合成生物学 ›› 2021, Vol. 2 ›› Issue (5): 697-715.DOI: 10.12211/2096-8280.2021-012
基因组挖掘在天然产物发现中的应用和前景
杨谦1, 程伯涛1, 汤志军1, 刘文1,2
- 1.中国科学院上海有机化学研究所,生命有机化学国家重点实验室,上海 200032
2.中国科学院上海有机化学研究所,湖州生物制造中心,浙江 湖州 313000
-
收稿日期:2021-01-27修回日期:2021-04-05出版日期:2021-10-31发布日期:2021-11-19 -
通讯作者:刘文 -
作者简介:杨谦 (1994—)女,博士,博士后。研究方向为天然产物化学及以基因组扫描为手段的新型天然产物发现。E-mail:yangqian117@sioc.ac.cn刘文 (1971—),男,研究员,博士生导师。研究方向为复杂天然产物的生物合成(遗传学、生物化学和化学),以产量提高和结构多样性为目的组合生物合成,以基因组扫描为手段的新型天然产物发现。E-mail:wliu@mail.sioc.ac.cn -
基金资助:国家重点研发计划(2019YFA0905400);中国科学院B类先导科技专项(XDB20020200);王宽诚率先人才计划
Applications and prospects of genome mining in the discovery of natural products
YANG Qian1, CHENG Botao1, TANG Zhijun1, LIU Wen1,2
- 1.State Key Laboratory of Bioorganic and Natural Products Chemistry,Shanghai Institute of Organic Chemistry,Chinese Academy of Sciences,Shanghai 200032,China
2.Huzhou Center of Bio-Synthetic Innovation,Shanghai Institute of Organic Chemistry,Chinese Academy of Sciences,Huzhou 313000,Zhejiang,China
-
Received:2021-01-27Revised:2021-04-05Online:2021-10-31Published:2021-11-19 -
Contact:LIU Wen
摘要:
天然产物一直以来都是药物先导化合物的重要来源。在药物发现领域,基因组数据常用来识别潜在的药物靶点或寻找先前被忽视的天然产物的生物合成基因簇。尽管基因组测序发现了微生物和植物中存在大量未开发的化学多样性,然而,仅仅利用传统的分离分析方法获取新的天然产物已经无法满足药物发展的需求。随着基因组时代的到来,数字化的基因组挖掘已经成为天然产物发现的重要组成部分。伴随着高通量测序方法的发展和DNA数据的丰富,各种基因组挖掘方法和工具被开发出来,以指导发现和表征这些天然产物。本文综述了近年来基因组挖掘的网络工具、数据库和方法,着重介绍次级代谢产物生物合成基因簇的挖掘手段,从经典的基因组挖掘到基于抗性基因挖掘、基于系统进化发育的挖掘,并对基因组挖掘在天然产物发现中的地位和前景进行了展望。
中图分类号:
引用本文
杨谦, 程伯涛, 汤志军, 刘文. 基因组挖掘在天然产物发现中的应用和前景[J]. 合成生物学, 2021, 2(5): 697-715.
YANG Qian, CHENG Botao, TANG Zhijun, LIU Wen. Applications and prospects of genome mining in the discovery of natural products[J]. Synthetic Biology Journal, 2021, 2(5): 697-715.
| 数据库或web工具 | 网址(URL) | 参考文献 |
|---|---|---|
| 天然产物数据库 | ||
| Dictionary of Natural Products(DNP) | http://dnp.chemnetbase.com | [ |
| The Natural Products Atlas | https://www.npatlas.org | [ |
| PubMed | https://pubmed.ncbi.nlm.nih.gov/ | |
| NPASS | http://bidd2.nus.edu.sg/NPASS | [ |
| StreptomeDB | http://132.230.56.4/streptomedb2/ | [ |
| MarinLit | http://pubs.rsc.org/marinlit/ | |
| AntiBase | https://sciencesolutions.wiley.com | |
| KNApSAcK | http://kanaya.naist.jp/KNApSAcK/ | [ |
| Norine | https://bioinfo.lifl.fr/norine/ | [ |
| MacrolactoneDB | https://macrolact.collaborationspharma.com/ | [ |
| ChEBI | http://www.ebi.ac.uk/chebi/ | [ |
| ChEMBl | https://www.ebi.ac.uk/chembl/ | [ |
| ChemSpider | http://www.chemspider.com/ | [ |
| COCONUT | https://doi.org/10.5281/zenod | [ |
| 生物合成基因簇数据库 | ||
| ClusterMine360 | http://www.clustermine360.ca/ | [ |
| DoBISCUIT | http://www.bio.nite.go.jp/pks/ | [ |
| MIBiG | https://mibig.secondarymetabolites.org/ | [ |
| IMG-ABC | https://img.jgi.doe.gov/cgi-bin/abc/main.cgi | [ |
| antiSMASH Database | https://antismash.secondarymetabolites.org/ | [ |
| ClustScan Database | http://csdb.bioserv.pbf.hr/csdb/ | [ |
| BiG-FAM | https://bigfam.bioinformatics.nl/ | [ |
| 蛋白家族数据库 | ||
| UniProtKB | https://www.uniprot.org/ | [ |
| Pfam | http://pfam.xfam.org/ | [ |
| InterPro | http://www.ebi.ac.uk/interpro/ | [ |
| 识别生物合成基因簇的网络工具 | ||
| BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | [ |
| HMMer | http://hmmer.org/ | [ |
| ClustScan | http://bioserv.pbf.hr/cms/ | [ |
| np.searcher | http://dna.sherman.lsi.umich.edu/ | [ |
| SMURF | http://jcvi.org/smurf/index.php | [ |
| antiSMASH | http://antismash.secondarymetabolites.org | [ |
| ClusterFinder | https://github.com/petercim/ClusterFinder | [ |
| RODEO | http://rodeo.scs.illinois.edu/ | [ |
表1 基因组挖掘的数据库及网络工具
Tab. 1 Database and web tools of genome mining
| 数据库或web工具 | 网址(URL) | 参考文献 |
|---|---|---|
| 天然产物数据库 | ||
| Dictionary of Natural Products(DNP) | http://dnp.chemnetbase.com | [ |
| The Natural Products Atlas | https://www.npatlas.org | [ |
| PubMed | https://pubmed.ncbi.nlm.nih.gov/ | |
| NPASS | http://bidd2.nus.edu.sg/NPASS | [ |
| StreptomeDB | http://132.230.56.4/streptomedb2/ | [ |
| MarinLit | http://pubs.rsc.org/marinlit/ | |
| AntiBase | https://sciencesolutions.wiley.com | |
| KNApSAcK | http://kanaya.naist.jp/KNApSAcK/ | [ |
| Norine | https://bioinfo.lifl.fr/norine/ | [ |
| MacrolactoneDB | https://macrolact.collaborationspharma.com/ | [ |
| ChEBI | http://www.ebi.ac.uk/chebi/ | [ |
| ChEMBl | https://www.ebi.ac.uk/chembl/ | [ |
| ChemSpider | http://www.chemspider.com/ | [ |
| COCONUT | https://doi.org/10.5281/zenod | [ |
| 生物合成基因簇数据库 | ||
| ClusterMine360 | http://www.clustermine360.ca/ | [ |
| DoBISCUIT | http://www.bio.nite.go.jp/pks/ | [ |
| MIBiG | https://mibig.secondarymetabolites.org/ | [ |
| IMG-ABC | https://img.jgi.doe.gov/cgi-bin/abc/main.cgi | [ |
| antiSMASH Database | https://antismash.secondarymetabolites.org/ | [ |
| ClustScan Database | http://csdb.bioserv.pbf.hr/csdb/ | [ |
| BiG-FAM | https://bigfam.bioinformatics.nl/ | [ |
| 蛋白家族数据库 | ||
| UniProtKB | https://www.uniprot.org/ | [ |
| Pfam | http://pfam.xfam.org/ | [ |
| InterPro | http://www.ebi.ac.uk/interpro/ | [ |
| 识别生物合成基因簇的网络工具 | ||
| BLAST | https://blast.ncbi.nlm.nih.gov/Blast.cgi | [ |
| HMMer | http://hmmer.org/ | [ |
| ClustScan | http://bioserv.pbf.hr/cms/ | [ |
| np.searcher | http://dna.sherman.lsi.umich.edu/ | [ |
| SMURF | http://jcvi.org/smurf/index.php | [ |
| antiSMASH | http://antismash.secondarymetabolites.org | [ |
| ClusterFinder | https://github.com/petercim/ClusterFinder | [ |
| RODEO | http://rodeo.scs.illinois.edu/ | [ |
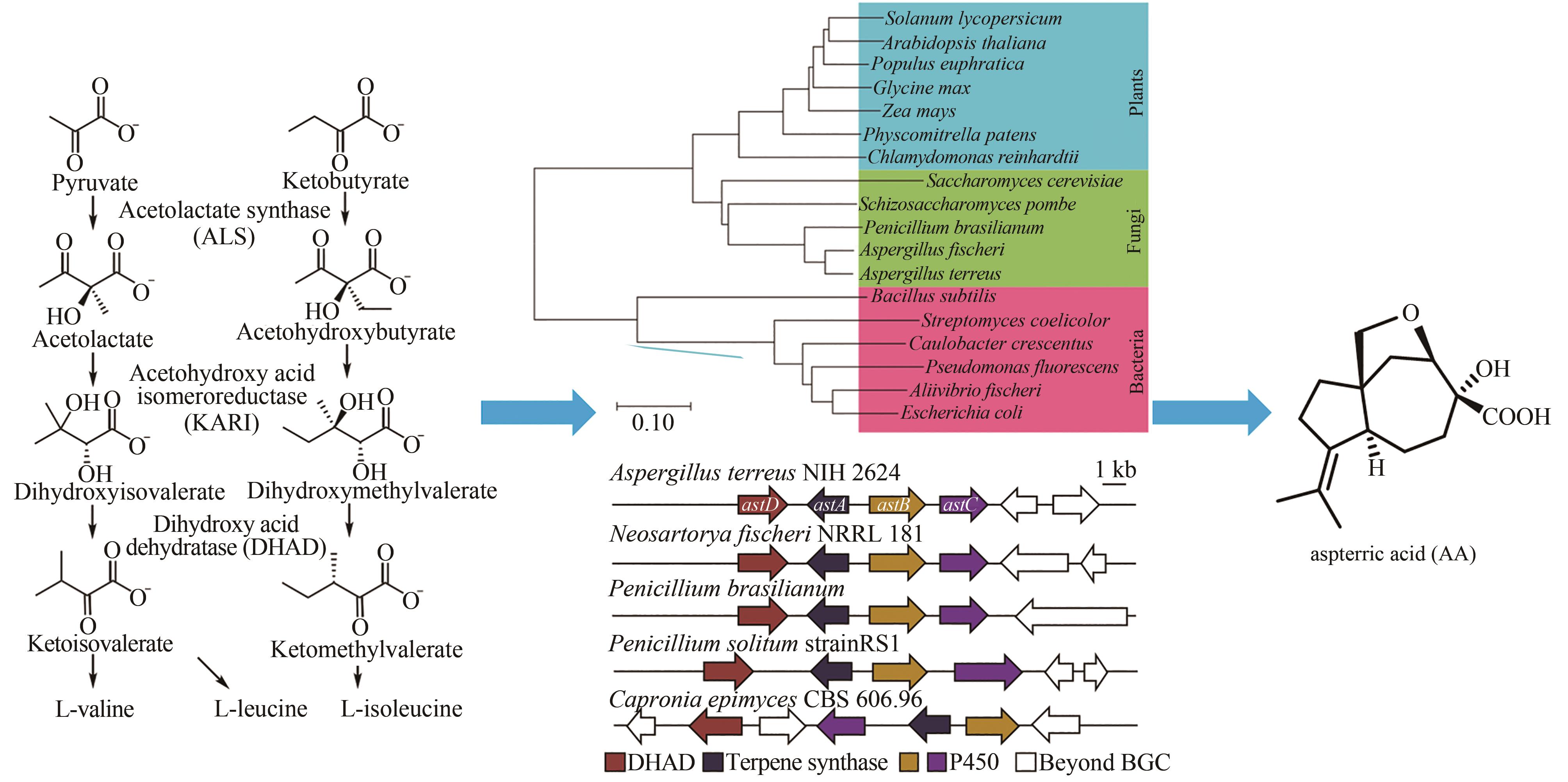
图7 从BCAA生物合成路径中关键的DHAD酶出发挖掘天然除草剂AA[102]
Fig. 7 Genome mining of a natural herbicide aspterric acid (AA) from the critical DHAD enzyme in the BCAA biosynthesis pathways
| 50 | BLIN K, SHAW S, STEINKE K, et al. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline[J]. Nucleic Acids Research, 2019, 47(W1): W81-W87. |
| 51 | MICHAEL A, FISCHBACH C T W. Antibiotics for emerging pathogens[J]. Science, 2009, 325: 1089-1093. |
| 52 | STREIT W R, SCHMITZ R A. Metagenomics-the key to the uncultured microbes[J]. Current Opinion in Microbiology, 2004, 7(5): 492-498. |
| 53 | CIMERMANCIC P, MEDEMA M H, CLAESEN J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters[J]. Cell, 2014, 158(2): 412-421. |
| 54 | CRUZ-MORALES P, KOPP J F, MARTINEZ-GUERRERO C, et al. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes[J]. Genome Biology and Evolution, 2016, 8(6):1906-1916. |
| 55 | TAKEDA I, UMEMURA M, KOIKE H, et al. Motif-Independent prediction of a secondary metabolism gene cluster using comparative genomics: Application to sequenced genomes of Aspergillus and ten other filamentous fungal species[J]. DNA Research, 2014, 21(4): 447-457. |
| 56 | SANTOS-ABERTURAS J, CHANDRA G, FRATTARUOLO L, et al. Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool[J]. Nucleic Acids Research, 2019, 47(9): 4624-4637. |
| 57 | KLOOSTERMAN A M, SHELTON K E, WEZEL G P VAN, et al. RRE-Finder: A genome-mining tool for class-independent RiPP discovery[J]. mSystems, 2020, 5(5): e00267-00220. |
| 58 | MERWIN N J, MOUSA W K, DEJONG C A, et al. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(1): 371-380. |
| 59 | TIETZ J I, SCHWALEN C J, PATEL P S, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape[J]. Nature Chemical Biology, 2017, 13(5): 470-478. |
| 60 | HASTINGS J, DE MATOS P, DEKKER A, et al. The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013[J]. Nucleic Acids Research, 2013, 41(D1): D456-D463. |
| 61 | BENTO A P, GAULTON A, HERSEY A, et al. The ChEMBL bioactivity database: an update[J]. Nucleic Acids Research, 2014, 42(D1): D1083-D1090. |
| 62 | PENCE H E, WILLIAMS A. ChemSpider: an online chemical information resource[J]. Journal of Chemical Education, 2010, 87(11): 1123-1124. |
| 63 | KAUTSAR S A, BLIN K, SHAW S, et al. BiG-FAM: the biosynthetic gene cluster families database[J]. Nucleic Acids Research, 2020, 49(D1): D490-D497. |
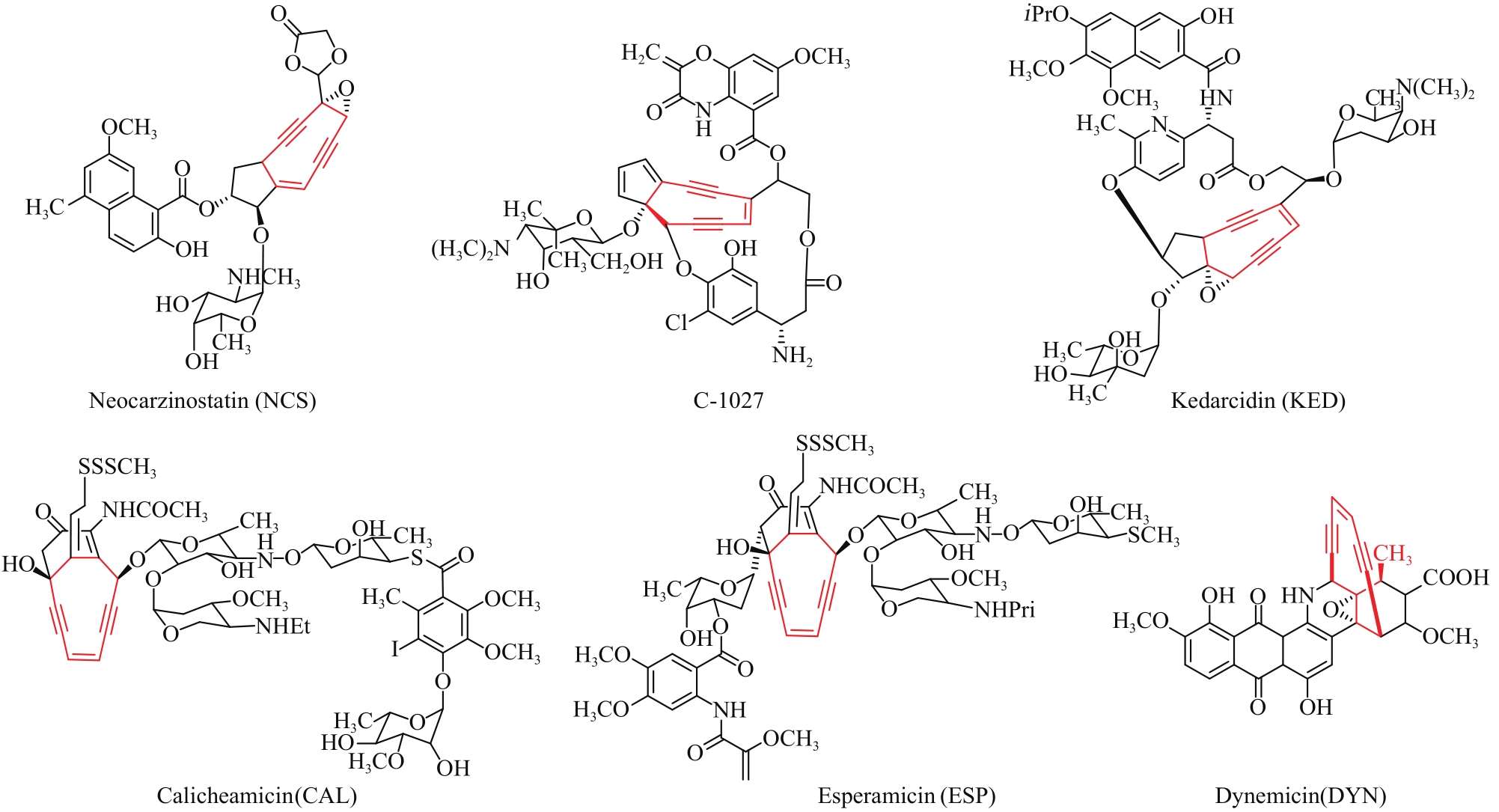
| 64 | WEN L. Progress of biosynthesis of enediyne antitumor antibiotics[J]. World Sci-Tech R & D, 2005, 27: 3. |
| 65 | ZAZOPOULOS E, HUANG K, STAFFA A, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways[J]. Nature Biotechnology, 2003, 21(2): 187-190. |
| 66 | ADHIKARI A, TEIJARO C N, TOWNSEND C A, et al. 1.12-biosynthesis of enediyne natural products[M]//LIU H-W, BEGLEY T P. Comprehensive natural products iii. Oxford: Elsevier, 2020: 365-414. |
| 67 | SHEN B, HINDRA, YAN X, et al. Enediynes: exploration of microbial genomics to discover new anticancer drug leads[J]. Bioorganic & Medicinal Chemistry Letters, 2015, 25(1): 9-15. |
| 68 | YAN X, GE H, HUANG T, et al. Strain prioritization and genome mining for enediyne natural products[J]. mBio, 2016, 7(6): e02104-02116. |
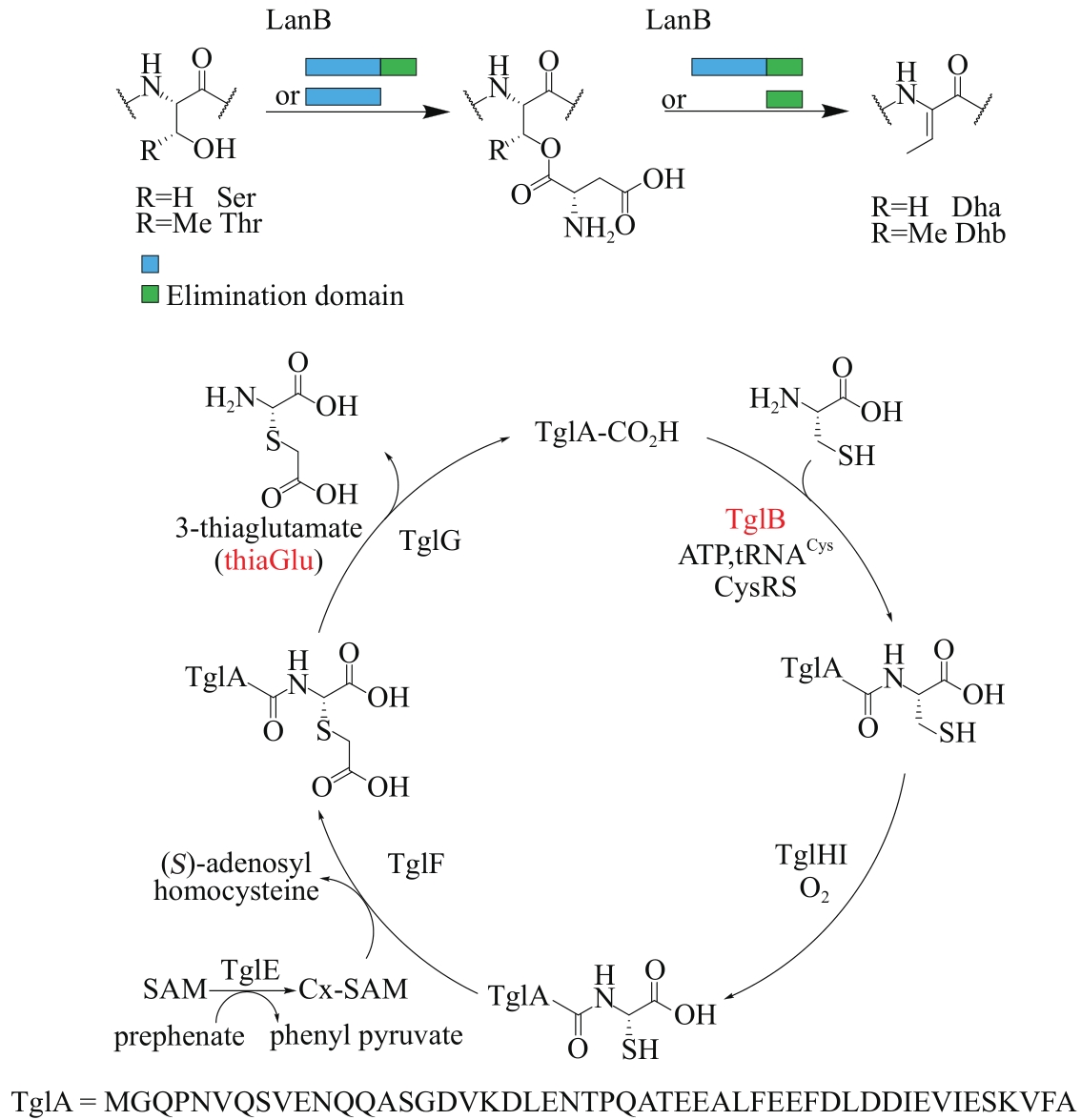
| 69 | ORTEGA M A, HAO Y, ZHANG Q, et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB[J]. Nature, 2015, 517(7535): 509-512. |
| 70 | HUDSON G A, ZHANG Z G, TIETZ J I, et al. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin[J]. Journal of the American Chemical Society, 2015, 137(51): 16012-16015. |
| 71 | TING C P, FUNK M A, HALABY S L, et al. Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products[J]. Science, 2019, 365(6450): 280-284. |
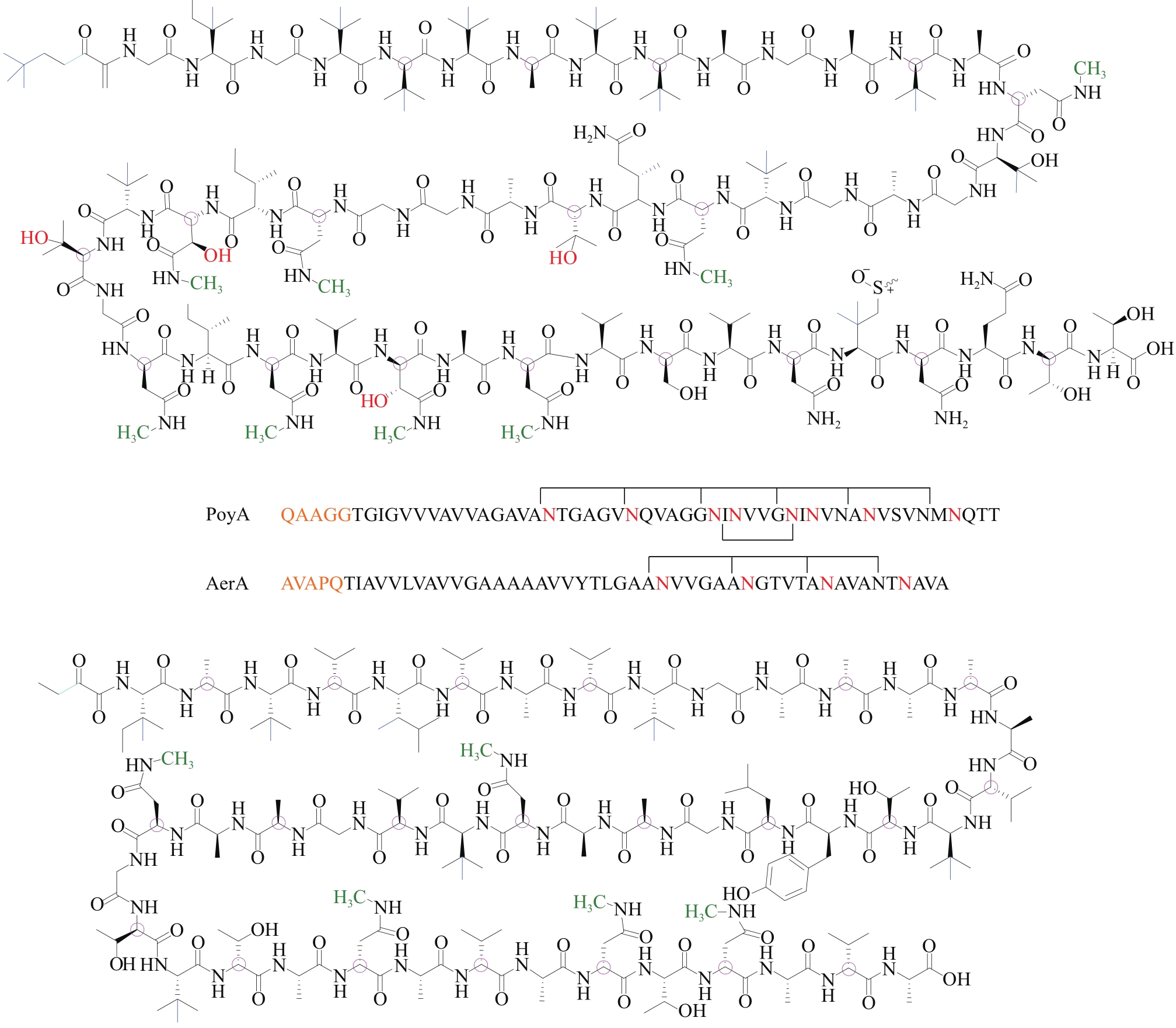
| 72 | HAMADA T, MATSUNAGA S, FUJIWARA M, et al. Solution structure of polytheonamide B, a highly cytotoxic nonribosomal polypeptide from marine sponge[J]. Journal of the American Chemical Society, 2010, 132(37): 12941-12945. |
| 73 | MORINAKA B I, VAGSTAD A L, HELF M J, et al. Radical S-adenosyl methionine epimerases: Regioselective introduction of diverse D-amino acid patterns into peptide natural products[J]. Angewandte Chemie International Edition, 2014, 53(32): 8503-8507. |
| 74 | FREEMAN M F, GURGUI C, HELF M J, et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides[J]. Science, 2012, 338(6105): 387-390. |
| 75 | BHUSHAN A, EGLI P J, PETERS E E, et al. Genome mining- and synthetic biology-enabled production of hypermodified peptides[J]. Nature Chemistry, 2019, 11(10): 931-939. |
| 76 | BRODERICK J B, DUFFUS B R, DUSCHENE K S, et al. Radical S-adenosylmethionine enzymes[J]. Chemical Reviews, 2014, 114(8): 4229-4317. |
| 77 | BENJDIA A, GUILLOT A, LEFRANC B, et al. Thioether bond formation by SPASM domain radical SAM enzymes: C α H-atom abstraction in subtilosin A biosynthesis[J]. Chemical Communications, 2016, 52(37): 6249-6252. |
| 78 | HUDSON G A, BURKHART B J, DICAPRIO A J, et al. Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new C α, C β, and C γ -linked thioether-containing peptides[J]. Journal of the American Chemical Society, 2019, 141(20): 8228-8238. |
| 79 | SCHRAMMA K R, BUSHIN L B, SEYEDSAYAMDOST M R. Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink[J]. Nature Chemistry, 2015, 7(5): 431-437. |
| 80 | GARDAN R, BESSET C, GUILLOT A, et al. The Oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9[J]. Journal of Bacteriology, 2009, 191(14): 4647-4655. |
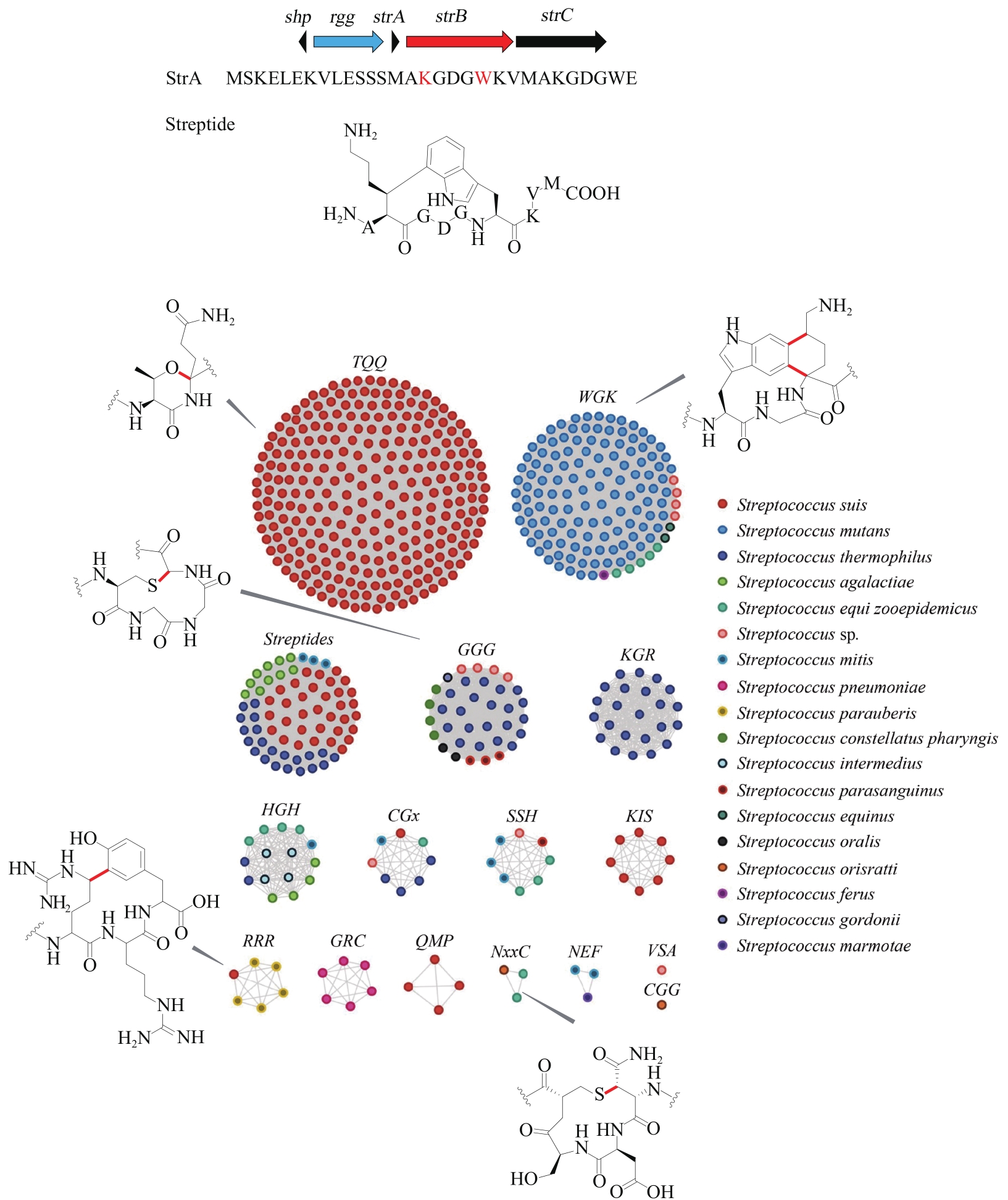
| 81 | BUSHIN L B, CLARK K A, PELCZER I, et al. Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif[J]. Journal of the American Chemical Society, 2018, 140(50): 17674-17684. |
| 82 | CARUSO A, MARTINIE R J, BUSHIN L B, et al. Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes[J]. Journal of the American Chemical Society, 2019, 141(42): 16610-16614. |
| 83 | CLARK K A, BUSHIN L B, SEYEDSAYAMDOST M R. Aliphatic ether bond formation expands the scope of radical SAM enzymes in natural product biosynthesis[J]. Journal of the American Chemical Society, 2019, 141(27): 10610-10615. |
| 84 | CARUSO A, BUSHIN L B, CLARK K A, et al. Radical approach to enzymatic β-thioether bond formation[J]. Journal of the American Chemical Society, 2019, 141(2): 990-997. |
| 85 | BUSHIN L B, COVINGTON B C, RUED B E, et al. Discovery and biosynthesis of streptosactin, a sactipeptide with an alternative topology encoded by commensal bacteria in the human microbiome[J]. Journal of the American Chemical Society, 2020, 142(38): 16265-16275. |
| 86 | LAUTRU S, DEETH R J, BAILEY L M, et al. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining[J]. Nature Chemical Biology, 2005, 1(5): 265-269. |
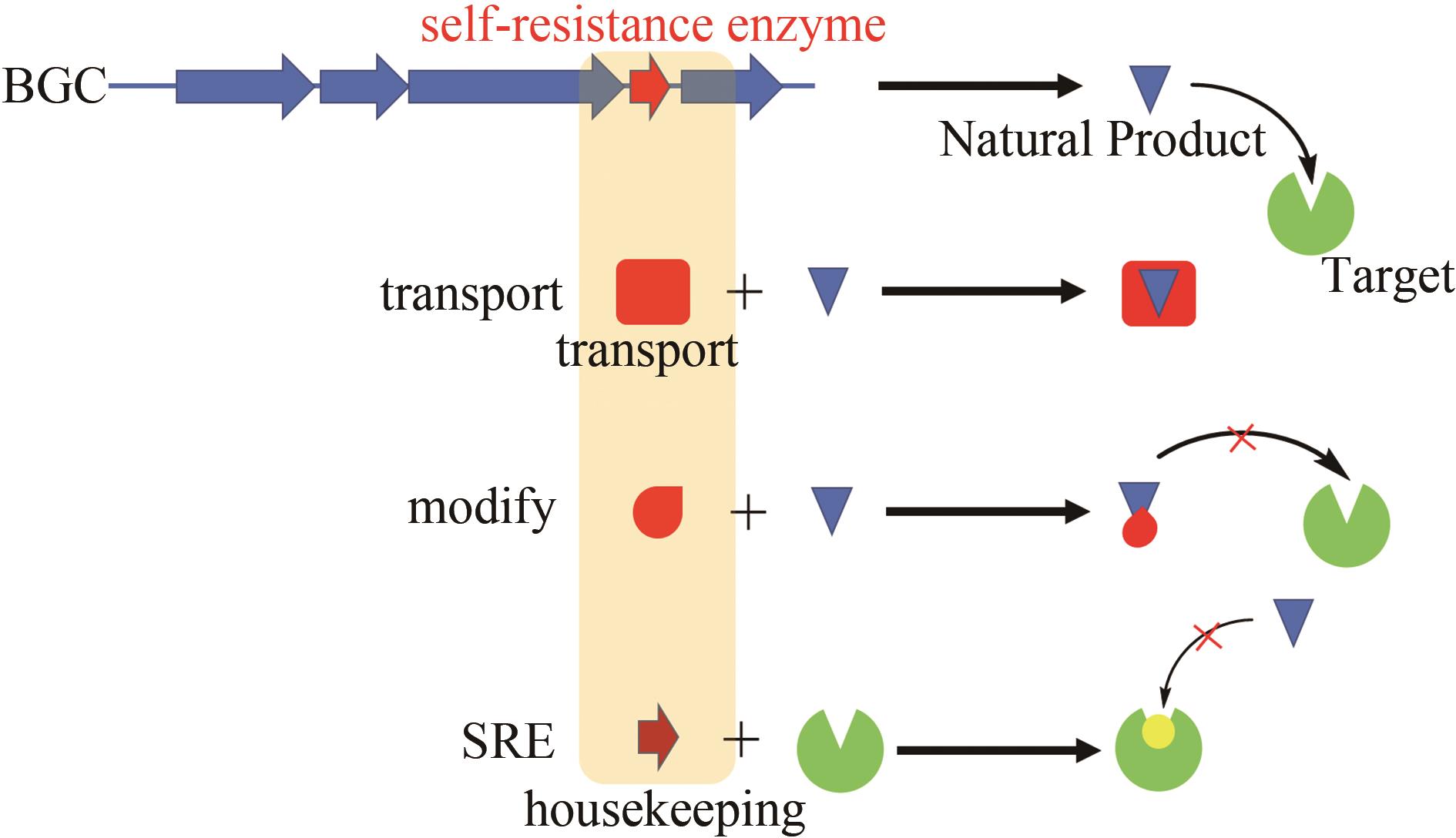
| 87 | YAN Y, LIU N, TANG Y. Recent developments in self-resistance gene directed natural product discovery[J]. Natural Product Reports, 2020, 37(7): 879-892. |
| 88 | GALM U, HAGER M H, LANEN S G VAN, et al. Antitumor antibiotics: bleomycin, enediynes, and mitomycin[J]. Chemical Reviews, 2005, 105(2): 739-758. |
| 89 | WEISBLUM B. Erythromycin resistance by ribosome modification[J]. Antimicrobial Agents and Chemotherapy, 1995, 39(3): 577-585. |
| 90 | O'NEILL E C, SCHORN M, LARSON C B, et al. Targeted antibiotic discovery through biosynthesis-associated resistance determinants: Target directed genome mining[J]. Critical Reviews in Microbiology, 2019, 45(3): 255-277. |
| 91 | ALMABRUK K H, DINH L K, PHILMUS B. Self-resistance of natural product producers: past, present, and future focusing on self-resistant protein variants[J]. ACS Chemical Biology, 2018, 13(6): 1426-1437. |
| 92 | MAXWELL A. The interaction between coumarin drugs and DNA gyrase[J]. Molecular Microbiology, 1993, 9(4): 681-686. |
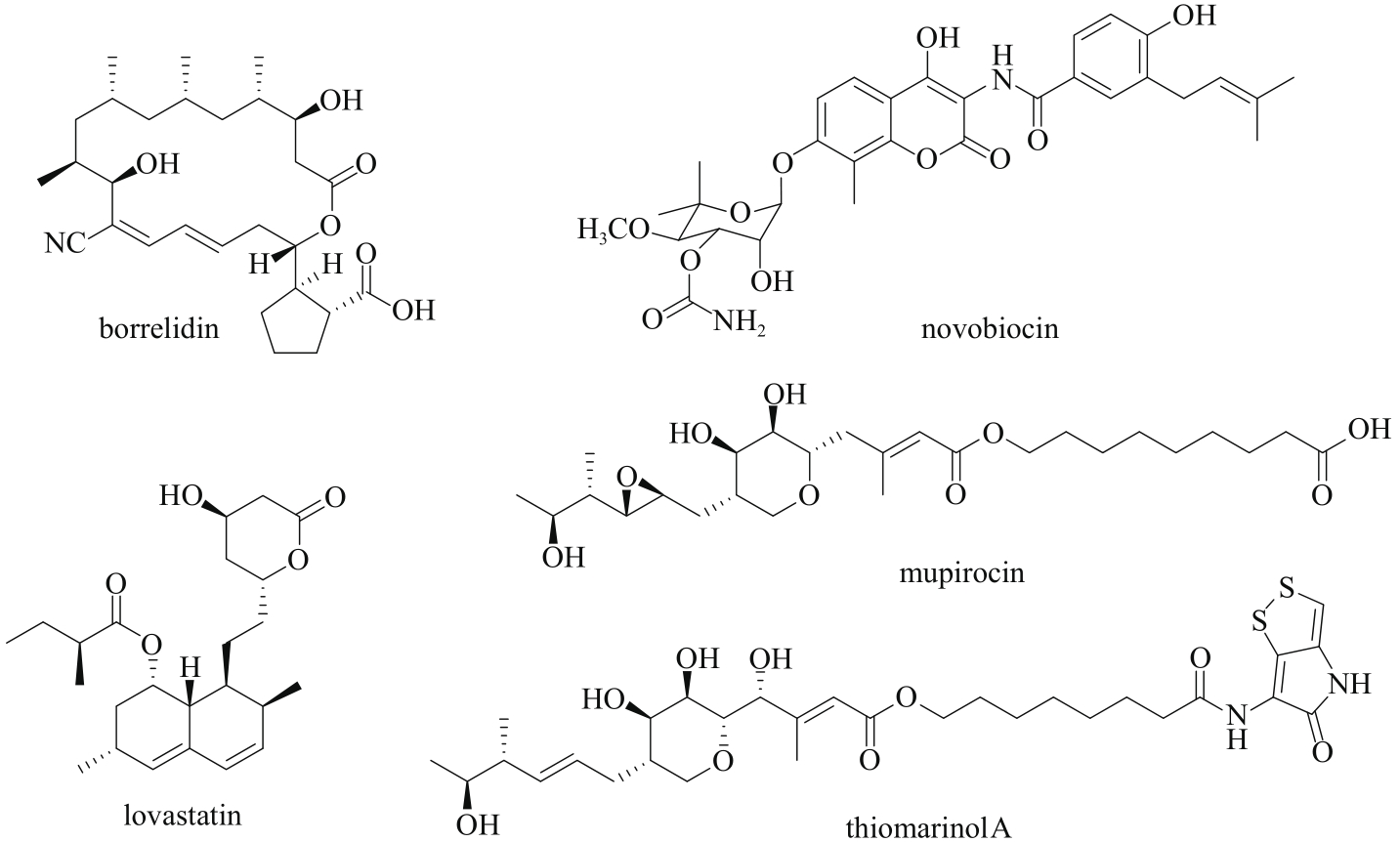
| 93 | THIARA A S, CUNDLIFFE E. Interplay of novobiocin-resistant and -sensitive DNA gyrase activities in self-protection of the novobiocin producer, Streptomyces sphaeroides [J]. Gene, 1989, 81(1): 65-72. |
| 94 | STEFFENSKY M, MÜHLENWEG A, WANG Z-X, et al. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891[J]. Antimicrobial Agents and Chemotherapy, 2000, 44(5): 1214-1222. |
| 95 | EL-SAYED A K, HOTHERSALL J, COOPER S M, et al. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586[J]. Chemistry & Biology, 2003, 10(5): 419-430. |
| 96 | FUKUDA D, HAINES A S, SONG Z, et al. A natural plasmid uniquely encodes two biosynthetic pathways creating a potent anti-MRSA antibiotic[J]. PLoS One, 2011, 6(3): e18031. |
| 97 | OLANO C, WILKINSON B, SÁNCHEZ C, et al. Biosynthesis of the angiogenesis inhibitor Borrelidin by Streptomyces parvulus Tü4055: Cluster Analysis and Assignment of Functions [J]. Chemistry & Biology, 2004, 11(1): 87-97. |
| 1 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs from 1981 to 2014[J]. Journal of Natural Products, 2016, 79(3): 629-661. |
| 2 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83 (3): 770-803. |
| 3 | LERNER C G, HAJDUK P J, WAGNER R, et al. From bacterial genomes to novel antibacterial agents: Discovery, characterization, and antibacterial activity of compounds that bind to HI0065 (YjeE) from Haemophilus influenzae [J]. Chemical Biology & Drug Design, 2007, 69 (6): 395-404. |
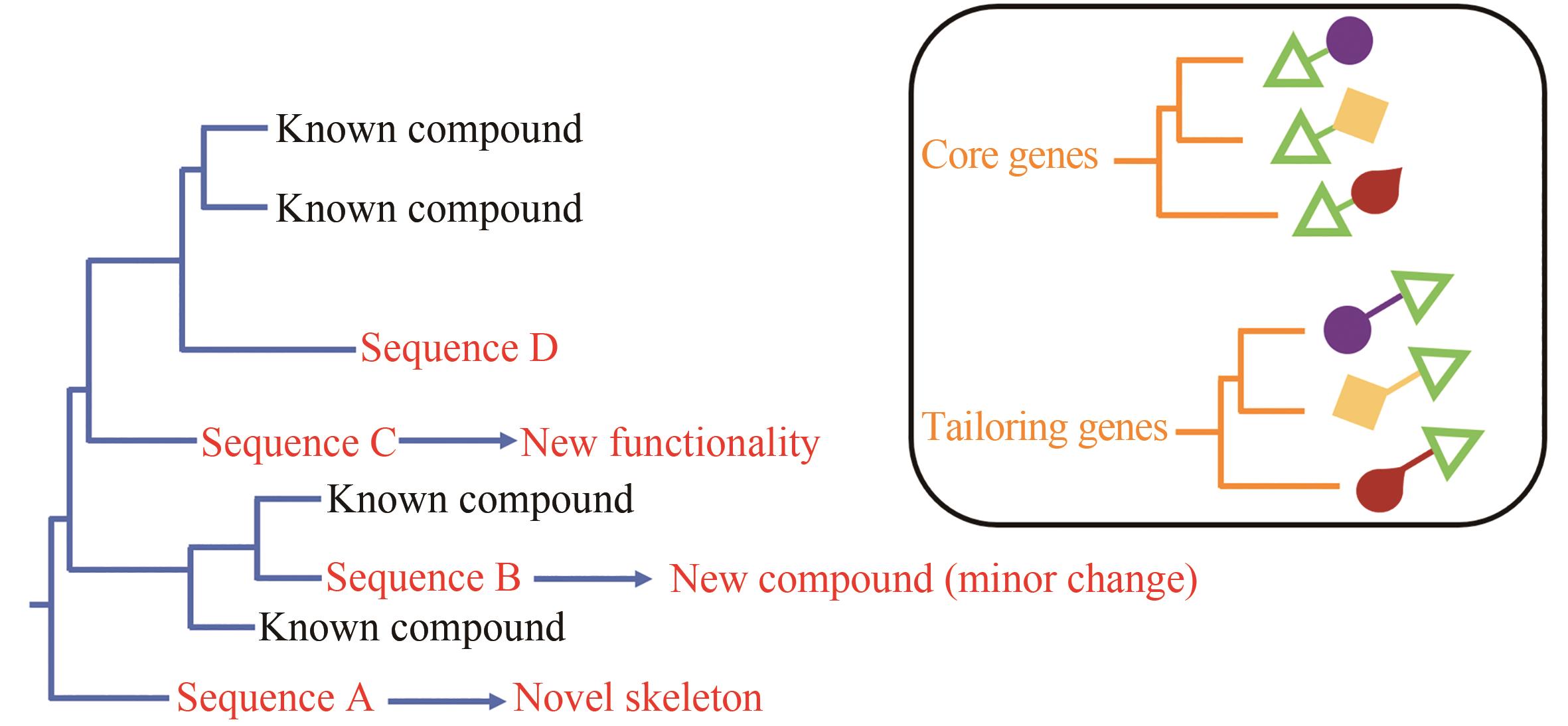
| 4 | CHALLIS G L. Genome mining for novel natural product discovery[J]. Journal of Medicinal Chemistry, 2008, 51(9): 2618-2628. |
| 5 | ZALLOT R, OBERG N O, GERLT J A. "Democratized" genomic enzymology web tools for functional assignment[J]. Current Opinion in Chemical Biology, 2018, 47: 77-85. |
| 6 | WEBER T. In silico tools for the analysis of antibiotic biosynthetic pathways[J]. International Journal of Medical Microbiology, 2014, 304(3/4): 230-235. |
| 7 | MEDEMA M H, FISCHBACH M A. Computational approaches to natural product discovery[J]. Nature Chemical Biology, 2015, 11(9): 639-648. |
| 8 | UMEMURA M, KOIKE H, MACHIDA M. Motif-independent de novo detection of secondary metabolite gene clusters-toward identification from filamentous fungi[J]. Frontiers in Microbiology, 2015, 6: 371. |
| 9 | WEBER T, KIM H U. The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production[J]. Synthetic and Systems Biotechnology, 2016, 1(2): 69-79. |
| 10 | BACHMANN B O, LANEN S G, BALTZ R H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making [J]? Journal of Industrial Microbiology & Biotechnology, 2014, 41(2): 175-184. |
| 11 | BODDY C N. Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides[J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(2): 443-450. |
| 12 | SCHEFFLER R J, COLMER S, TYNAN H, et al. Antimicrobials, drug discovery, and genome mining[J]. Applied Microbiology and Biotechnology, 2013, 97(3): 969-978. |
| 13 | YAEGASHI J, OAKLEY B R, WANG C C C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans [J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(2): 433-442. |
| 14 | SANTEN J A VAN, KAUTSAR S A, MEDEMA M H, et al. Microbial natural product databases: moving forward in the multi-omics era[J]. Natural Product Reports, 2021,38(1):264-278. |
| 98 | STANCU C, SIMA A. Statins: mechanism of action and effects[J]. Journal of Cellular and Molecular Medicine, 2001, 5(4): 378-387. |
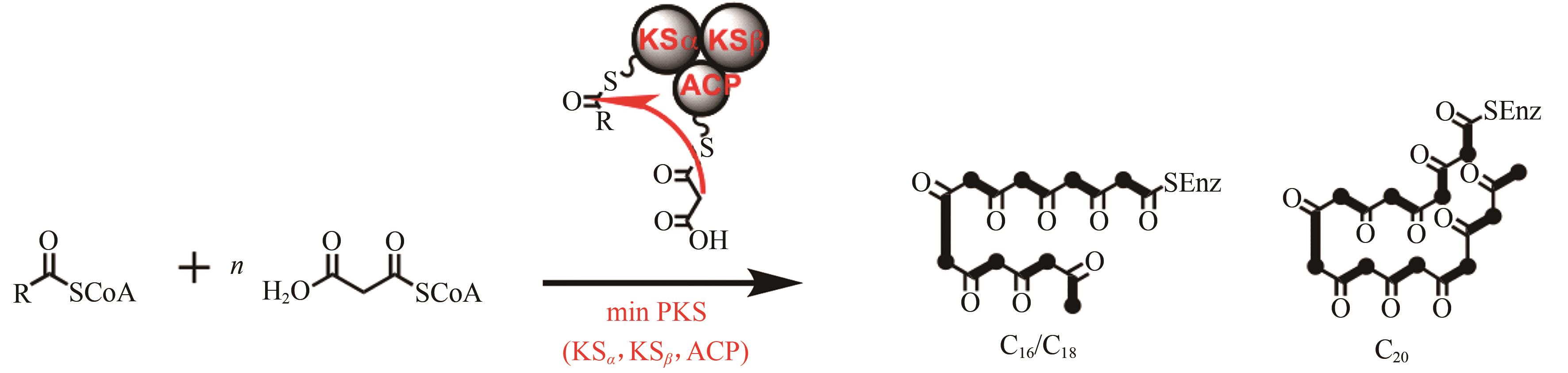
| 99 | HUTCHINSON C R, KENNEDY J, PARK C, et al. Aspects of the biosynthesis of non-aromatic fungal polyketides by iterative polyketide synthases[J]. Antonie van Leeuwenhoek, 2000, 78(3/4): 287-295. |
| 100 | CHAMILOS G, LEWIS R E, KONTOYIANNIS D P. Lovastatin has significant activity against zygomycetes and interacts synergistically with voriconazole[J]. Antimicrobial Agents and Chemotherapy, 2006, 50(1): 96-103. |
| 101 | AMORIM FRANCO T M, BLANCHARD J S. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability[J]. Biochemistry, 2017, 56(44): 5849-5865. |
| 102 | YAN Y, LIU Q, ZANG X, et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action[J]. Nature, 2018, 559(7714): 415-418. |
| 103 | TANG M C, ZOU Y, YEE D, et al. Identification of the pyranonigrin A biosynthetic gene cluster by genome mining in Penicillium thymicola IBT 5891[J]. AIChE Journal, 2018, 64(12): 4182-4186. |
| 104 | KANG H-S. Phylogeny-guided (meta) genome mining approach for the targeted discovery of new microbial natural products[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(2): 285-293. |
| 105 | FENG Z, KALLIFIDAS D, BRADY S F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(31): 12629-12634. |
| 106 | HERTWECK C. The biosynthetic logic of polyketide diversity[J]. Angewandte Chemie International Edition, 2009, 48(26): 4688-4716. |
| 107 | HILLENMEYER M E, VANDOVA G A, BERLEW E E, et al. Evolution of chemical diversity by coordinated gene swaps in type II polyketide gene clusters[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(45): 13952-13957. |
| 108 | KANG H-S, BRADY S F. Mining soil metagenomes to better understand the evolution of natural product structural diversity: Pentangular polyphenols as a case study[J]. Journal of the American Chemical Society, 2014, 136(52): 18111-18119. |
| 109 | MINOTTI G, MENNA P, SALVATORELLI E, et al. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity[J]. Pharmacological Reviews, 2004, 56(2): 185. |
| 15 | SOROKINA M, STEINBECK C. Review on natural products databases: where to find data in 2020[J]. Journal of Cheminformatics, 2020, 12(1): 20. |
| 16 | HARBORNE J B. Dictionary of natural products[EB/OL].2015, . |
| 17 | BOLTON EVAN E, WANG Y L, THIESSEN P A, et al. PubChem: integrated platform of small molecules and biological activities[J]. Annual Reports in Computational Chemistry, 2010, 4: 217-241. |
| 18 | AFENDI F M, OKADA T, YAMAZAKI M, et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research[J]. Plant and Cell Physiology, 2012, 53(2): e1. |
| 19 | CABOCHE S, PUPIN M, LECLÈRE V, al et, Norine : a database of nonribosomal peptides[J]. Nucleic Acids Research, 2008, 36(1): D326-D331. |
| 20 | ZIN P P K, WILLIAMS G J, EKINS S. Cheminformatics analysis and modeling with MacrolactoneDB[J]. Scientific Reports, 2020, 10(1): 6284. |
| 21 | ZENG X, ZHANG P, HE W D, et al. NPASS: natural product activity and species source database for natural product research, discovery and tool development[J]. Nucleic Acids Research, 2018, 46(D1): D1217-D1222. |
| 22 | KLEMENTZ D, DÖRING K, LUCAS X, et al. StreptomeDB 2.0—an extended resource of natural products produced by Streptomycetes [J]. Nucleic Acids Research, 2016, 44(D1): D509-D514. |
| 23 | SANTEN J A VAN, JACOB G, SINGH A L, et al. The natural products atlas: an open access knowledge base for microbial natural products discovery[J]. ACS Central Science, 2019, 5(11): 1824-1833. |
| 24 | KAUTSAR S A, BLIN K, SHAW S, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function[J]. Nucleic Acids Research, 2020, 48(D1): D454-D458. |
| 25 | WANG M, CARVER J J, PHELAN V V, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking[J]. Nature Biotechnology, 2016, 34(8): 828-837. |
| 26 | BENSON D A, CAVANAUGH M, CLARK K, et al. GenBank[J]. Nucleic Acids Research, 2013, 41(D1): D36-D42. |
| 27 | CONWAY K R, BODDY C N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis[J]. Nucleic Acids Research, 2013, 41(D1): D402-D407. |
| 28 | ICHIKAWA N, SASAGAWA M, YAMAMOTO M, et al. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters[J]. Nucleic Acids Research, 2013, 41(D1): D408-D414. |
| 29 | CHEN I M A, CHU K, PALANIAPPAN K, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes[J]. Nucleic Acids Research, 2019, 47(D1): D666-D677. |
| 30 | BLIN K, PASCAL ANDREU V, DE LOS SANTOS E L C, et al. The antiSMASH database version 2: a comprehensive resource on secondary metabolite biosynthetic gene clusters[J]. Nucleic Acids Research, 2019, 47(D1): D625-D630. |
| 31 | DIMINIC J, ZUCKO J, RUZIC I T, et al. Databases of the thiotemplate modular systems (CSDB) and their in silico recombinants (R-CSDB)[J]. Journal of Industrial Microbiology & Biotechnology, 2013, 40(6): 653-659. |
| 32 | MEDEMA M H, KOTTMANN R, YILMAZ P, et al. Minimum information about a biosynthetic gene cluster[J]. Nature Chemical Biology, 2015, 11(9): 625-631. |
| 33 | GRIGORIEV I V, NIKITIN R, HARIDAS S, et al. MycoCosm portal: gearing up for 1000 fungal genomes[J]. Nucleic Acids Research, 2014, 42(D1): D699-D704. |
| 34 | BLIN K, MEDEMA M H, KOTTMANN R, et al. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters[J]. Nucleic Acids Research, 2017, 45(D1): D555-D559. |
| 35 | O′LEARY N A, WRIGHT M W, BRISTER J R, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation[J]. Nucleic Acids Research, 2016, 44(D1): D733-D745. |
| 36 | The UniProt Consortium. UniProt: The universal protein knowledgebase[J]. Nucleic Acids Research, 2017, 45(D1): D158-D169. |
| 37 | RADIVOJAC P, CLARK W T, ORON T R, et al. A large-scale evaluation of computational protein function prediction[J]. Nature Methods, 2013, 10(3): 221-227. |
| 38 | EL-GEBALI S, MISTRY J, BATEMAN A, et al. The Pfam protein families database in 2019[J]. Nucleic Acids Research, 2019, 47(D1): D427-D432. |
| 110 | TACAR O, SRIAMORNSAK P, DASS C R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems[J]. Journal of Pharmacy and Pharmacology, 2013, 65(2): 157-170. |
| 111 | KANG H S, BRADY S F. Arimetamycin A: improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes[J]. Angewandte Chemie International Edition, 2013, 52(42): 11063-11067. |
| 112 | KANEHISA M, FURUMICHI M, TANABE M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs[J]. Nucleic Acids Research, 2017, 45(D1): D353-D361. |
| 39 | BLUM M, CHANG H-Y, CHUGURANSKY S, et al. The InterPro protein families and domains database: 20 years on[J]. Nucleic Acids Research, 2021:49(D1):D344-D354. |
| 40 | FINN R D, COGGILL P, EBERHARDT R Y, et al. The Pfam protein families database: towards a more sustainable future[J]. Nucleic Acids Research, 2016, 44(D1): D279-D285. |
| 41 | BENTLEY S D, CHATER K F, CERDEÑO-TÁRRAGA A M, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2)[J]. Nature, 2002, 417(6885): 141-147. |
| 42 | CAMACHO C, COULOURIS G, AVAGYAN V, et al. BLAST+: architecture and applications[J]. BMC Bioinformatics, 2009, 10: 421. |
| 43 | EDDY S R. Accelerated profile HMM searches[J]. PLoS Computational Biology, 2011, 7(10): e1002195. |
| 44 | BUCHFINK B, XIE C, HUSON D H. Fast and sensitive protein alignment using DIAMOND[J]. Nature Methods, 2015, 12(1): 59-60. |
| 45 | STARCEVIC A, ZUCKO J, SIMUNKOVIC J, et al. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures[J]. Nucleic Acids Research, 2008, 36(21): 6882-6892. |
| 46 | WEBER T, RAUSCH C, LOPEZ P, et al. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters[J]. Journal of Biotechnology, 2009, 140(1/2): 13-17. |
| 47 | LI M H, UNG P M, ZAJKOWSKI J, et al. Automated genome mining for natural products[J]. BMC Bioinformatics, 2009, 10(1): 185. |
| 48 | KHALDI N, SEIFUDDIN F T, TURNER G, et al. SMURF: genomic mapping of fungal secondary metabolite clusters[J]. Fungal Genetics and Biology, 2010, 47(9): 736-741. |
| 49 | MEDEMA M H, BLIN K, CIMERMANCIC P, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences[J]. Nucleic Acids Research, 2011, 39(2): W339-W346. |
| [1] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| [2] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| [3] | 谢向前, 郭雯, 王欢, 李进. 含氨基乙烯半胱氨酸核糖体肽的生物合成与化学合成[J]. 合成生物学, 2024, 5(5): 981-996. |
| [4] | 张俊, 金诗雪, 云倩, 瞿旭东. 聚酮化合物非天然延伸单元的生物合成与结构改造应用[J]. 合成生物学, 2024, 5(3): 561-570. |
| [5] | 虞旭昶, 吴辉, 李雷. 文库构建与基因簇靶向筛选驱动的微生物天然产物高效发现[J]. 合成生物学, 2024, 5(3): 492-506. |
| [6] | 冯金, 潘海学, 唐功利. 近十年天然产物药物的生物合成研究进展[J]. 合成生物学, 2024, 5(3): 408-446. |
| [7] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| [8] | 雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526. |
| [9] | 施鑫杰, 杜艺岭. 双嵌入家族抗肿瘤非核糖体肽的生物合成研究进展[J]. 合成生物学, 2024, 5(3): 593-611. |
| [10] | 张瑞, 金文铮, 陈依军. 细菌聚酮合酶间的杂合方式及聚酮化合物生物合成逻辑[J]. 合成生物学, 2024, 5(3): 548-560. |
| [11] | 宋永相, 张秀凤, 李艳芹, 肖华, 闫岩. 自抗性基因导向的活性天然产物挖掘[J]. 合成生物学, 2024, 5(3): 474-491. |
| [12] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [13] | 胡哲辉, 徐娟, 卞光凯. 自动化高通量技术在天然产物生物合成中的应用[J]. 合成生物学, 2023, 4(5): 932-946. |
| [14] | 张凡忠, 相长君, 张骊駻. 进化与大数据导向生物信息学在天然产物研究中的发展及应用[J]. 合成生物学, 2023, 4(4): 629-650. |
| [15] | 吕靖伟, 邓子新, 张琪, 丁伟. 基于深度学习识别RiPPs前体肽及裂解位点[J]. 合成生物学, 2022, 3(6): 1262-1276. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||