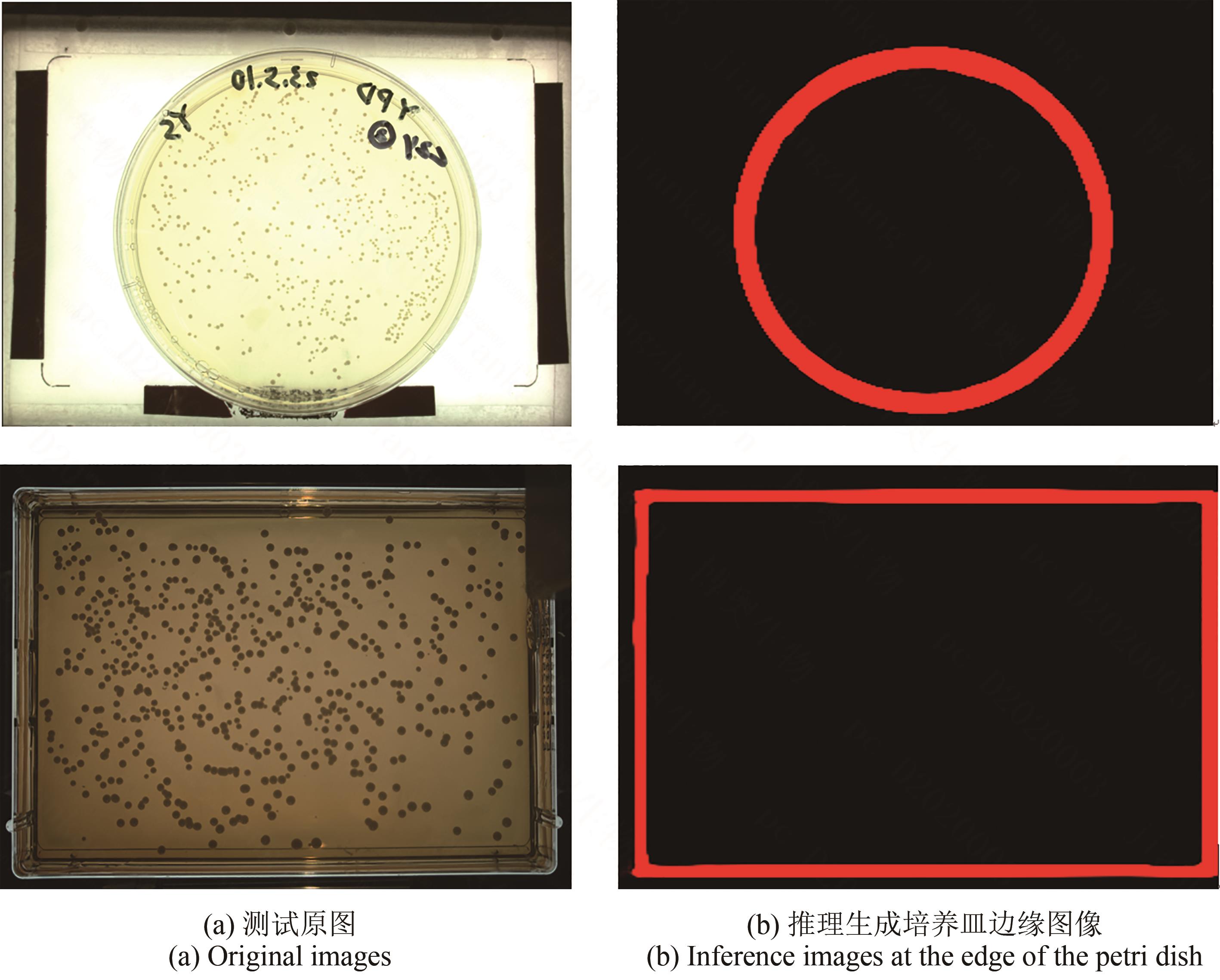
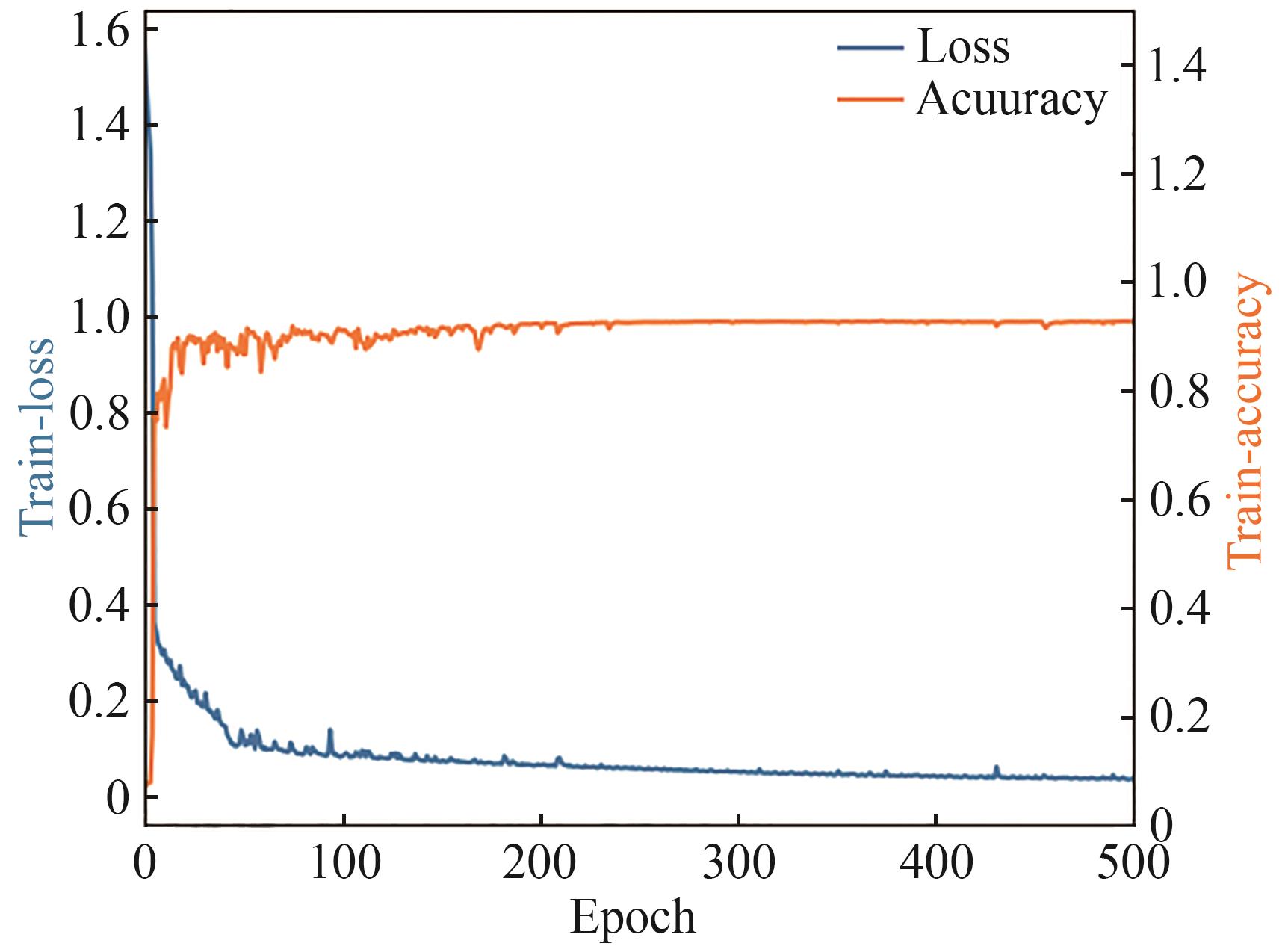
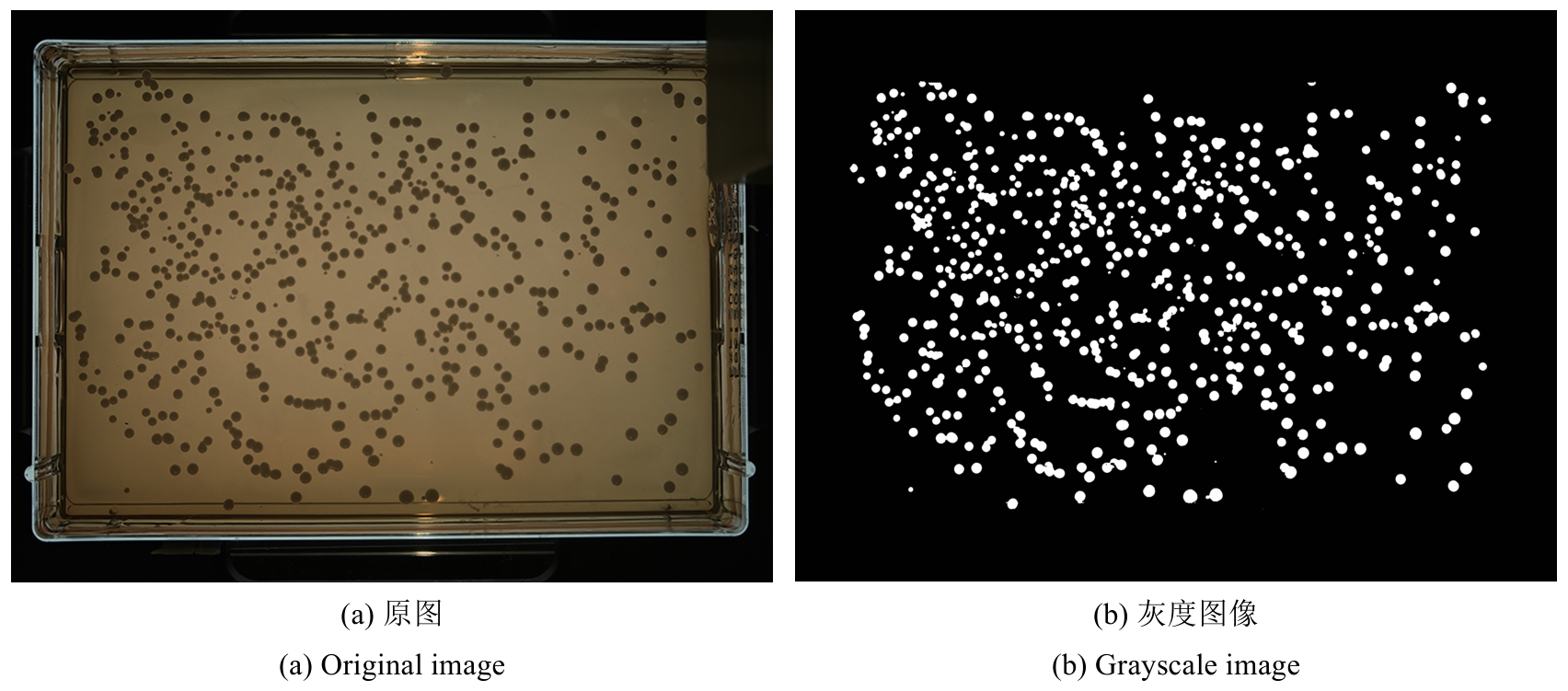
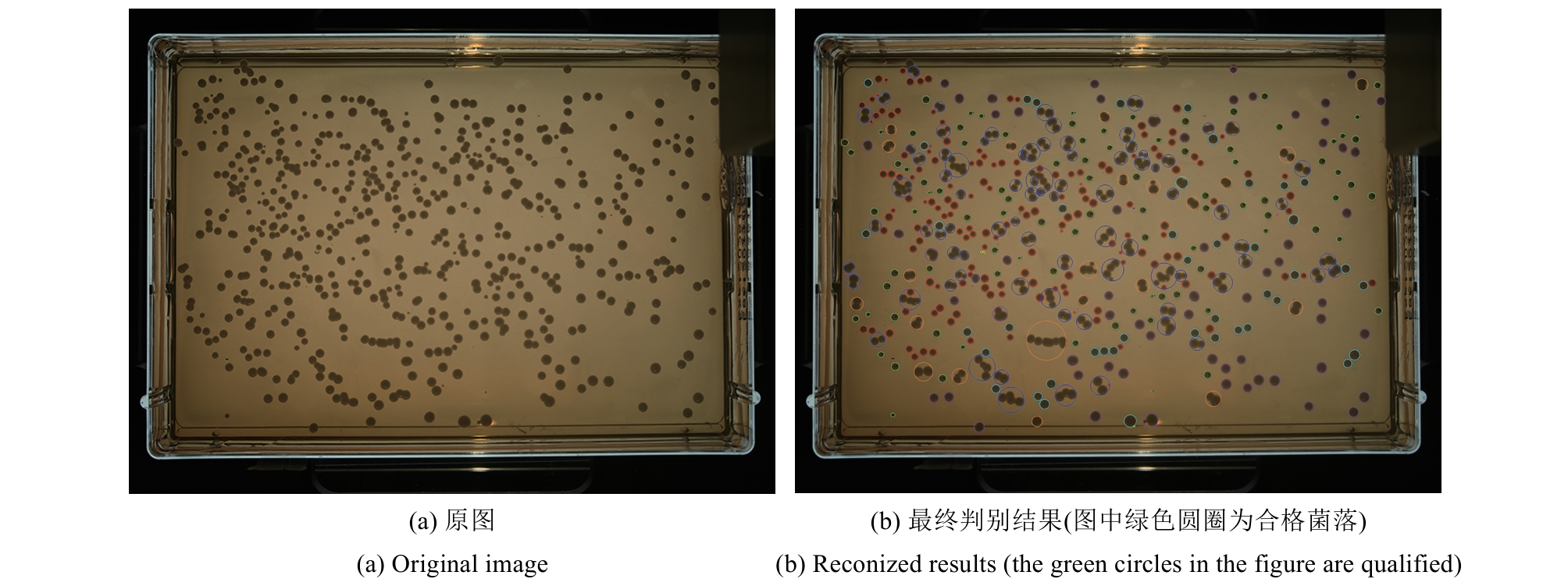
合成生物学 ›› 2025, Vol. 6 ›› Issue (4): 956-971.DOI: 10.12211/2096-8280.2025-038
基于机器视觉的高通量微生物克隆挑选工作站研制及应用
张建康1,2, 王文君1,2, 郭洪菊1,2, 白北辰1,2, 张亚飞1,2, 袁征1,2, 李彦辉1,2, 李航1,2
- 1.生物芯片北京国家工程研究中心,北京 102206
2.博奥生物集团有限公司,北京 102206
-
收稿日期:2025-04-29修回日期:2025-06-25出版日期:2025-08-31发布日期:2025-09-03 -
通讯作者:李航 -
作者简介:张建康 (1986—),男,高级工程师。研究方向为实验室自动化系统开发、机器人技术应用、智能装备研发。 E-mail:jiankangzhang@capitalbio.com李航 (1971—),男,技术负责人,工程师。研究方向为工业自动化系统集成、精密机电设备研发、智能检测技术工程化应用。 E-mail:hli@capitalbio.com -
基金资助:国家重点研发计划(2018YFA0902302);国家重点研发计划(2018YFA0902304)
Development and application of a high-throughput microbial clone picking workstation based on machine vision
ZHANG Jiankang1,2, WANG Wenjun1,2, GUO Hongju1,2, BAI Beichen1,2, ZHANG Yafei1,2, YUAN Zheng1,2, LI Yanhui1,2, LI Hang1,2
- 1.National Engineering Research Center for Beijing Biochip Technology,Beijing 102206,China
2.CapitalBio Corporation,Beijing 102206,China
-
Received:2025-04-29Revised:2025-06-25Online:2025-08-31Published:2025-09-03 -
Contact:LI Hang
摘要:
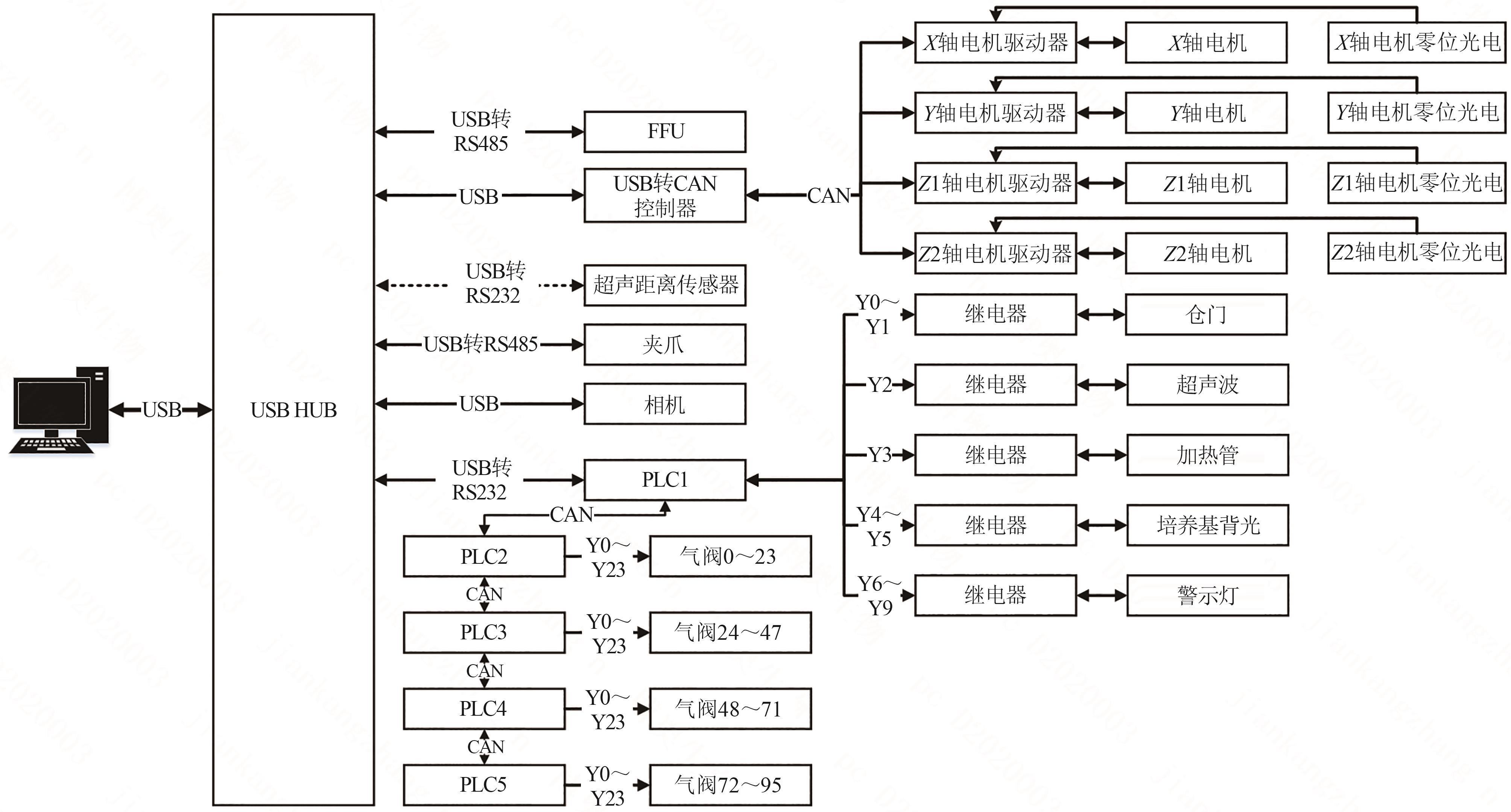
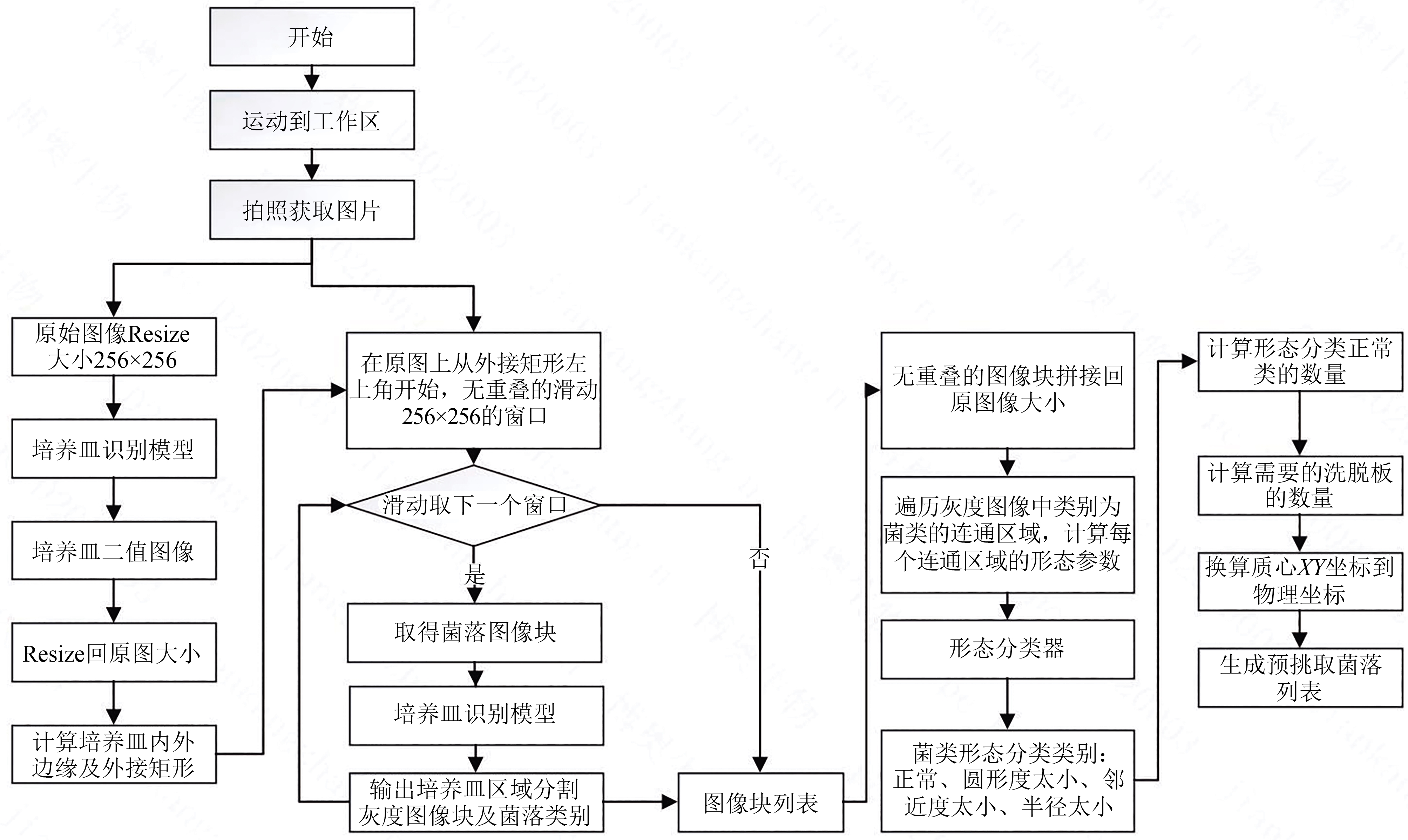
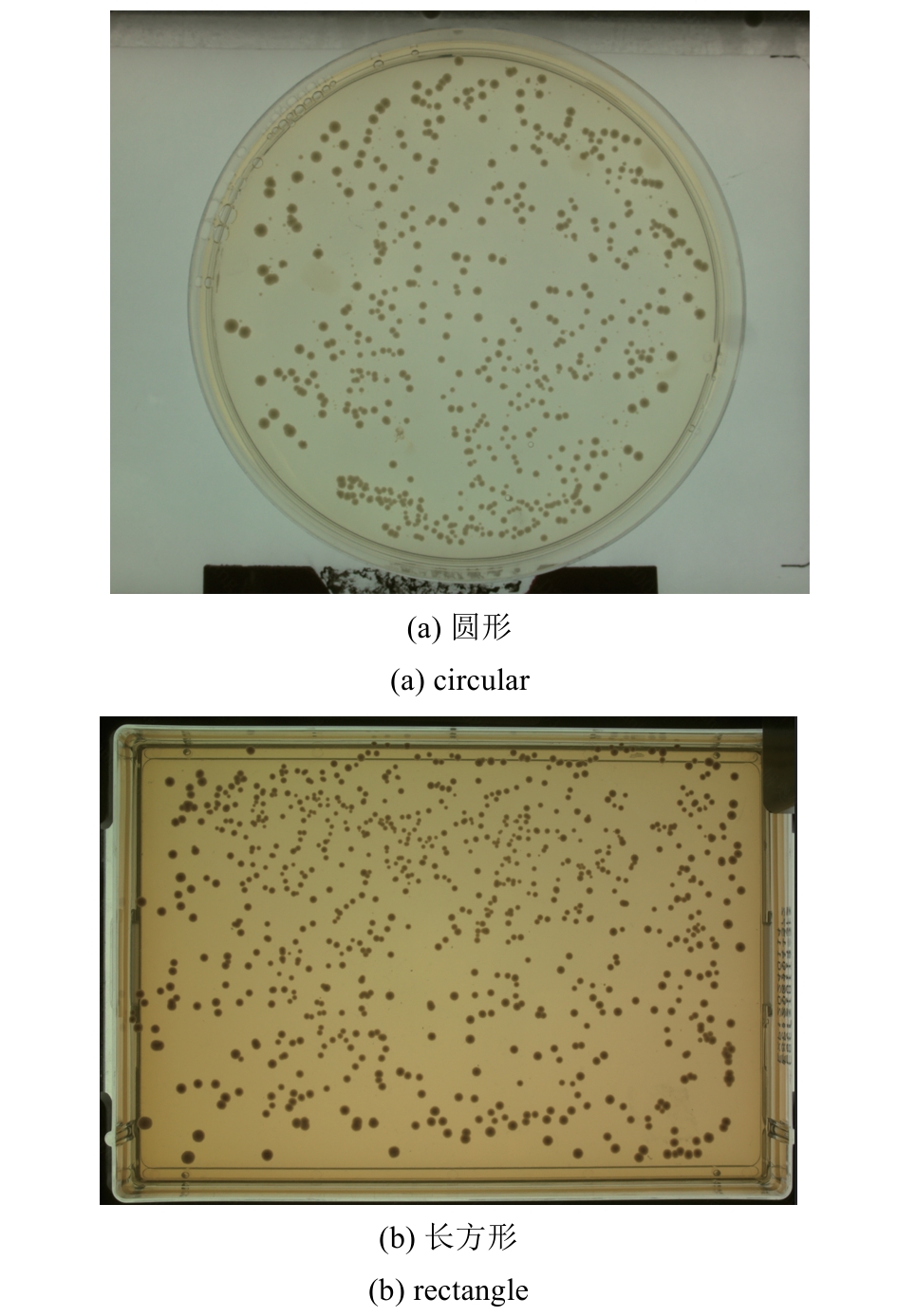
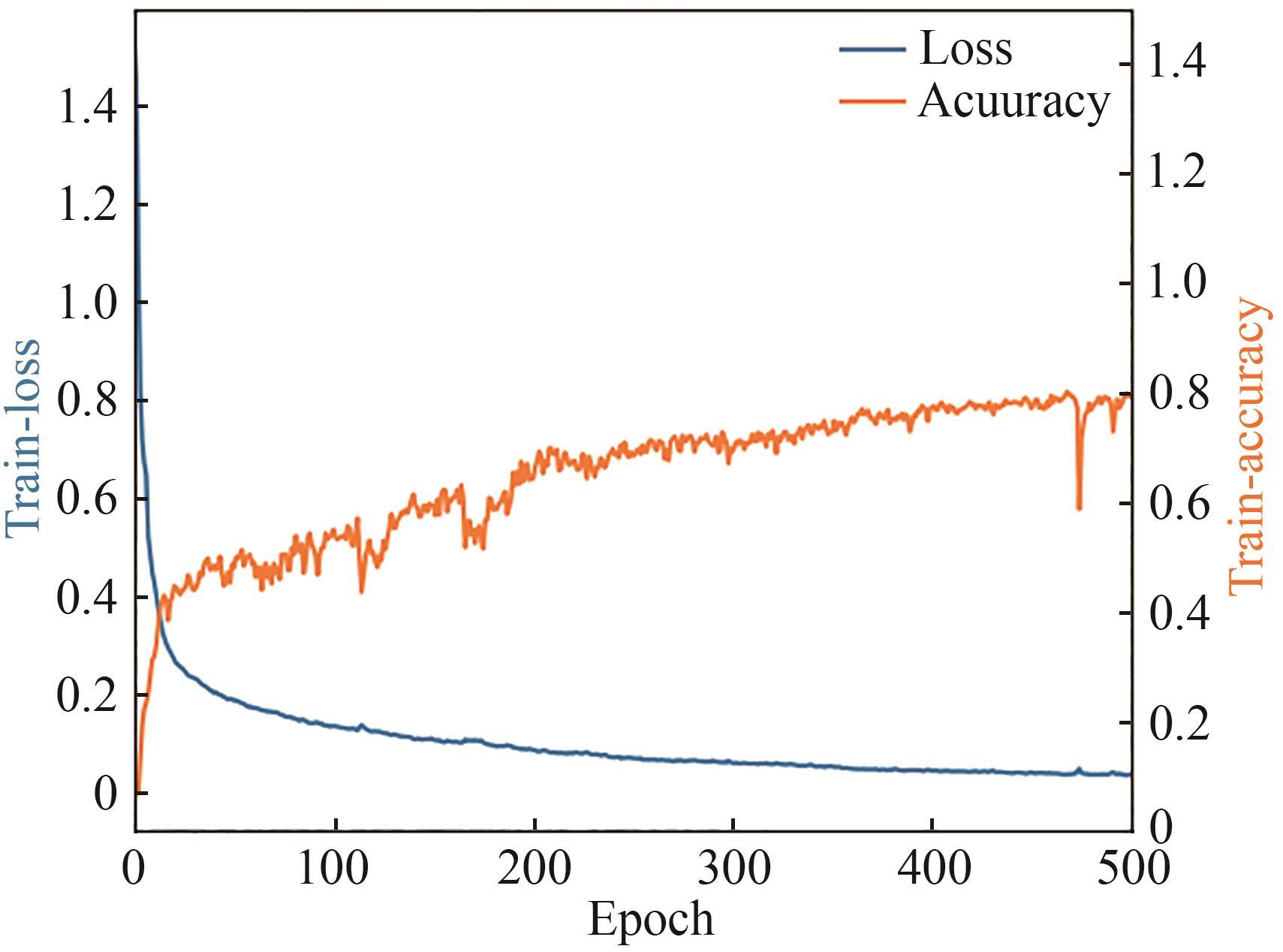
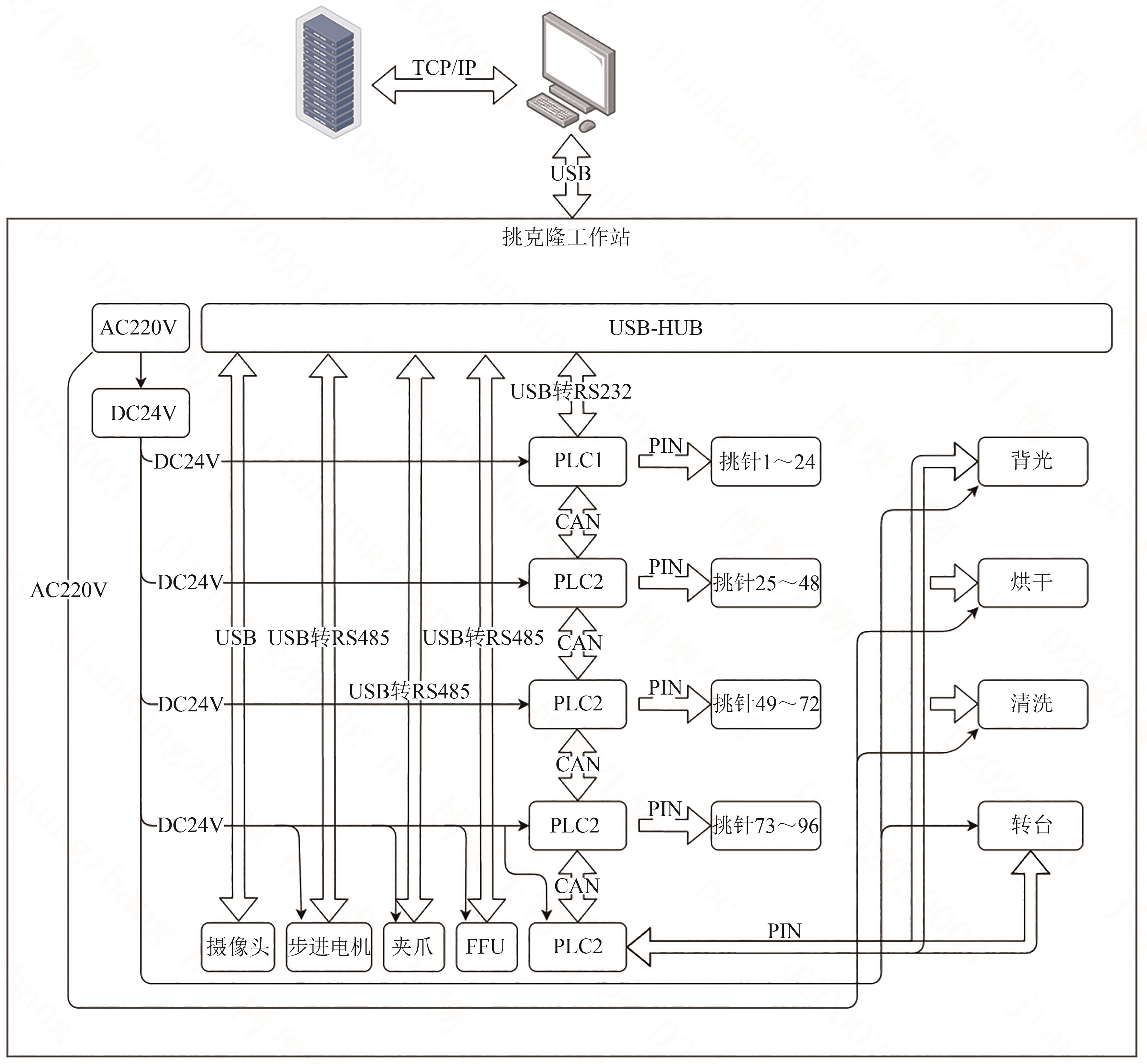
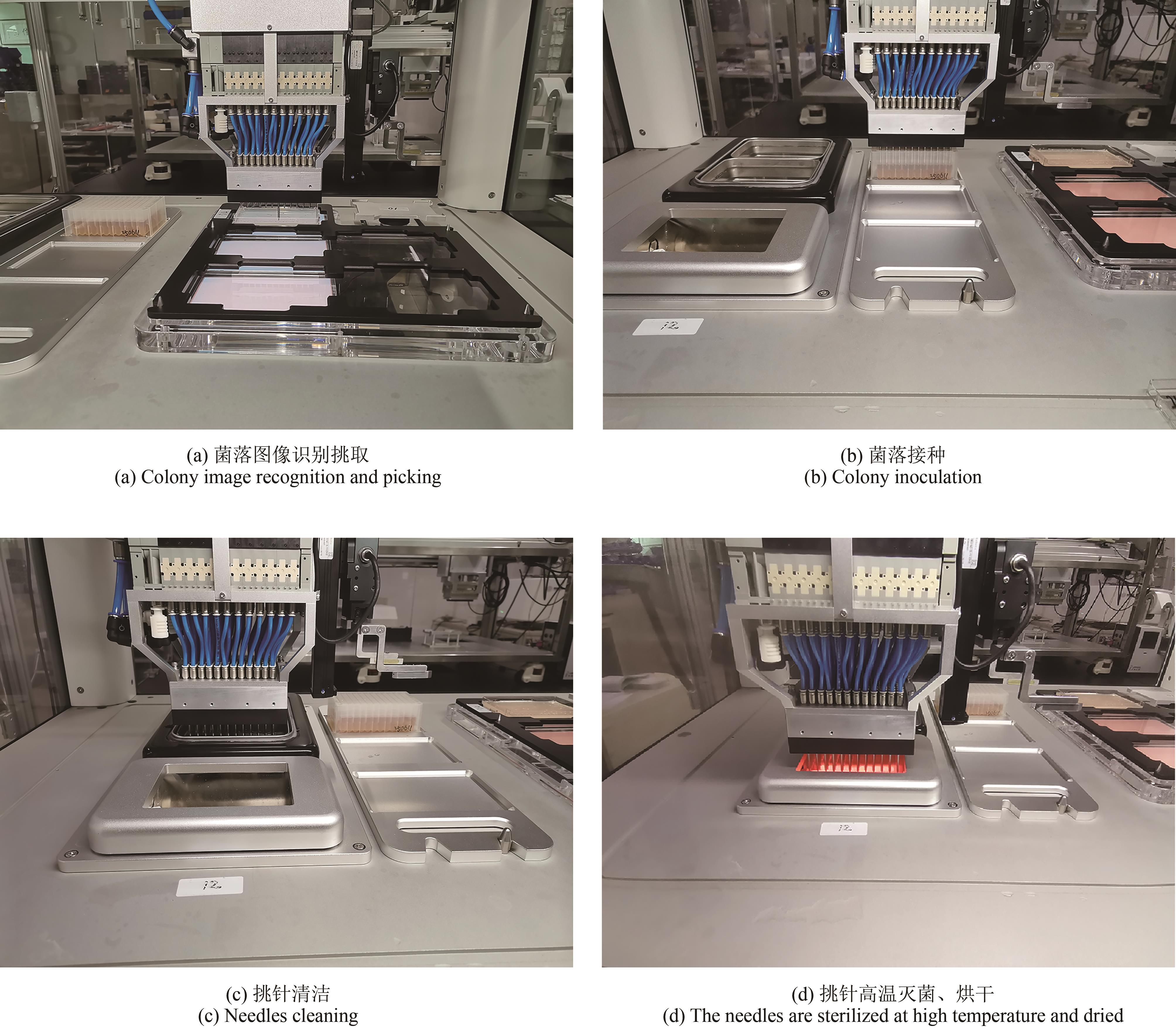
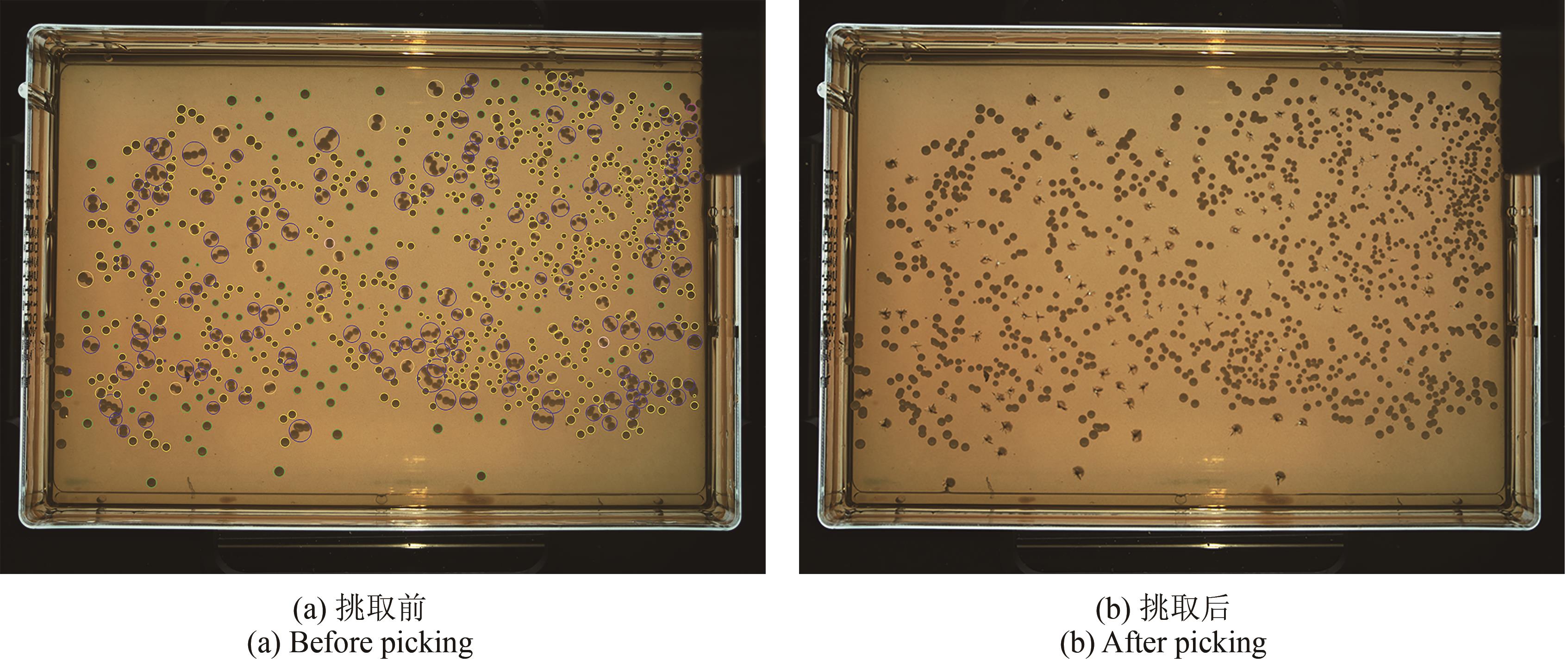
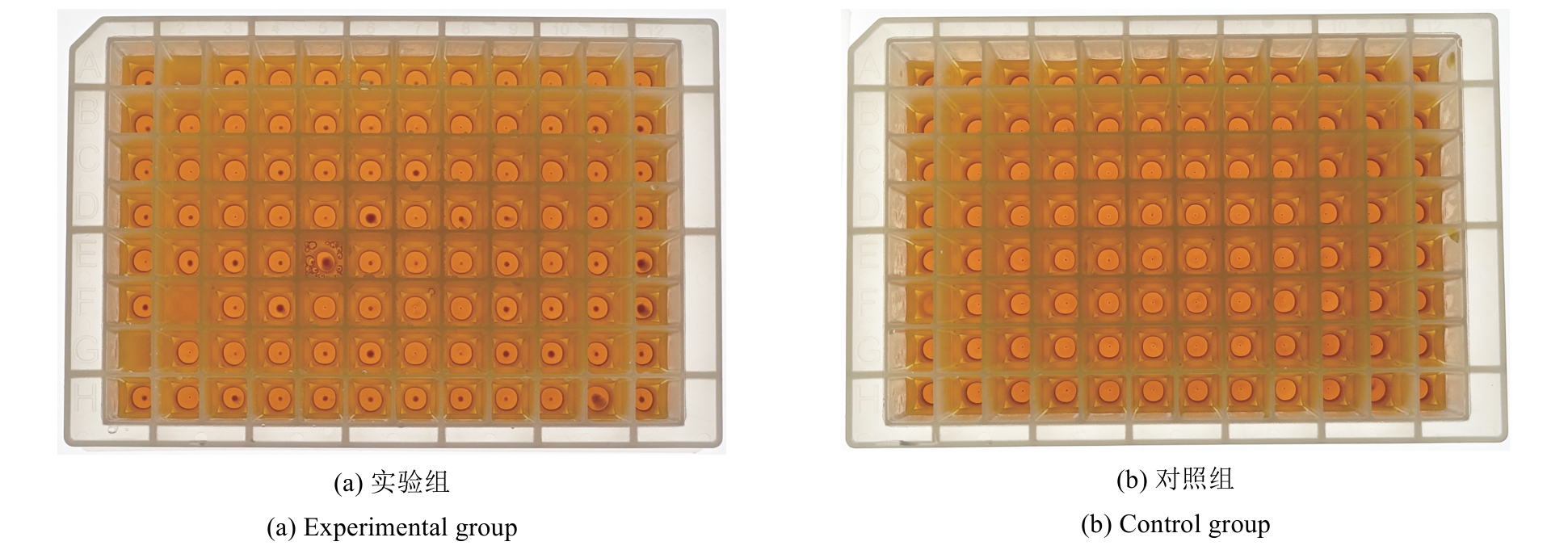
微生物克隆挑选是基因工程生物实验中的关键环节,需要从生长有大量菌落的培养皿中将符合质量要求的单克隆菌落准确、快速挑取出来并接种到培养基中,以便进一步扩大培养或检测。在高通量实验中,克隆挑选环节任务量大、记录繁复、容易交叉污染,依靠人工操作难以在短时间准确完成。针对这一难题,本项目成功研制出一种自动化克隆挑选工作站,通过菌落图像的深度学习实现克隆定位和筛选,并利用机器人技术完成挑取-接种-清洗-高温灭菌过程。在所研制的可视化工作界面中,工作站系统能够个性化编辑适用于多种微生物克隆的多项实验操作流程。通过样机验证实验结果,证明了所研制系统和方法的可行性和有效性,为高通量实验室自动化发展提供了有效工具和有益实践。
中图分类号:
引用本文
张建康, 王文君, 郭洪菊, 白北辰, 张亚飞, 袁征, 李彦辉, 李航. 基于机器视觉的高通量微生物克隆挑选工作站研制及应用[J]. 合成生物学, 2025, 6(4): 956-971.
ZHANG Jiankang, WANG Wenjun, GUO Hongju, BAI Beichen, ZHANG Yafei, YUAN Zheng, LI Yanhui, LI Hang. Development and application of a high-throughput microbial clone picking workstation based on machine vision[J]. Synthetic Biology Journal, 2025, 6(4): 956-971.
数据集 Data set | 训练集 Training set | 测试集 Test set |
|---|---|---|
完整菌落图像 Complete colony images | 34 | 4 |
挑选出小图像块 Small image blocks selected | 5160 | 1466 |
表1 数据集分布情况
Table 1 Distribution of the dataset
数据集 Data set | 训练集 Training set | 测试集 Test set |
|---|---|---|
完整菌落图像 Complete colony images | 34 | 4 |
挑选出小图像块 Small image blocks selected | 5160 | 1466 |
| 设备型号 | 通道数量 | 挑取 方式 | CCD分辨率/(Pixel/mm) | 菌落识 别尺寸/mm | 菌落识别准确度/% | 挑取准确率/% | 有效像素/万 |
|---|---|---|---|---|---|---|---|
| QPix 420 | 96 | 挑针 | 22 | ≥ 0.1 | 0.5~0.7mm菌落,>97% | > 98% | 500 |
| Hedylax T200智能微生物菌落挑选工作站 | 单通道或8通道 | 吸头 | — | — | — | ≥ 98% | 2500 |
| 全自动菌落挑选工作站G3000 | 2或4 | 挑针 | — | ≥ 0.5 | — | 1 mm以上98% | 600~2000 |
| 本设备 | 96 | 挑针 | 30 | ≥0.2 | ≥ 98% | ≥ 98% | 1000 |
表2 本设备与其他3款克隆挑选工作站的技术参数对比
Table 2 A comparison of technical parameters of this device with other three clone picking workstations
| 设备型号 | 通道数量 | 挑取 方式 | CCD分辨率/(Pixel/mm) | 菌落识 别尺寸/mm | 菌落识别准确度/% | 挑取准确率/% | 有效像素/万 |
|---|---|---|---|---|---|---|---|
| QPix 420 | 96 | 挑针 | 22 | ≥ 0.1 | 0.5~0.7mm菌落,>97% | > 98% | 500 |
| Hedylax T200智能微生物菌落挑选工作站 | 单通道或8通道 | 吸头 | — | — | — | ≥ 98% | 2500 |
| 全自动菌落挑选工作站G3000 | 2或4 | 挑针 | — | ≥ 0.5 | — | 1 mm以上98% | 600~2000 |
| 本设备 | 96 | 挑针 | 30 | ≥0.2 | ≥ 98% | ≥ 98% | 1000 |
| [1] | MACARRÓN R, HERTZBERG R P. Design and implementation of high throughput screening assays[J]. Molecular Biotechnology, 2011, 47(3): 270-285. |
| [2] | HUGHES S R, BUTT T R, BARTOLETT S, et al. Design and construction of a first-generation high-throughput integrated robotic molecular biology platform for bioenergy applications[J]. SLAS Technology, 2011, 16(4): 292-307. |
| [3] | 孙梦楚, 陆亮宇, 申晓林, 等. 基于荧光检测的高通量筛选技术和装备助力细胞工厂构建[J]. 合成生物学, 2023, 4(5): 947-965. |
| SUN M C, LU L Y, SHEN X L, et al. Fluorescence detection-based high-throughput screening systems and devices facilitate cell factories construction[J]. Synthetic Biology Journal, 2023, 4(5): 947-965. | |
| [4] | 卢挥, 张芳丽, 黄磊. 合成生物学自动化装置iBioFoundry的构建与应用[J]. 合成生物学, 2023, 4(5): 877-891. |
| LU H, ZHANG F L, HUANG L. Establishment of iBioFoundry for synthetic biology applications[J]. Synthetic Biology Journal, 2023, 4(5): 877-891. | |
| [5] | STEPHENSON A, LASTRA L, NGUYEN B, et al. Physical laboratory automation in synthetic biology[J]. ACS Synthetic Biology, 2023, 12(11): 3156-3169. |
| [6] | PAVAN M. Setting up an automated biomanufacturing laboratory[J]. Methods in Molecular Biology, 2021, 2229: 137-155. |
| [7] | MARTIN V J J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids[J]. Nature Biotechnology, 2003, 21(7): 796-802. |
| [8] | WIN M N, SMOLKE C D. Higher-order cellular information processing with synthetic RNA devices[J]. Science, 2008, 322(5900): 456-460. |
| [9] | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| [10] | CHAO R, MISHRA S, SI T, et al. Engineering biological systems using automated biofoundries[J]. Metabolic Engineering, 2017, 42: 98-108. |
| [11] | 唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台[J]. 科学通报, 2021, 66(3): 300-309. |
| TANG T, FU L H, GUO E P, et al. Automation in synthetic biology using biological foundries[J]. Chinese Science Bulletin, 2021, 66(3): 300-309. | |
| [12] | ZIMMERMANN M, ZIMMERMANN-KOGADEEVA M, WEGMANN R, et al. Mapping human microbiome drug metabolism by gut bacteria and their genes[J]. Nature, 2019, 570(7762): 462-467. |
| [13] | CERBINI T, LUO Y Q, RAO M S, et al. Transfection, selection, and colony-picking of human induced pluripotent stem cells TALEN-targeted with a GFP gene into the AAVS1 safe harbor[J]. Journal of Visualized Experiments, 2015(96): 52504. |
| [14] | DJATNA T, HADI M Z. Implementation of an ant colony approach to solve multi-objective order picking problem in beverage warehousing with drive-in rack system[C]//2017 International Conference on Advanced Computer Science and Information Systems (ICACSIS), 28-29 October 2017, Bali, Indonesia, 2017: 137-142. |
| [15] | HUANG C, HE K, LIU C L, et al. A colony picking robot with multi-pin synchronous manipulator[C]//2018 IEEE International Conference on Information and Automation(ICIA), 11-13 August 2018, Wuyi Mountain, Fujian, China, 2018: 7-12. |
| [16] | 张依朗. 基于机器视觉的高通量自动菌落挑取系统研究[D]. 北京: 北京化工大学, 2021. |
| ZHANG Y L. Research on high throughput automatic colony picking sysetem based on machine vision[D]. Beijing: Beijing University of Chemical Technology, 2021. | |
| [17] | CHEN W B, ZHANG C C. Automated bacterial colony counting for clonogenic assay[M]//Dental Computing and Applications. New York: IGI Global, 2009: 134-145. |
| [18] | GOROKHOV O, FAZYLOV R, KAZACHUK M, et al. Bacterial colony detection method for microbiological photographic images[C]//2022 International Joint Conference on Neural Networks (IJCNN). July 18-23, 2022, Padua, Italy. IEEE, 2022: 1-8. |
| [19] | ZHANG L. Phase contrast microscopy cell population segmentation: a survey[EB/OL]. arXiv, 2019: 1911.11111. (2019-11-29)[2025-03-01]. . |
| [20] | LAWLESS C, WILKINSON D J, YOUNG A, et al. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar[J]. BMC Bioinformatics, 2010, 11: 287. |
| [21] | REHMAN A, SALEEM Z, AMJAD J, et al. A comparison of bacterial colonies count from petri dishes utilizing hough transform and traditional manual counting[EB/OL].arXiv, 2025: 2505.20365. (2025-05-26)[2025-06-01]. . |
| [22] | RUSS J C, NEAL F B. The image processing handbook[M/OL]. 7th ed. Boca Raton: CRC Press, 2016. (2018-09-03)[2025-06-25]. . |
| [23] | MOEN E, BANNON D, KUDO T, et al. Deep learning for cellular image analysis[J]. Nature Methods, 2019, 16(12): 1233-1246. |
| [24] | WAKABAYASHI R, AOYANAGI A, TOMINAGA T. Rapid counting of coliforms and Escherichia coli by deep learning-based classifier[J]. Journal of Food Safety, 2024, 44(4): e13158. |
| [25] | SHEN B Y, CHEN X, CAI D L, et al. Real-Space Imaging of the Ordered Small Molecule Orientations in Porous Frameworks by Electron Microscopy[EB/OL]. arXiv, 2020: 2001.09588.(2020-01-27)[2025-06-01]. . |
| [26] | AKBAR S ALI, GHAZALI K H, HASAN H, et al. Rapid bacterial colony classification using deep learning[J]. Indonesian Journal of Electrical Engineering and Computer Science, 2022, 26(1): 352. |
| [27] | ANDREINI P, BONECHI S, BIANCHINI M, et al. a deep learning approach to bacterial colony segmentation[C/OL]//KŮRKOVÁ V, MANOLOPOULOS Y, HAMMER B, et al. Artificial neural networks and machine learning-ICANN 2018. 27th International Conference on Artificial Neural Networks, Rhodes, Greece, October 4-7, 2018 , Proceedings, Part III. Cham: Springer, 2018. (2018-09-18)[2025-06-01]. . |
| [28] | RONNEBERGER O, FISCHER P, BROXET T, et al. U-Net: convolutional networks for biomedical image segmentation[EB/OL]. arXiv, 2015: 1505.04597. (2015-05-18)[2025-06-01]. . |
| [29] | OKTAY O, SCHLEMPER J, LE FOLGOC L, et al. Attention U-Net: learning where to look for the pancreas[EB/OL]. arXiv, 2018: 1804.03999. (2018-05-20)[2025-06-01]. . |
| [30] | HE K M, GKIOXARI G, DOLLÁRET P, et al. Mask R-CNN for object detection and instance segmentation[EB/OL]. arXiv, 2017: 1703.06870. (2018-01-24)[2025-06-01]. . |
| [31] | JHA D, SMEDSRUD P H, JOHANSEN D, et al. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation[J]. IEEE Journal of Biomedical and Health Informatics, 2021, 25(6): 2029-2040. |
| [32] | 曹淑艳, 陈雪景, 张洵洋. 中国塑料行业绿色低碳发展研究报告[R/OL]. [2025-06-01]. . |
| CAO S Y, CHEN X J, ZHANG X Y. Green and low carbon development of the plastics industry in China[R/OL]. [2025-06-01]. . |
| [1] | 方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898. |
| [2] | 朱欣悦, 陈恬恬, 邵恒煊, 唐曼玉, 华威, 程艳玲. 益生菌辅助防治恶性肿瘤的研究进展[J]. 合成生物学, 2025, 6(4): 899-919. |
| [3] | 吴晓燕, 宋琪, 许睿, 丁陈君, 陈方, 郭勍, 张波. 合成生物学研发竞争态势对比分析[J]. 合成生物学, 2025, 6(4): 940-955. |
| [4] | 李全飞, 陈乾, 刘浩, 贺坤东, 潘亮, 雷鹏, 谷益安, 孙良, 李莎, 邱溢彬, 王瑞, 徐虹. 高黏性蛋白材料的合成生物学及应用[J]. 合成生物学, 2025, 6(4): 806-828. |
| [5] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [6] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [7] | 章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700. |
| [8] | 张成辛. 基于文本数据挖掘的蛋白功能预测:机遇与挑战[J]. 合成生物学, 2025, 6(3): 603-616. |
| [9] | 李明辰, 钟博子韬, 余元玺, 姜帆, 张良, 谭扬, 虞慧群, 范贵生, 洪亮. DeepSeek模型分析及其在AI辅助蛋白质工程中的应用[J]. 合成生物学, 2025, 6(3): 636-650. |
| [10] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [11] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [12] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [13] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [14] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [15] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||