051
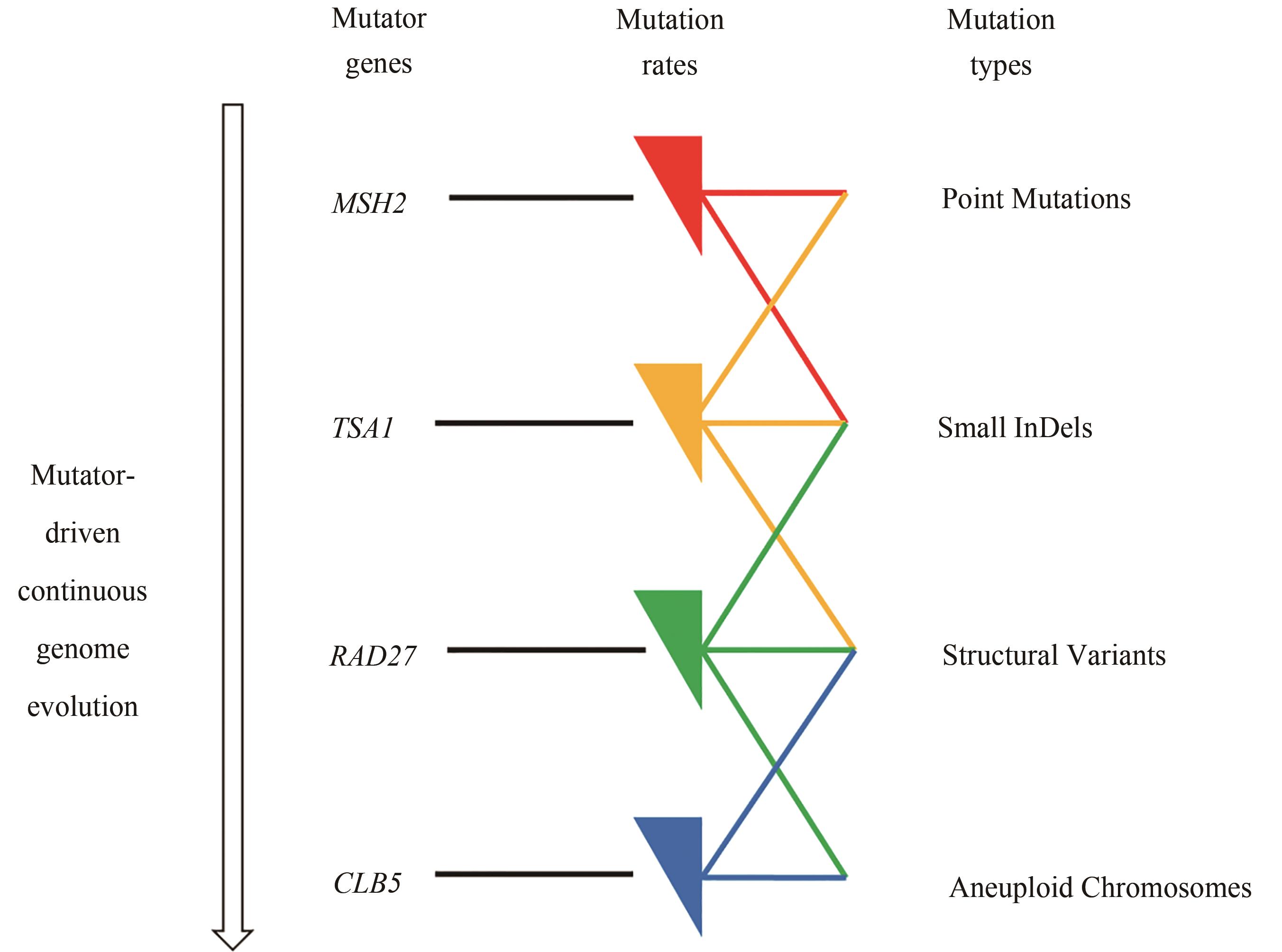
Fig. 1 Continuous genome evolution of S. cerevisiae through mutator genes(There are mutator genes in S. cerevisiae to maintain its genetic stability, and their knockdown or suppression can increase the genome mutation rate, and also result in different types of mutations. Using the CRISPRi system to inhibit the expression of mutator genes, four mutators were constructed to induce different types of mutations in the genome of S. cerevisiae.)
| Strains | Description | Source |
|---|---|---|
| BY4741-iAID6 | BY4741 with CRISPRi machinery (dSpCas9-RD1152) integrated | Our lab[ |
| y426 | BY4741-iAID6/p426 | This study |
| yMM | BY4741-iAID6/MM | This study |
| yTM | BY4741-iAID6/TM | This study |
| yRM | BY4741-iAID6/RM | This study |
| yCM | BY4741-iAID6/CM | This study |
| BY4741-iAID6-CrtIEYB | BY4741-iAID6-CrtE-CrtYB-CrtI | Our lab[ |
| yCar426 | BY4741-iAID6-CrtE-CrtYB-CrtI/p426 | This study |
| yCarMM | BY4741-iAID6-CrtE-CrtYB-CrtI/MM | This study |
| yCarTM | BY4741-iAID6-CrtE-CrtYB-CrtI/TM | This study |
| yCarRM | BY4741-iAID6-CrtE-CrtYB-CrtI/RM | This study |
| yCarCM | BY4741-iAID6-CrtE-CrtYB-CrtI/CM | This study |
| BY4741-iAID6-psXP | BY4741-iAID6-XR-XDH-XKS | This study |
| yX426 | BY4741-iAID6-XR-XDH-XKS/p426 | This study |
| yXMM | BY4741-iAID6-XR-XDH-XKS/MM | This study |
| yXRM | BY4741-iAID6-XR-XDH-XKS/RM | This study |
| yXTM | BY4741-iAID6-XR-XDH-XKS/TM | This study |
| yXCM | BY4741-iAID6-XR-XDH-XKS/CM | This study |
| yXMTRC | BY4741-iAID6-XR-XDH-XKS/MTRC | This study |
| yXMTRC*2 | BY4741-iAID6-XR-XDH-XKS/MTRC-MTRC | This study |
| CT | CEN-PK2-1C; TEF1p-mVenus-PGK1t; URA3 | Our lab[ |
| MSH2p-mVenus | BY4741-iAID6/p415-MSH2p-mVenus | This study |
| TSA1p-mVenus | BY4741-iAID6/p415-TSA1p-mVenus | This study |
| RAD27p-mVenus | BY4741-iAID6/p415-RAD27p-mVenus | This study |
| CLB5p-mVenus | BY4741-iAID6/p415-CLB5p-mVenus | This study |
Table 1 S. cerevisiae strains used in this study
| Strains | Description | Source |
|---|---|---|
| BY4741-iAID6 | BY4741 with CRISPRi machinery (dSpCas9-RD1152) integrated | Our lab[ |
| y426 | BY4741-iAID6/p426 | This study |
| yMM | BY4741-iAID6/MM | This study |
| yTM | BY4741-iAID6/TM | This study |
| yRM | BY4741-iAID6/RM | This study |
| yCM | BY4741-iAID6/CM | This study |
| BY4741-iAID6-CrtIEYB | BY4741-iAID6-CrtE-CrtYB-CrtI | Our lab[ |
| yCar426 | BY4741-iAID6-CrtE-CrtYB-CrtI/p426 | This study |
| yCarMM | BY4741-iAID6-CrtE-CrtYB-CrtI/MM | This study |
| yCarTM | BY4741-iAID6-CrtE-CrtYB-CrtI/TM | This study |
| yCarRM | BY4741-iAID6-CrtE-CrtYB-CrtI/RM | This study |
| yCarCM | BY4741-iAID6-CrtE-CrtYB-CrtI/CM | This study |
| BY4741-iAID6-psXP | BY4741-iAID6-XR-XDH-XKS | This study |
| yX426 | BY4741-iAID6-XR-XDH-XKS/p426 | This study |
| yXMM | BY4741-iAID6-XR-XDH-XKS/MM | This study |
| yXRM | BY4741-iAID6-XR-XDH-XKS/RM | This study |
| yXTM | BY4741-iAID6-XR-XDH-XKS/TM | This study |
| yXCM | BY4741-iAID6-XR-XDH-XKS/CM | This study |
| yXMTRC | BY4741-iAID6-XR-XDH-XKS/MTRC | This study |
| yXMTRC*2 | BY4741-iAID6-XR-XDH-XKS/MTRC-MTRC | This study |
| CT | CEN-PK2-1C; TEF1p-mVenus-PGK1t; URA3 | Our lab[ |
| MSH2p-mVenus | BY4741-iAID6/p415-MSH2p-mVenus | This study |
| TSA1p-mVenus | BY4741-iAID6/p415-TSA1p-mVenus | This study |
| RAD27p-mVenus | BY4741-iAID6/p415-RAD27p-mVenus | This study |
| CLB5p-mVenus | BY4741-iAID6/p415-CLB5p-mVenus | This study |
| Mutators | gRNA plasmids | ||||
|---|---|---|---|---|---|
| MM | p426-Sg11 | p426-Sg12 | p426-Sg15 | p426-Sg17 | p426-SpSgH |
| TM | p426-Sg21 | p426-Sg22 | p426-Sg24 | p426-Sg26 | p426-SpSgH |
| RM | p426-Sg32 | p426-Sg33 | p426-Sg34 | p426-Sg36 | p426-SpSgH |
| CM | p426-Sg41 | p426-Sg42 | p426-Sg44 | p426-Sg45 | p426-SpSgH |
Table 2 gRNA plasmid libraries for mutators
| Mutators | gRNA plasmids | ||||
|---|---|---|---|---|---|
| MM | p426-Sg11 | p426-Sg12 | p426-Sg15 | p426-Sg17 | p426-SpSgH |
| TM | p426-Sg21 | p426-Sg22 | p426-Sg24 | p426-Sg26 | p426-SpSgH |
| RM | p426-Sg32 | p426-Sg33 | p426-Sg34 | p426-Sg36 | p426-SpSgH |
| CM | p426-Sg41 | p426-Sg42 | p426-Sg44 | p426-Sg45 | p426-SpSgH |
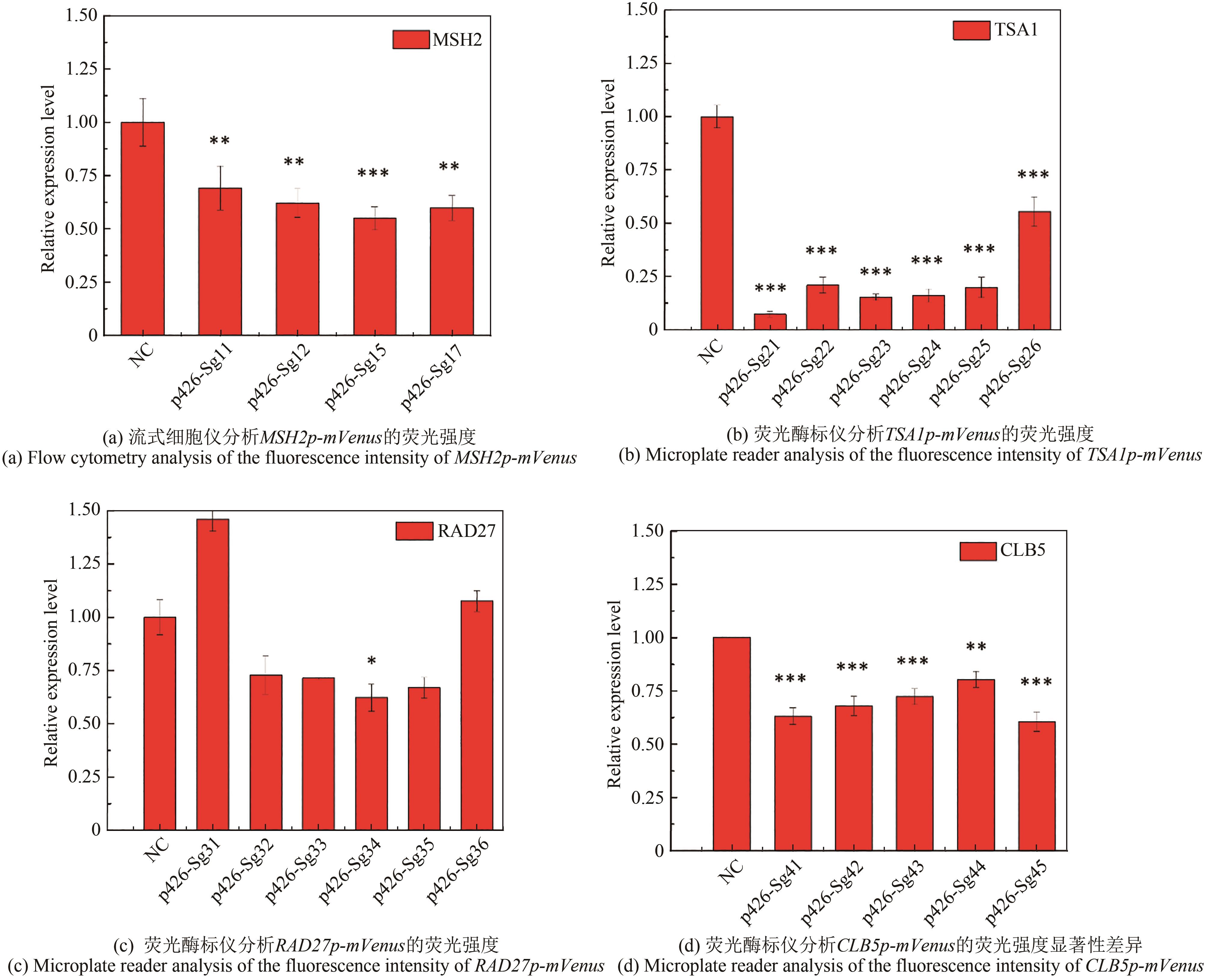
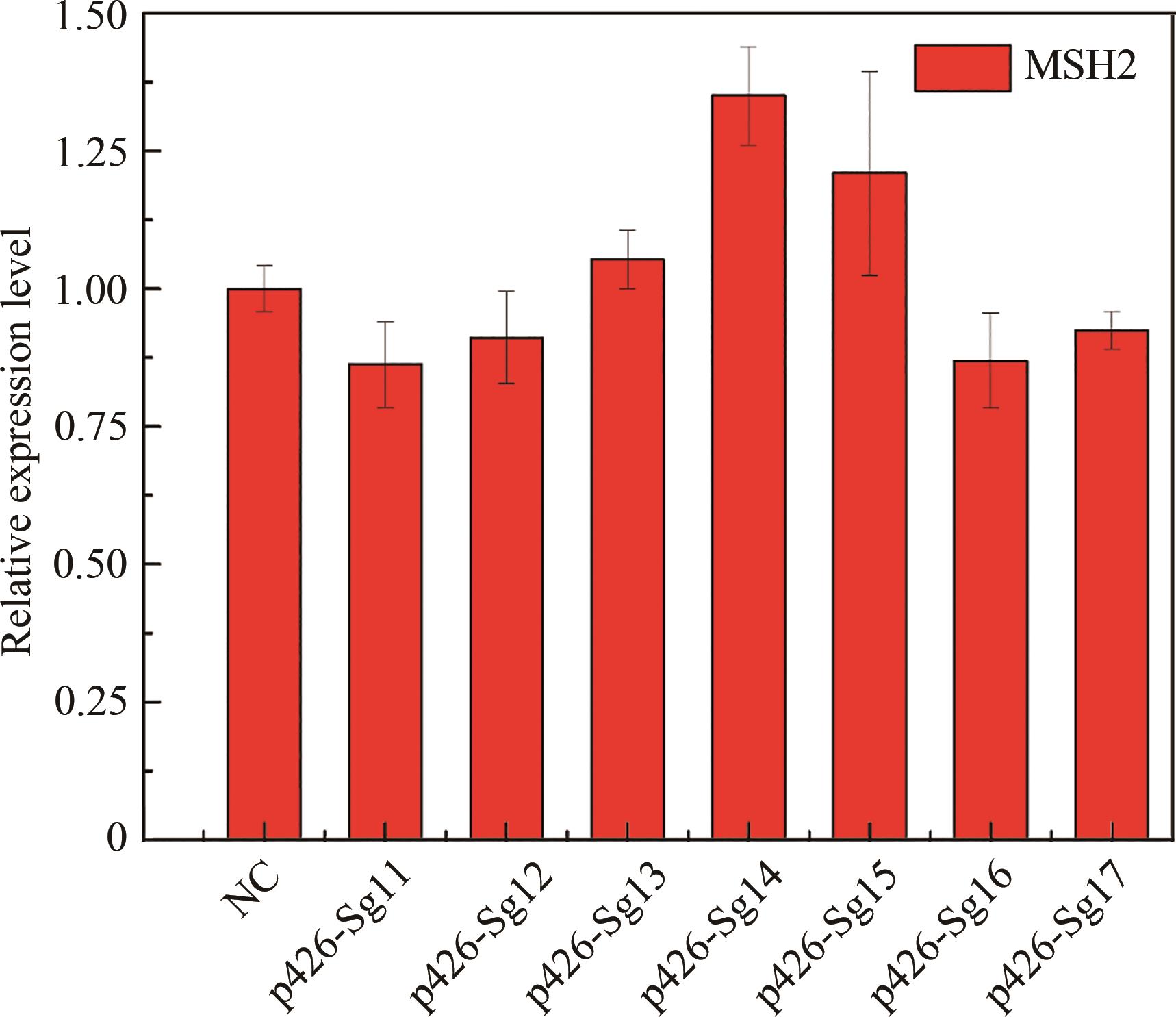
Fig. 2 Inhibition efficiency of different gRNAs on the expression of mutator genes with mVenus as a reporter(*, ** and *** for the significance p < 0.05, 0.01 and 0.001, respectively)

Fig. 3 Mutation rates and types in the genome of S. cerevisiae induced by mutators(a) Relative mutation rates of the genome of S. cerevisiae induced by mutators (* for significance p < 0.05); (b) Mutation type of the genome of S. cerevisiae induced by mutators. All 4 mutators induced SNPs, TM and RM induced small Indels, and RM and CM also induced other mutation types. * and ** for significance p < 0.05 and 0.01, respectively.
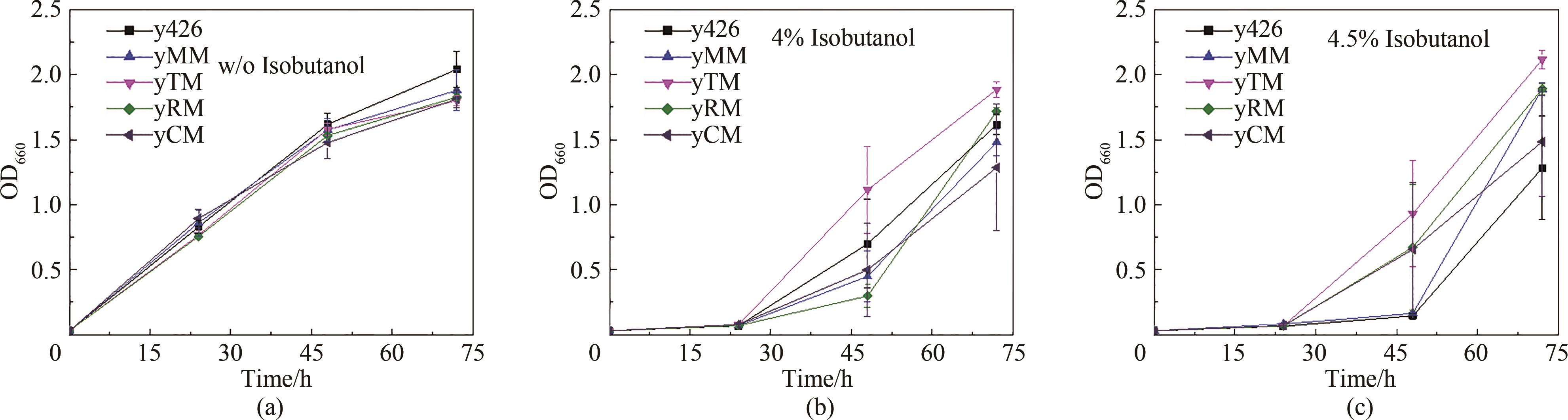
Fig. 4 Evaluation of the mutators for improved isobutanol tolerance(a) No significant difference in the growth profiles of the mutated and control strains when isobutanol was not present in the medium; (b) When the concentration of isobutanol in the medium reached 4%, the growth rate of yTM was significantly higher than that of other strains; (c) When isobutanol concentration reached 4.5%, all mutated strains showed significant growth advantages over the control, with yTM as the best strain.
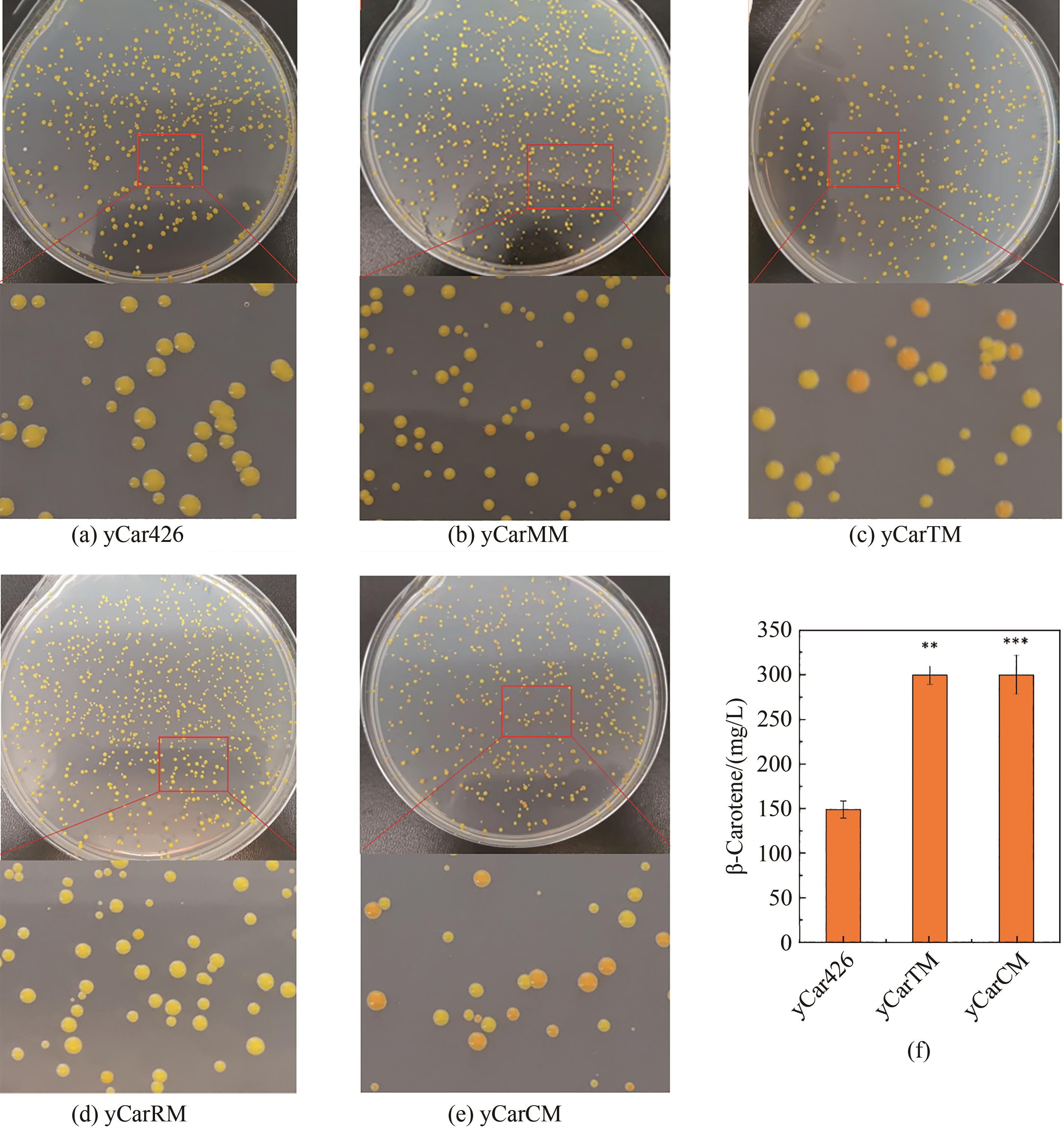
Fig. 5 Evaluation of the mutators in increasing β-carotene biosynthesis(a)~(e) Mutation resulted in the formation of colonies with different color densities; (f) Production of β-carotene in the mutated and control strains determined by HPLC. ** and *** for significance p < 0.01 and 0.001, respectively.

Fig. 6 Evaluation of the mutators in improving xylose utilization[Profiles for growth (a) and xylose consumption (b) of the mutated and control strains with xylose as the sole carbon source. (c) Profiles for growth (hollow symbols) and xylose consumption (solid symbol) of the single (MTRC) and double (MTRC*2) mutated strains. (d) Distribution of gRNA abundance when the MTRC mutated strain was continuously subcultured in the liquid medium containing xylose.]
| Plasmid | Genotype | Source |
|---|---|---|
| pRS415 | CEN/ARS; Amp; LEU2 | Our lab |
| pRS406-RV500 | Amp; URA3; mVenus; mCherry | Our lab |
| p415-MSH2p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-TSA1p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-RAD27p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-CLB5p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p426-SpSgH | 2 μ; Amp; URA3 | Our lab |
| p426-Sg11 | p426-SpSgH target on MSH2 | This study |
| p426-Sg12 | p426-SpSgH target on MSH2 | This study |
| p426-Sg13 | p426-SpSgH target on MSH2 | This study |
| p426-Sg14 | p426-SpSgH target on MSH2 | This study |
| p426-Sg15 | p426-SpSgH target on MSH2 | This study |
| p426-Sg16 | p426-SpSgH target on MSH2 | This study |
| p426-Sg17 | p426-SpSgH target on MSH2 | This study |
| p426-Sg18 | p426-SpSgH target on MSH2 | This study |
| p426-Sg21 | p426-SpSgH target on TSA1 | This study |
| p426-Sg22 | p426-SpSgH target on TSA1 | This study |
| p426-Sg23 | p426-SpSgH target on TSA1 | This study |
| p426-Sg24 | p426-SpSgH target on TSA1 | This study |
| p426-Sg25 | p426-SpSgH target on TSA1 | This study |
| p426-Sg26 | p426-SpSgH target on TSA1 | This study |
| p426-Sg31 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg32 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg33 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg34 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg35 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg36 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg41 | p426-SpSgH target on CLB5 | This study |
| p426-Sg42 | p426-SpSgH target on CLB5 | This study |
| p426-Sg43 | p426-SpSgH target on CLB5 | This study |
| p426-Sg44 | p426-SpSgH target on CLB5 | This study |
| p426-Sg45 | p426-SpSgH target on CLB5 | This study |
Supplementary Table S1 Plasmids used in this study
| Plasmid | Genotype | Source |
|---|---|---|
| pRS415 | CEN/ARS; Amp; LEU2 | Our lab |
| pRS406-RV500 | Amp; URA3; mVenus; mCherry | Our lab |
| p415-MSH2p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-TSA1p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-RAD27p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p415-CLB5p-mVenus | CEN/ARS; Amp; URA3; mVenus | This study |
| p426-SpSgH | 2 μ; Amp; URA3 | Our lab |
| p426-Sg11 | p426-SpSgH target on MSH2 | This study |
| p426-Sg12 | p426-SpSgH target on MSH2 | This study |
| p426-Sg13 | p426-SpSgH target on MSH2 | This study |
| p426-Sg14 | p426-SpSgH target on MSH2 | This study |
| p426-Sg15 | p426-SpSgH target on MSH2 | This study |
| p426-Sg16 | p426-SpSgH target on MSH2 | This study |
| p426-Sg17 | p426-SpSgH target on MSH2 | This study |
| p426-Sg18 | p426-SpSgH target on MSH2 | This study |
| p426-Sg21 | p426-SpSgH target on TSA1 | This study |
| p426-Sg22 | p426-SpSgH target on TSA1 | This study |
| p426-Sg23 | p426-SpSgH target on TSA1 | This study |
| p426-Sg24 | p426-SpSgH target on TSA1 | This study |
| p426-Sg25 | p426-SpSgH target on TSA1 | This study |
| p426-Sg26 | p426-SpSgH target on TSA1 | This study |
| p426-Sg31 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg32 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg33 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg34 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg35 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg36 | p426-SpSgH target on TRAD27 | This study |
| p426-Sg41 | p426-SpSgH target on CLB5 | This study |
| p426-Sg42 | p426-SpSgH target on CLB5 | This study |
| p426-Sg43 | p426-SpSgH target on CLB5 | This study |
| p426-Sg44 | p426-SpSgH target on CLB5 | This study |
| p426-Sg45 | p426-SpSgH target on CLB5 | This study |
| Oligo | Sequences(5′→3′) | Applications |
|---|---|---|
| SpSg11-for | gatcaatatactgaaaataaaaag | To construct gRNA plasmids targeting MSH2 |
| SpSg11-rev | aaacctttttattttcagtatatt | |
| SpSg12-for | gatcgaattttagctctggcctag | |
| SpSg12-rev | aaacctaggccagagctaaaattc | |
| SpSg13-for | gatcgcctttatccactaatctaa | |
| SpSg13-rev | aaacttagattagtggataaaggc | |
| SpSg14-for | gatcggaagttctttgccgttaca | |
| SpSg14-rev | aaactgtaacggcaaagaacttcc | |
| SpSg15-for | gatctatgtatatatagaatatta | |
| SpSg15-rev | aaactaatattctatatatacata | |
| SpSg16-for | gatcttaaaagtatgtcctccact | |
| SpSg16-rev | aaacagtggaggacatacttttaa | |
| SpSg17-for | gatcaaaattctctgatgtatcag | |
| SpSg17-rev | aaacctgatacatcagagaatttt | |
| SpSg18-for | gatcacttctataagaagtataca | |
| SpSg18-rev | aaactgtatacttcttatagaagt | |
| SpSg21-for | gatcaagtcggctggcaacaaacc | To construct gRNA plasmids targeting TSA1 |
| SpSg21-rev | aaacggtttgttgccagccgactt | |
| SpSg22-for | gatcaccaggacatatataaaggg | |
| SpSg22-rev | aaacccctttatatatgtcctggt | |
| SpSg23-for | gatcaaggcccgttgagaacggtt | |
| SpSg23-rev | aaacaaccgttctcaacgggcctt | |
| SpSg24-for | gatcaggggaaggcccgttgagaa | |
| SpSg24-rev | aaacttctcaacgggccttcccct | |
| SpSg25-for | gatcggttgtgagcaattgaacga | |
| SpSg25-rev | aaactcgttcaattgctcacaacc | |
| SpSg26-for | gatcacatacacatacatacacaa | |
| SpSg26-rev | aaacttgtgtatgtatgtgtatgt | |
| SpSg31-for | gatcaaagctttaaacgcgttagg | To construct gRNA plasmids targeting RAD27 |
| SpSg31-rev | aaaccctaacgcgtttaaagcttt | |
| SpSg32-for | gatcgcgtaacatcgcgcaaatga | |
| SpSg32-rev | aaactcatttgcgcgatgttacgc | |
| SpSg33-for | gatcaaagcgttgacagcatacat | |
| SpSg33-rev | aaacatgtatgctgtcaacgcttt | |
| SpSg34-for | gatcaagaaataggaaacggacac | |
| SpSg34-rev | aaacgtgtccgtttcctatttctt | |
| SpSg35-for | gatcgacaccggaagaaaaaatat | |
| SpSg35-rev | aaacatattttttcttccggtgtc | |
| SpSg36-for | gatcaggtttgaatgcaattatat | |
| SpSg36-rev | aaacatataattgcattcaaacct | |
| SpSg41-for | gatcaattggccgtgaaaagcttt | To construct gRNA plasmids targeting CLB5 |
| SpSg41-rev | aaacaaagcttttcacggccaatt | |
| SpSg42-for | gatcctttgtgtgagacaactaat | |
| SpSg42-rev | aaacattagttgtctcacacaaag | |
| SpSg43-for | gatcgttcagcggctttaaataca | |
| SpSg43-rev | aaactgtatttaaagccgctgaac | |
| SpSg44-for | gatctgaacacctttactgaacaa | |
| SpSg44-rev | aaacttgttcagtaaaggtgttca | |
| SpSg45-for | gatctaatactctgctcatggtcg | |
| SpSg45-rev | aaaccgaccatgagcagagtatta | |
| MSH2p-for | NNNNNctcgagattagaattaaaatgtgtag | Used to amplify MSH2p |
| MSH2p-rev | NNNNNggatcctctctcctctgatacatcag | |
| mVenus-for | NNNNNggatccggcggcagcggcggcagcatggaattcgtgagcaagg | Used to amplify mVenus |
| mVenus-rev | NNNNNgagctccaggaagaatacac | |
| TSA1p-for | NNNNNctcgagacagcaggaaaacgaagatg | Used to amplify TSA1p |
| TSA1p-rev | NNNNNggatccgacggcagttttcttaaaag | |
| RAD27p-for | NNNNNctcgagaaacaaaaagaacagggaaag | Used to amplify RAD27p |
| RAD27p-rev | NNNNNggatccagcagagggaacatgttccg | |
| CLB5p-for | NNNNNctcgagtgaagacgcgcccttgatgg | Used to amplify CLB5p |
| CLB5p-rev | NNNNNggatccaatcatagaatttcttttaatac | |
| SpSg-GJ-for | cttcctgggtctcagtcccctcgagcacagggtaataact | To construct gRNA plasmid library MTRC*2 |
| SpSg-GJ-rev | tatctctaggtctcagtgggagctccctgcaggcatg | |
| 2gRNA-F1 | NNNNNggtctccggactctttgaaaagataatgtatg | |
| 2gRNA-R1 | NNNNNggtctcccggacttgcatgcctgcagggagctc | |
| 2gRNA-F2 | NNNNNggtctcctccgtctttgaaaagataatgtatg | |
| 2gRNA-R2 | NNNNNggtctcccaaccttgcatgcctgcagggagctc |
Supplementary Table S2 Primers used in this study
| Oligo | Sequences(5′→3′) | Applications |
|---|---|---|
| SpSg11-for | gatcaatatactgaaaataaaaag | To construct gRNA plasmids targeting MSH2 |
| SpSg11-rev | aaacctttttattttcagtatatt | |
| SpSg12-for | gatcgaattttagctctggcctag | |
| SpSg12-rev | aaacctaggccagagctaaaattc | |
| SpSg13-for | gatcgcctttatccactaatctaa | |
| SpSg13-rev | aaacttagattagtggataaaggc | |
| SpSg14-for | gatcggaagttctttgccgttaca | |
| SpSg14-rev | aaactgtaacggcaaagaacttcc | |
| SpSg15-for | gatctatgtatatatagaatatta | |
| SpSg15-rev | aaactaatattctatatatacata | |
| SpSg16-for | gatcttaaaagtatgtcctccact | |
| SpSg16-rev | aaacagtggaggacatacttttaa | |
| SpSg17-for | gatcaaaattctctgatgtatcag | |
| SpSg17-rev | aaacctgatacatcagagaatttt | |
| SpSg18-for | gatcacttctataagaagtataca | |
| SpSg18-rev | aaactgtatacttcttatagaagt | |
| SpSg21-for | gatcaagtcggctggcaacaaacc | To construct gRNA plasmids targeting TSA1 |
| SpSg21-rev | aaacggtttgttgccagccgactt | |
| SpSg22-for | gatcaccaggacatatataaaggg | |
| SpSg22-rev | aaacccctttatatatgtcctggt | |
| SpSg23-for | gatcaaggcccgttgagaacggtt | |
| SpSg23-rev | aaacaaccgttctcaacgggcctt | |
| SpSg24-for | gatcaggggaaggcccgttgagaa | |
| SpSg24-rev | aaacttctcaacgggccttcccct | |
| SpSg25-for | gatcggttgtgagcaattgaacga | |
| SpSg25-rev | aaactcgttcaattgctcacaacc | |
| SpSg26-for | gatcacatacacatacatacacaa | |
| SpSg26-rev | aaacttgtgtatgtatgtgtatgt | |
| SpSg31-for | gatcaaagctttaaacgcgttagg | To construct gRNA plasmids targeting RAD27 |
| SpSg31-rev | aaaccctaacgcgtttaaagcttt | |
| SpSg32-for | gatcgcgtaacatcgcgcaaatga | |
| SpSg32-rev | aaactcatttgcgcgatgttacgc | |
| SpSg33-for | gatcaaagcgttgacagcatacat | |
| SpSg33-rev | aaacatgtatgctgtcaacgcttt | |
| SpSg34-for | gatcaagaaataggaaacggacac | |
| SpSg34-rev | aaacgtgtccgtttcctatttctt | |
| SpSg35-for | gatcgacaccggaagaaaaaatat | |
| SpSg35-rev | aaacatattttttcttccggtgtc | |
| SpSg36-for | gatcaggtttgaatgcaattatat | |
| SpSg36-rev | aaacatataattgcattcaaacct | |
| SpSg41-for | gatcaattggccgtgaaaagcttt | To construct gRNA plasmids targeting CLB5 |
| SpSg41-rev | aaacaaagcttttcacggccaatt | |
| SpSg42-for | gatcctttgtgtgagacaactaat | |
| SpSg42-rev | aaacattagttgtctcacacaaag | |
| SpSg43-for | gatcgttcagcggctttaaataca | |
| SpSg43-rev | aaactgtatttaaagccgctgaac | |
| SpSg44-for | gatctgaacacctttactgaacaa | |
| SpSg44-rev | aaacttgttcagtaaaggtgttca | |
| SpSg45-for | gatctaatactctgctcatggtcg | |
| SpSg45-rev | aaaccgaccatgagcagagtatta | |
| MSH2p-for | NNNNNctcgagattagaattaaaatgtgtag | Used to amplify MSH2p |
| MSH2p-rev | NNNNNggatcctctctcctctgatacatcag | |
| mVenus-for | NNNNNggatccggcggcagcggcggcagcatggaattcgtgagcaagg | Used to amplify mVenus |
| mVenus-rev | NNNNNgagctccaggaagaatacac | |
| TSA1p-for | NNNNNctcgagacagcaggaaaacgaagatg | Used to amplify TSA1p |
| TSA1p-rev | NNNNNggatccgacggcagttttcttaaaag | |
| RAD27p-for | NNNNNctcgagaaacaaaaagaacagggaaag | Used to amplify RAD27p |
| RAD27p-rev | NNNNNggatccagcagagggaacatgttccg | |
| CLB5p-for | NNNNNctcgagtgaagacgcgcccttgatgg | Used to amplify CLB5p |
| CLB5p-rev | NNNNNggatccaatcatagaatttcttttaatac | |
| SpSg-GJ-for | cttcctgggtctcagtcccctcgagcacagggtaataact | To construct gRNA plasmid library MTRC*2 |
| SpSg-GJ-rev | tatctctaggtctcagtgggagctccctgcaggcatg | |
| 2gRNA-F1 | NNNNNggtctccggactctttgaaaagataatgtatg | |
| 2gRNA-R1 | NNNNNggtctcccggacttgcatgcctgcagggagctc | |
| 2gRNA-F2 | NNNNNggtctcctccgtctttgaaaagataatgtatg | |
| 2gRNA-R2 | NNNNNggtctcccaaccttgcatgcctgcagggagctc |
| Strains | SNPs | Small InDels | Other mutation types |
|---|---|---|---|
| yMM | 16 | 0 | 0 |
| yTM | 12 | 4 | 0 |
| yRM | 12 | 2 | 2 |
| yCM | 4 | 0 | 12 |
Supplementary Table S3 Mutation types of the S. cerevisiae genome induced by mutators
| Strains | SNPs | Small InDels | Other mutation types |
|---|---|---|---|
| yMM | 16 | 0 | 0 |
| yTM | 12 | 4 | 0 |
| yRM | 12 | 2 | 2 |
| yCM | 4 | 0 | 12 |
| 1 | 李祎, 林振泉, 刘子鹤. 酿酒酵母适应性实验室进化工具的最新进展[J]. 合成生物学, 2021, 2(2): 287-301. |
| LI Y, LIN Z Q, LIU Z H. Advances in yeast based adaptive laboratory evolution[J]. Synthetic Biology Journal, 2021, 2(2): 287-301. | |
| 2 | 夏思杨, 江丽红, 蔡谨, 等. 酿酒酵母基因组进化的研究进展[J]. 合成生物学, 2020, 1(5): 556-569. |
| XIA S Y, JIANG L H, CAI J, et al. Advances in genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2020, 1(5): 556-569. | |
| 3 | SI T, XIAO H, ZHAO H M. Rapid prototyping of microbial cell factories via genome-scale engineering[J]. Biotechnology Advances, 2015, 33(7): 1420-1432. |
| 4 | LUAN G D, CAI Z, LI Y, et al. Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production[J]. Biotechnology for Biofuels, 2013, 6(1): 137. |
| 5 | BADRAN A H, LIU D R. In vivo continuous directed evolution[J]. Current Opinion in Chemical Biology, 2015, 24: 1-10. |
| 6 | ARNOLD F H, VOLKOV A A. Directed evolution of biocatalysts[J]. Current Opinion in Chemical Biology, 1999, 3(1): 54-59. |
| 7 | WANG Y J, XUE P, CAO M F, et al. Directed evolution: methodologies and applications[J]. Chemical Reviews, 2021, 121(20): 12384-12444. |
| 8 | 祁延萍, 朱晋, 张凯, 等. 定向进化在蛋白质工程中的应用研究进展 [J]. 合成生物学, 2022, 3(6): 1-29. |
| QI Y P, ZHU J, ZHANG K, et al. Recent development of directed evolution in protein engineering [J]. Synthetic Biology Journal, 2022, 3(6): 1-29. | |
| 9 | SCHELLER J, SCHÜRER A, RUDOLPH C, et al. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage[J]. Genetics, 2000, 155(3): 1069-1081. |
| 10 | SPEYER J F. Mutagenic DNA polymerase[J]. Biochemical and Biophysical Research Communications, 1965, 21(1): 6-8. |
| 11 | CHOU H H, KEASLING J D. Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4: 2595. |
| 12 | DROTSCHMANN K, CLARK A B, TRAN H T, et al. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(6): 2970-2975. |
| 13 | SERERO A, JUBIN C, LOEILLET S, et al. Mutational landscape of yeast mutator strains[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(5): 1897-1902. |
| 14 | REENAN R A, KOLODNER R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions[J]. Genetics, 1992, 132(4): 975-985. |
| 15 | KATJU V, KONRAD A, DEISS T C, et al. Mutation rate and spectrum in obligately outcrossing Caenorhabditis elegans mutation accumulation lines subjected to RNAi-induced knockdown of the mismatch repair gene msh-2 [J]. G3 Genes|Genomes|Genetics, 2021, 12(1): jkab364. |
| 16 | GOTO M, SASAKI M, KOBAYASHI T. The S-phase cyclin Clb5 promotes rRNA gene (rDNA) stability by maintaining replication initiation efficiency in rDNA[J]. Molecular and Cellular Biology, 2021, 41(5): e00324-e00320. |
| 17 | TISHKOFF D X, FILOSI N, GAIDA G M, et al. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair[J]. Cell, 1997, 88(2): 253-263. |
| 18 | MCCULLEY J L, PETES T D. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(25): 11465-11470. |
| 19 | PENNANEACH V, KOLODNER R D. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast[J]. Nature Genetics, 2004, 36(6): 612-617. |
| 20 | PARK H, KIM S. Gene-specific mutagenesis enables rapid continuous evolution of enzymes in vivo [J]. Nucleic Acids Research, 2021, 49(6): e32. |
| 21 | YONA A H, MANOR Y S, HERBST R H, et al. Chromosomal duplication is a transient evolutionary solution to stress[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(51): 21010-21015. |
| 22 | GILBERT L A, LARSON M H, MORSUT L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes[J]. Cell, 2013, 154(2): 442-451. |
| 23 | YAO Z, WANG Q H, DAI Z J. Recent advances in directed yeast genome evolution[J]. Journal of Fungi, 2022, 8(6): 635. |
| 24 | LIAN J Z, HAMEDIRAD M, HU S M, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nature Communications, 2017, 8: 1688. |
| 25 | GIETZ R D, SCHIESTL R H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method[J]. Nature Protocols, 2007, 2(1): 31-34. |
| 26 | PATTERSON M N, MAXWELL P H. Combining magnetic sorting of mother cells and fluctuation tests to analyze genome instability during mitotic cell aging in Saccharomyces cerevisiae [J]. Journal of Visualized Experiments: JoVE, 2014(92): e51850. |
| 27 | BOEKE J D, LACROUTE F, FINK G R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance[J]. Molecular & General Genetics: MGG, 1984, 197(2): 345-346. |
| 28 | WHELAN W L, GOCKE E, MANNEY T R. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates[J]. Genetics, 1979, 91(1): 35-51. |
| 29 | QI M M, ZHANG B, JIANG L H, et al. PCR & Go: a pre-installed expression chassis for facile integration of multi-gene biosynthetic pathways[J]. Frontiers in Bioengineering and Biotechnology, 2021, 8: 613771. |
| 30 | TSAI C S, KONG I I, LESMANA A, et al. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR[J]. Biotechnology and Bioengineering, 2015, 112(11): 2406-2411. |
| 31 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. |
| [1] | Wentao SUN, Xinzhe ZHANG, Shengtong WAN, Ruwen WANG, Chun LI. Regulation on oxidation selectivity for β-amyrin by Class Ⅱ cytochrome P450 enzymes [J]. Synthetic Biology Journal, 2021, 2(5): 804-814. |
| [2] | Xiaodong LI, Chengshuai YANG, Pingping WANG, Xing YAN, Zhihua ZHOU. Production of sesquiterpenoids α-neoclovene and β-caryophyllene by engineered Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2021, 2(5): 792-803. |
| [3] | Yi LI, Zhenquan LIN, Zihe LIU. Advances in yeast based adaptive laboratory evolution [J]. Synthetic Biology Journal, 2021, 2(2): 287-301. |
| [4] | Yue SHENG, Genlin ZHANG. Yeast terminator engineering: from mechanism exploration to artificial design [J]. Synthetic Biology Journal, 2020, 1(6): 709-721. |
| [5] | Siyang XIA, Lihong JIANG, Jin CAI, Lei HUANG, Zhinan XU, Jiazhang LIAN. Advances in genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2020, 1(5): 556-569. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||