|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
CRISPR/Cas systems and their applications in gene editing with filamentous fungi
Synthetic Biology Journal
2024, 5 (3):
672-693.
DOI: 10.12211/2096-8280.2023-097
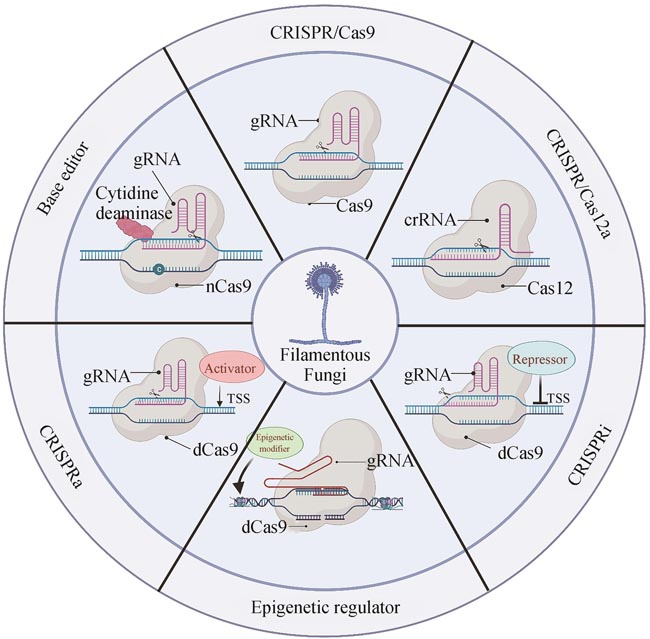
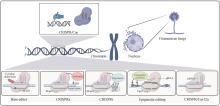
Filamentous fungi, which present distinct morphology and cell structure, play a critical role in human health as well as industrial and agricultural production. However, the unique characteristics of filamentous fungi make them difficult to be manipulated with traditional genetic engineering methods. Thus, the development of an efficient gene editing system is essential for exploring biological resources and understanding metabolic processes in filamentous fungi. The development of the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein (CRISPR/Cas) system promotes more efficient and effective gene editing in different species, and brings a revolutionary breakthrough in fungal fundamental research and applications. In this review, we first briefly introduce the history, working mechanism, and classifications of the CRISPR/Cas mediated gene editing system. Next, we comment the functional components of CRISPR/Cas9 such as selective marker, Cas9 and gRNA and the delivery methods of these components in various filamentous fungi. Furthermore, we systematically discuss the applications of CRISPR related technologies, including CRISPR/Cas12, base-editor, CRISPRa, CRISPRi and CRISPR mediated epigenetic regulation, in the genetic engineering of filamentous fungi, particularly in marine-derived filamentous fungi. Finally, we address challenges with relative low gene editing efficiency and off-targets effects in engineering filamentous fungi, and highlight the potential solutions for developing novel CRISPR/Cas-based gene editing systems. This review can provide guidance for developing an efficient gene editing platform in filamentous fungi and pave the way for further exploration of the secondary metabolites and establishment of robust fungal cell factories.
Table 2
Screening markers applied in CRISPR/Cas systems for engineering filamentous fungi
Extracts from the Article
丝状真菌,尤其是各种特殊生境来源的野生真菌,选择性标记的缺乏是突变体筛选的主要障碍之一。常用的筛选标记包括:抗性标记、营养标记和表型报告标记。表型报告基因通常来源于特定物种中的独特表型,如一些色素合成基因、生长关键基因等,这些基因通常不具有通用性。综合现有的综述数据和研究论文[41],现将丝状真菌中应用于CRISPR/Cas9的筛选标记总结于表2中。
表观遗传修饰是指染色体DNA和组蛋白上的化学修饰,包括DNA甲基化、组蛋白甲基化、组蛋白乙酰化、组蛋白磷酸化等,在不改变DNA序列的情况下,这些修饰可以通过改变染色质状态来控制基因的表达,并具有可遗传性。CRISPR表观遗传编辑是指将失去切割活性的Cas蛋白与表观遗传修饰因子融合来靶向改变目标区域的表观遗传修饰标记,从而影响基因的转录[135]。与传统的作用于全基因组的表观遗传调控手段相比,CRISPR表观遗传编辑因其可实现高效的靶向特异性表观遗传修饰,在生命科学研究中具有极大的应用前景。CRISPR表观遗传编辑系统的改造对象主要包括DNA的甲基化修饰和染色质上组蛋白的表观修饰。基于人源E1A相关蛋白p300的组蛋白乙酰转移酶(HAT)核心结构域与dCas9融合的基因激活系统,在人类细胞和多个物种表现出精准且高效的基因诱导活性[136-137]。2021年,研究人员首次将dCas9-p300表观编辑系统应用于丝状真菌中[138]。在黑曲霉中,dCas9-p300可诱导多个次级代谢产物合成基因的表达,并提高了伏马毒素B2的产量。此外,研究人员还将dCas9分别与黑曲霉内源性组蛋白乙酰化酶GcnE、组蛋白去乙酰化酶HosA和RpdA融合,构建了多个CRISPR表观遗传编辑系统。测试结果发现dCas9-GcnE/HosA/RpdA编辑体系均可抑制次级代谢产物合成基breF的表达,dCas9-HosA还可上调色素合成基因fwnA的表达[138]。这些研究结果表明丝状真菌中还存在其他因素可影响内源性表观遗传调控因子的调控效果,在推广应用丝状真菌来源的各种表观调控因子之前,还需更深入地研究这些表观调控因子的作用机制。
Other Images/Table from this Article
|