合成生物学 ›› 2022, Vol. 3 ›› Issue (4): 690-708.DOI: 10.12211/2096-8280.2021-076
可基因编码点击化学在材料合成生物学中的应用
易琪昆1, 孙晨博1, 杨中光1, 王日1, 寇松姿2, 李朝霞3, 孙飞1,2
- 1.香港科技大学化学及生物工程学系,中国 香港 000000
2.深圳湾实验室粤港澳生物医学创新中心,广东 深圳 518000
3.盐城工学院海洋与生物工程学院,江苏 盐城 224000
-
收稿日期:2021-07-23修回日期:2022-02-21出版日期:2022-08-31发布日期:2022-09-08 -
通讯作者:孙飞 -
作者简介:易琪昆 (1995—),博士研究生。研究方向为工程活体材料、材料合成生物学、化学生物学。E-mail:qyi@connect.ust.hk孙飞 (1987—),博士,副教授。研究方向为蛋白质工程、材料合成生物学、化学生物学。E-mail:kefsun@ust.hk -
基金资助:国家重点研发计划(2020YFA0908100);广东省自然科学基金(2019A1515011691)
Genetically encoded click chemistry, an enabling tool for materials synthetic biology
YI Qikun1, SUN Chenbo1, YANG Zhongguang1, WANG Ri1, KOU Songzi2, LI Zhaoxia3, SUN Fei1,2
- 1.Department of Chemical and Biological Engineering,Hong Kong University of Science and Technology,Hong Kong SAR 000000,China
2.Shenzhen Bay Laboratory,Shenzhen 518000,Guangdong,China
3.College of Marine and Biological Engineering,Yancheng Institute of Technology,Yancheng 224000,Jiangsu,China
-
Received:2021-07-23Revised:2022-02-21Online:2022-08-31Published:2022-09-08 -
Contact:SUN Fei
摘要:
对材料的结构与功能实现精准控制是材料学领域的根本挑战之一。很多生物大分子之间具有多种特异性的反应,利用这些特异性反应将生物大分子乃至活体细胞直接组装成更高级的结构提供了一种可能的解决方案。为了实现这种特异有效的组装,借鉴于传统的点击化学,可基因编码点击化学应运而生。这套新的蛋白质化学源于自然界微生物中的异肽键化学,通过基因编辑的方式控制功能化蛋白质分子的自组装反应,同时具有点击化学的高效等优点,经过多种优化之后在合成生物学中被广泛运用。其发展和应用有力促进了蛋白质工程、材料学与合成生物学的交叉融合。本文简要介绍了可基因编码点击化学在近年的应用,包括以多种非线性蛋白质分子为代表的蛋白质拓扑工程产物、多种具有不同功能的全蛋白质水凝胶材料、重组疫苗及工程活体材料等,并总结了其发展潜力。
中图分类号:
引用本文
易琪昆, 孙晨博, 杨中光, 王日, 寇松姿, 李朝霞, 孙飞. 可基因编码点击化学在材料合成生物学中的应用[J]. 合成生物学, 2022, 3(4): 690-708.
YI Qikun, SUN Chenbo, YANG Zhongguang, WANG Ri, KOU Songzi, LI Zhaoxia, SUN Fei. Genetically encoded click chemistry, an enabling tool for materials synthetic biology[J]. Synthetic Biology Journal, 2022, 3(4): 690-708.
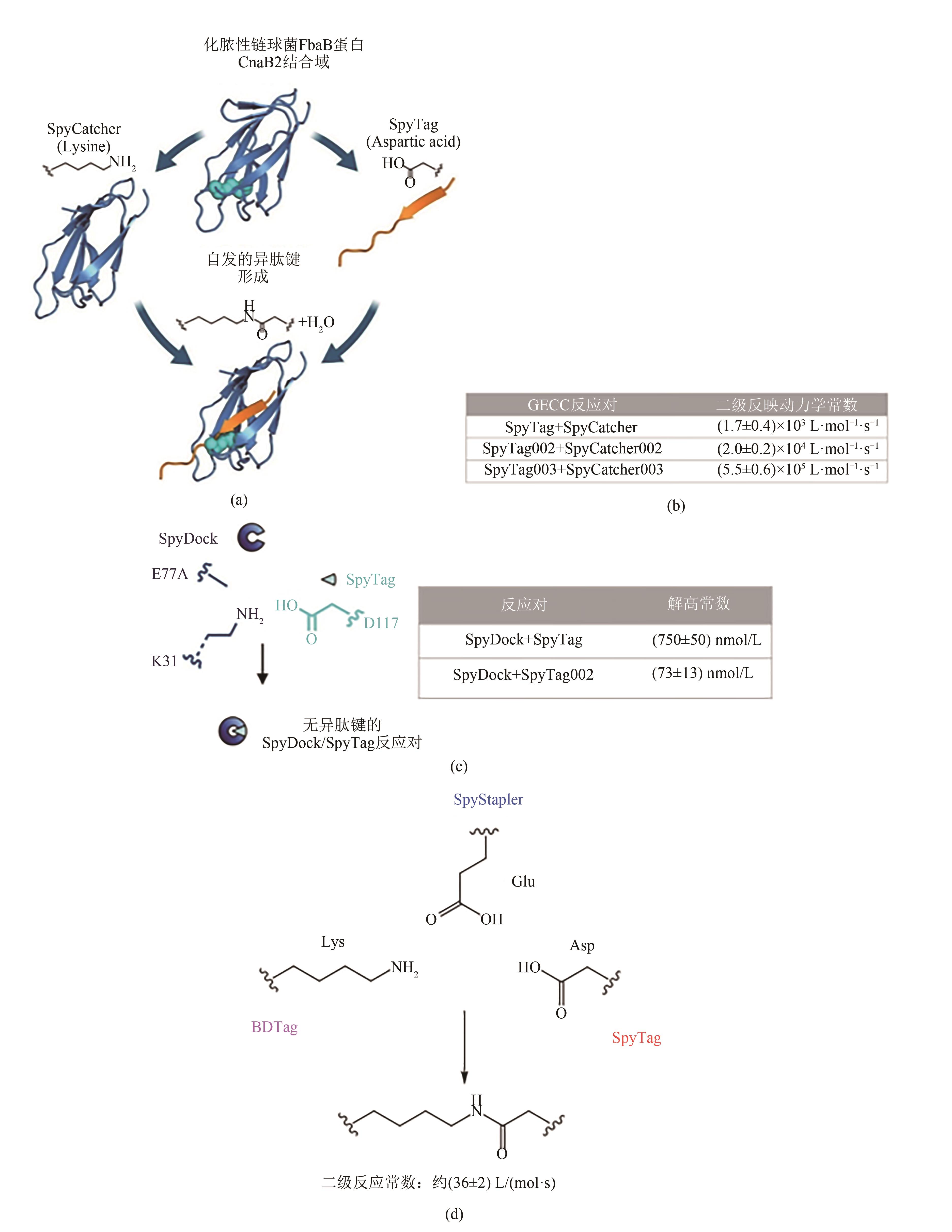
图1 可基因编码点击化学(GECC)概览(a)代表性的可基因编码点击化学——谍化学(SpyTag/SpyCatcher chemistry)。伴随着SpyTag与SpyCatcher的特异性的分子识别与结合,天冬氨酸与赖氨酸的侧链自发形成异肽键[5-7]。SpyTag/SpyCatcher复合物结构的PDB编号为4MLI。(b)谍化学及其优化版本(Spy002与Spy003)的二级反应动力学常数[8, 9]。(c)无异肽键的SpyTag/SpyDock可逆反应体系[10]。SpyDock为SpyCatcher-E77A突变体,失去了形成异肽键的能力。(d)三组分SpyStapler/SpyTag/BDTag反应体系及其二级反应动力学常数[11]

图3 利用GECC构建全蛋白水凝胶材料LIF—白血病抑制因子;A-LIF-A—SpyTag-ELP-LIF-ELP-SpyTag
Fig. 3 Development of entirely protein-based hydrogels based on GECC(Adapted from Ref. 49 with permission from National Academy of Sciences, copyright 2014.)LIF—Leukemia inhibitory factor; A-LIF-A—SpyTag-ELP-LIF-LEP-SpyTag
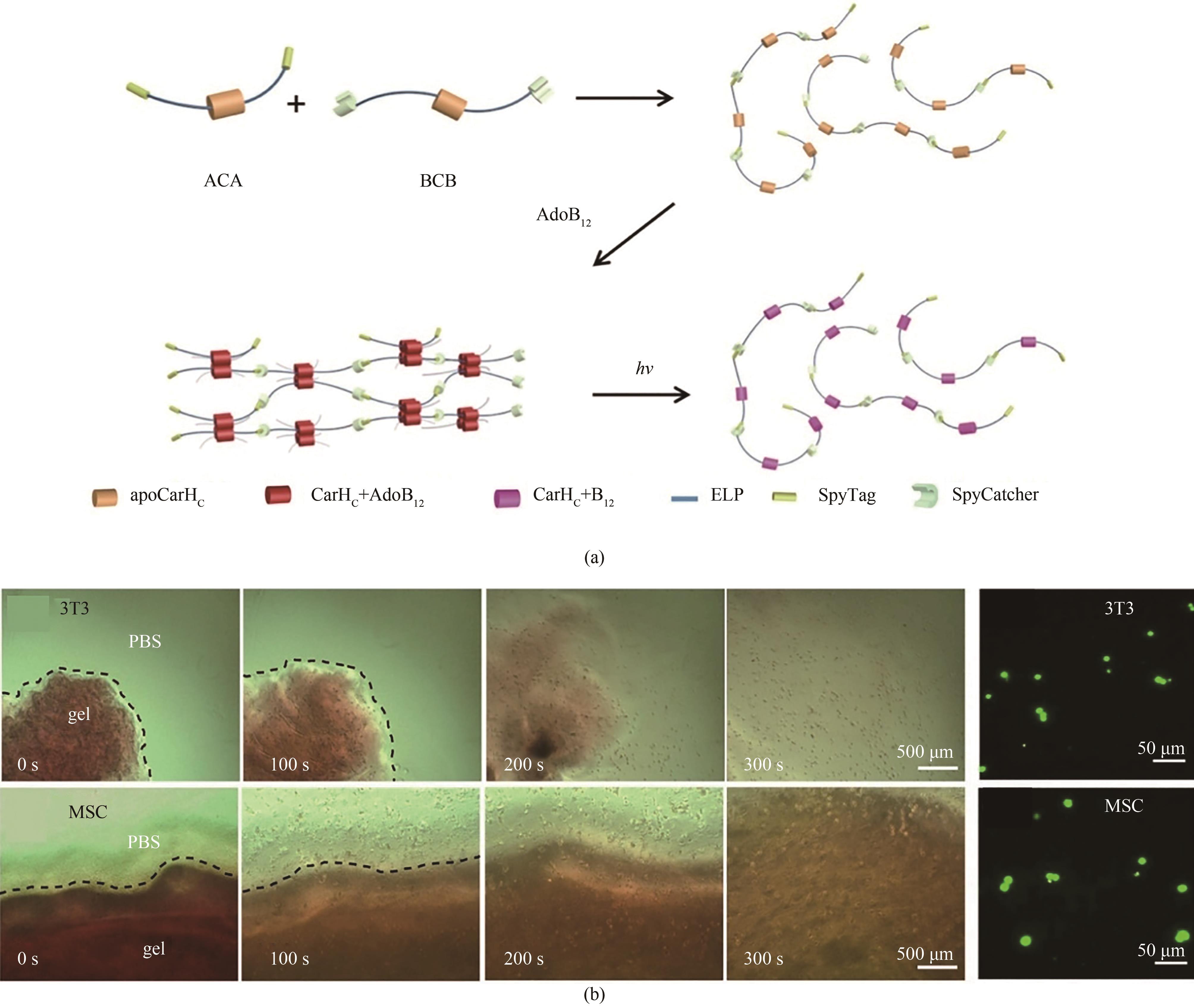
图4 利用GECC制备“智能”光响应水凝胶(a)利用谍化学将CarHc组装成线性聚合物,后者在黑暗条件下经历AdoB12诱导的液固相变与光诱导的固液相变。(b)光诱导的细胞释放。3T3—成纤维细胞;hMSCs—人间充质干细胞。
Fig. 4 Development of smart photo-responsive hydrogel based on GECCAdapted from Ref. 52 with permission from National Academy of Sciences, copyright 2017.(a) Constructing CarHc into linear polymer utilizing SpyChemistry, which can undergo liquid-to-solid phase transition induced by AdoB12, and solid-to-liquid phase transition induced by light. (b) Photo-induced release of 3T3 cells or hMSCs.

图5 利用GECC合成重金属结合蛋白水凝胶(a)超级铀酰吸附蛋白材料与铬酸根吸附蛋白材料的合成。(b)SUP水凝胶与拥有更强铀酰结合能力的突变体SUP(E64D)水凝胶从天然海水中富集铀元素。富集指数是指富集后蛋白材料中铀的浓度与水体中铀的浓度的比值。(c)ModA水凝胶去除自来水样品中的铬酸盐。去除比例是指吸附后水体中铬酸盐的减少比例。去除效率是指蛋白材料吸附铬酸盐的实际质量与理论上最大可能质量的比例
Fig. 5 Applying GECC to develop heavy-metal binding protein hydrogelAdapted from Ref. 51 with permission from The Royal Society of Chemistry, copyright 2017.(a) Synthesis of super uranyl-binding protein (SUP) materials and chromate-binding protein materials. (b) Uranium extraction from seawater using the hydrogels comprising SUP or its mutant, SUP-E64D, which exhibits stronger uranyl binding. Enrichment index was calculated as the ratio of uranium concentration in protien material to that in water body after enrichment. (c) Removal of chromate from tap water using ModA hydrogels. Removal percentage was calculated as the reduced percentage of chromate in water body after removal. Removal efficiency was calculated as the ratio of the actual amount of chromate absorbed by the material to the theoretical maximum by the material.
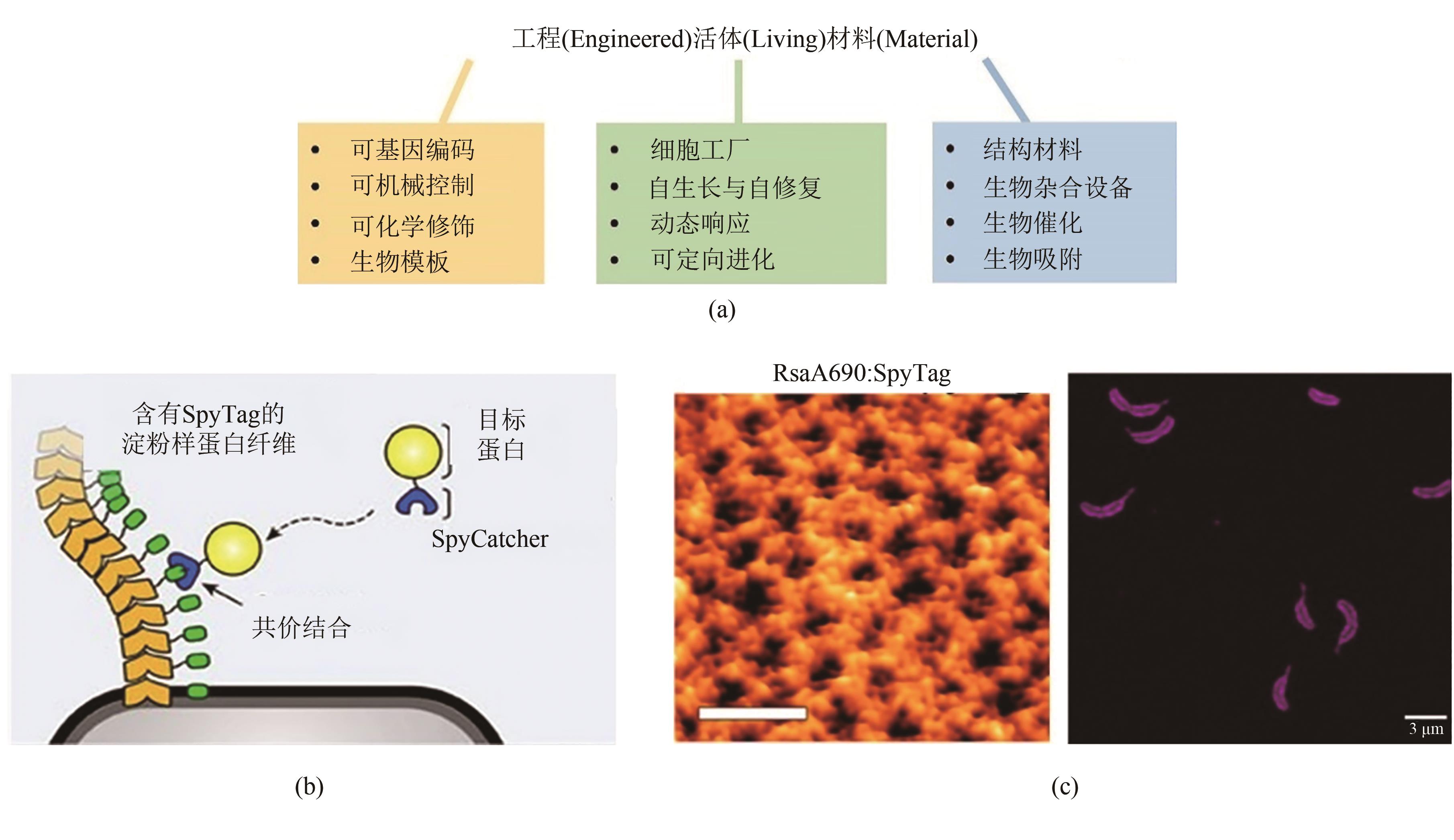
图7 GECC与工程活体材料(a)工程活体材料的特点[102]。(b)利用GECC修饰大肠杆菌生物被膜[111]。(c)利用GECC修饰新月柄杆菌表层蛋白晶格。左图:原子力显微镜下修饰有SpyTag的新月柄杆菌表层蛋白晶格结构,比例尺40 nm;右图:用SpyCatcher-mRFP1荧光蛋白特异性标记的含SpyTag的新月柄杆菌材料,比例尺3 μm[114]
Fig. 7 GECC and engineered living materials(a) Features of engineered living materials. (b) Decoration of E. coli biofilm via GECC. (c) Decoration of 2D hexameric protein lattice (S-layer) on the surface of C. crescentus via GECC. Left: SpyTagged S-layer visualized by AFM. Scale bar: 40 nm. Right: the C. crescentus material labelled with SpyCatcher-mRFP1 imaged via fluorescence microscopy. Scale bar: 3 μm.
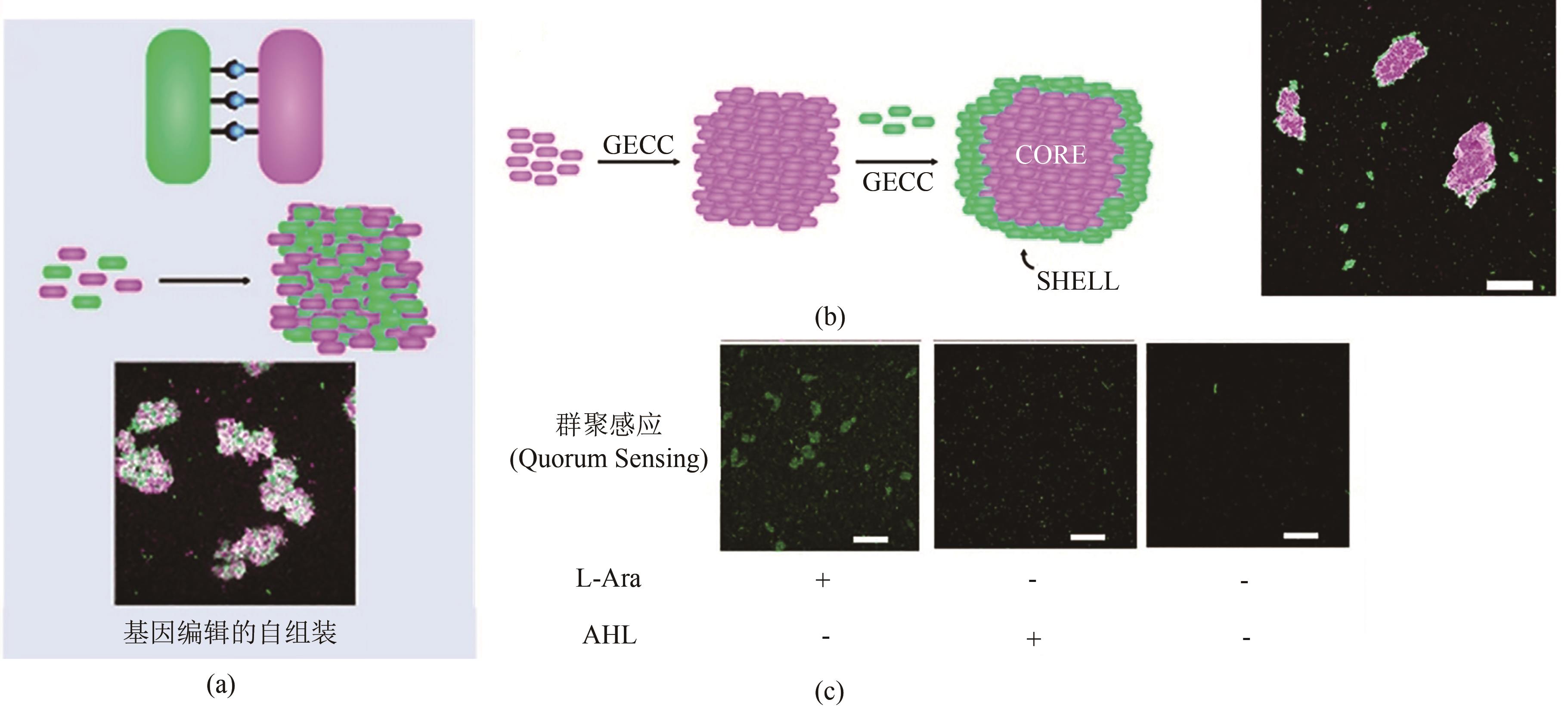
图9 基于GECC自组装的大肠杆菌活体材料及其特点(a)基于GECC自组装的大肠杆菌活体材料示意图[118]。(b)自组装形成的壳层结构工程活体材料,比例尺100 μm。(c)基于SpyTag/SpyCatcher的组装能够激活细菌的群聚感应。AHL—N-Acyl-Homoserine Lactone,群聚感应(quorum sensing)的自诱导剂。群聚感应的报告基因为绿色荧光蛋白mWasabi。阿拉伯糖(L-Arabinose或者L-Ara)用于诱导SpyTag/SpyCatcher的表达。比例尺100 μm
Fig. 9 GECC-based self-assembled E. coli living material and its features(a) Diagram of GECC-based self-assembled E. coli living material[118]. (b) Self-assembled engineered living materials with core-shell structure, scale bar: 100 μm. (c) Intercellular assembly enabled by Spy chemistry activates quorum sensing. AHL, N-Acyl-Homoserine Lactone, an autoinducer of quorum sening. The green fluorescent protein, mWasabi, served as a reporter of quorum sensing. L-Arabinose (L-Ara) was used to induce the expression of SpyTag and SpyCatcher. Scale bar: 100 μm.
| 1 | KOLB H C, FINN M G, SHARPLESS K B. Click chemistry: diverse chemical function from a few good reactions[J]. Angewandte Chemie International Edition, 2001, 40(11): 2004-2021. |
| 2 | BARNER-KOWOLLIK C, DU PREZ F E, ESPEEL P, et al. “Clicking” polymers or just efficient linking: what is the difference? [J]. Angewandte Chemie International Edition, 2011, 50(1): 60-62. |
| 3 | KANG H J, COULIBALY F, CLOW F, et al. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure[J]. Science, 2007, 318(5856): 1625-1628. |
| 4 | ZAKERI B, HOWARTH M. Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting[J]. Journal of the American Chemical Society, 2010, 132(13): 4526-4527. |
| 5 | ZAKERI B, FIERER J O, CELIK E, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(12): E690-E697. |
| 6 | TAN L, HOON S S, WONG F T. Kinetic controlled tag-catcher interactions for directed covalent protein assembly[J]. PLoS One, 2016, 11(10): e0165074. |
| 7 | HATLEM D, TRUNK T, LINKE D, et al. Catching a SPY: using the SpyCatcher-SpyTag and related systems for labeling and localizing bacterial proteins[J]. International Journal of Molecular Sciences, 2019, 20(9): 2129. |
| 8 | KEEBLE A H, TURKKI P, STOKES S, et al. Approaching infinite affinity through engineering of peptide-protein interaction[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(52): 26523-26533. |
| 9 | SUN F, ZHANG W B. Genetically encoded click chemistry [J]. Chinese Journal of Chemistry, 2020, 38(8): 894-896. |
| 10 | KHAIRIL ANUAR I N A, BANERJEE A, KEEBLE A H, et al. Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox[J]. Nature Communications, 2019, 10: 1734. |
| 11 | WU X L, LIU Y J, LIU D, et al. An intrinsically disordered peptide-peptide stapler for highly efficient protein ligation both in vivo and in vitro [J]. Journal of the American Chemical Society, 2018, 140(50): 17474-17483. |
| 12 | REDDINGTON S C, HOWARTH M. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher[J]. Current Opinion in Chemical Biology, 2015, 29: 94-99. |
| 13 | PRÖSCHEL M, DETSCH R, BOCCACCINI A R, et al. Engineering of metabolic pathways by artificial enzyme channels[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 168. |
| 14 | ZAKERI B. Synthetic biology: a new tool for the trade[J]. Chembiochem, 2015, 16(16): 2277-2282. |
| 15 | BEDBROOK C N, KATO M, RAVINDRA KUMAR S, et al. Genetically encoded spy peptide fusion system to detect plasma membrane-localized proteins in vivo [J]. Chemistry & Biology, 2015, 22(8): 1108-1121. |
| 16 | MOON H, BAE Y, KIM H, et al. Plug-and-playable fluorescent cell imaging modular toolkits using the bacterial superglue, SpyTag/SpyCatcher[J]. Chemical Communications, 2016, 52(97): 14051-14054. |
| 17 | PESSINO V, CITRON Y R, FENG S Y, et al. Covalent protein labeling by SpyTag-SpyCatcher in fixed cells for super-resolution microscopy[J]. ChemBioChem, 2017, 18(15): 1492-1495. |
| 18 | YOUNG P G, YOSAATMADJA Y, HARRIS P W R, et al. Harnessing ester bond chemistry for protein ligation[J]. Chemical Communications, 2017, 53(9): 1502-1505. |
| 19 | ZOTTIG X, CÔTÉ-CYR M, ARPIN D, et al. Protein supramolecular structures: from self-assembly to nanovaccine design[J]. Nanomaterials (Basel, Switzerland), 2020, 10(5): 1008. |
| 20 | VEGGIANI G, ZAKERI B, HOWARTH M. Superglue from bacteria: unbreakable bridges for protein nanotechnology[J]. Trends in Biotechnology, 2014, 32(10): 506-512. |
| 21 | ZHANG W B, SUN F, TIRRELL D A, et al. Controlling macromolecular topology with genetically encoded SpyTag-SpyCatcher chemistry[J]. Journal of the American Chemical Society, 2013, 135(37): 13988-13997. |
| 22 | SHAH N H, MUIR T W. Inteins: nature’s gift to protein chemists[J]. Chemical Science, 2014, 5(1): 446-461. |
| 23 | SCHMIDT M, TOPLAK A, QUAEDFLIEG P J, et al. Enzyme-mediated ligation technologies for peptides and proteins[J]. Current Opinion in Chemical Biology, 2017, 38: 1-7. |
| 24 | ZHANG F and ZHANG W B. Encrypting Chemical Reactivity in Protein Sequences toward Information-Coded Reactions[J]. Chinese Journal of Chemistry, 2020,38(8): 864-878. |
| 25 | ANDERSSON F I, PINA D G, MALLAM A L, et al. Untangling the folding mechanism of the 52-knotted protein UCH-L3[J]. The FEBS Journal, 2009, 276(9): 2625-2635. |
| 26 | BOUTZ D R, CASCIO D, WHITELEGGE J, et al. Discovery of a thermophilic protein complex stabilized by topologically interlinked chains[J]. Journal of Molecular Biology, 2007, 368(5): 1332-1344. |
| 27 | MALLAM A L, JACKSON S E. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins[J]. Nature Chemical Biology, 2012, 8(2): 147-153. |
| 28 | SÁNCHEZ-HIDALGO M, MONTALBÁN-LÓPEZ M, CEBRIÁN R, et al. AS-48 bacteriocin: close to perfection[J]. Cellular and Molecular Life Sciences, 2011, 68(17): 2845-2857. |
| 29 | TRABI M, SCHIRRA H J, CRAIK D J. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from Rhesus macaque Leukocytes[J]. Biochemistry, 2001, 40(14): 4211-4221. |
| 30 | VAN ELDIJK M B, VAN LEEUWEN I, MIKHAILOV V A, et al. Evidence that the catenane form of CS2 hydrolase is not an artefact[J]. Chemical Communications, 2013, 49(71): 7770-7772. |
| 31 | NGUYEN G K T, QIU Y B, CAO Y, et al. Butelase-mediated cyclization and ligation of peptides and proteins[J]. Nature Protocols, 2016, 11(10): 1977-1988. |
| 32 | WU Z M, GUO X Q, GUO Z W. Sortase A-catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides[J]. Chemical Communications, 2011, 47(32): 9218-9220. |
| 33 | XU M Q, EVANS T C JR. Intein-mediated ligation and cyclization of expressed proteins[J]. Methods, 2001, 24(3): 257-277. |
| 34 | WANG X W, ZHANG W B. Cellular synthesis of protein catenanes[J]. Angewandte Chemie International Edition, 2016, 55(10): 3442-3446. |
| 35 | SCHOENE C, FIERER J O, BENNETT S P, et al. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme[J]. Angewandte Chemie International Edition, 2014, 53(24): 6101-6104. |
| 36 | WANG Y D, TIAN J W, XIAO Y, et al. SpyTag/SpyCatcher cyclization enhances the thermostability and organic solvent tolerance of L-phenylalanine aldolase[J]. Biotechnology Letters, 2019, 41(8/9): 987-994. |
| 37 | XU C, XU Q, HUANG H, et al. Enhancing the stability of trehalose synthase via SpyTag/SpyCatcher cyclization to improve its performance in industrial biocatalysts[J]. Bioscience, Biotechnology, and Biochemistry, 2018, 82(9): 1473-1479. |
| 38 | ZHENG Q, WANG M J, ZHANG L, et al. Topology engineering via protein catenane construction to strengthen an industrial biocatalyst[J]. Journal of Biotechnology, 2021, 325: 271-279. |
| 39 | LEE K Y, MOONEY D J. Hydrogels for tissue engineering[J]. Chemical Reviews, 2001, 101(7): 1869-1879. |
| 40 | GANJI F, VASHEGHANI-FARAHANI S, VASHEGHANI-FARAHANI E. Theoretical description of hydrogel swelling: a review [J]. Iranian Polymer Journal, 2010,19(5): 375-398 |
| 41 | BURDICK J A, MURPHY W L. Moving from static to dynamic complexity in hydrogel design[J]. Nature Communications, 2012, 3: 1269. |
| 42 | BURDICK J A, MAUCK R L, GERECHT S. To serve and protect: hydrogels to improve stem cell-based therapies[J]. Cell Stem Cell, 2016, 18(1): 13-15. |
| 43 | TESSMAR J K, GÖPFERICH A M. Matrices and scaffolds for protein delivery in tissue engineering[J]. Advanced Drug Delivery Reviews, 2007, 59(4/5): 274-291. |
| 44 | DEFOREST C A, POLIZZOTTI B D, ANSETH K S. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments[J]. Nature Materials, 2009, 8(8): 659-664. |
| 45 | BANTA S, WHEELDON I R, BLENNER M. Protein engineering in the development of functional hydrogels[J]. Annual Review of Biomedical Engineering, 2010, 12: 167-186. |
| 46 | LI Y, XUE B, CAO Y. 100th anniversary of macromolecular science viewpoint: synthetic protein hydrogels[J]. ACS Macro Letters, 2020, 9(4): 512-524. |
| 47 | SUN F, ZHANG W B. Unleashing chemical power from protein sequence space toward genetically encoded “click” chemistry[J]. Chinese Chemical Letters, 2017, 28(11): 2078-2084. |
| 48 | HUANG P S, BOYKEN S E, BAKER D. The coming of age of de novo protein design[J]. Nature, 2016, 537(7620): 320-327. |
| 49 | SUN F, ZHANG W B, MAHDAVI A, et al. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11269-11274. |
| 50 | YANG Z G, KOU S Z, WEI X, et al. Genetically programming stress-relaxation behavior in entirely protein-based molecular networks[J]. ACS Macro Letters, 2018, 7(12): 1468-1474. |
| 51 | KOU S Z, YANG Z G, LUO J R, et al. Entirely recombinant protein-based hydrogels for selective heavy metal sequestration[J]. Polymer Chemistry, 2017, 8(39): 6158-6164. |
| 52 | WANG R, YANG Z G, LUO J R, et al. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(23): 5912-5917. |
| 53 | KOU S Z, YANG X, YANG Z G, et al. Cobalt-cross-linked, redox-responsive spy network protein hydrogels[J]. ACS Macro Letters, 2019, 8(7): 773-778. |
| 54 | CAO Y J, WEI X, LIN Y, et al. Synthesis of bio-inspired viscoelastic molecular networks by metal-induced protein assembly[J]. Molecular Systems Design & Engineering, 2020, 5(1): 117-124. |
| 55 | YANG Z G, YANG Y, WANG M, et al. Dynamically tunable, macroscopic molecular networks enabled by cellular synthesis of 4-arm star-like proteins[J]. Matter, 2020, 2(1): 233-249. |
| 56 | GAO X Y, FANG J, XUE B, et al. Engineering protein hydrogels using SpyCatcher-SpyTag chemistry[J]. Biomacromolecules, 2016, 17(9): 2812-2819. |
| 57 | ORTIZ-GUERRERO J M, POLANCO M C, MURILLO F J, et al. Light-dependent gene regulation by a coenzyme B12-based photoreceptor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(18): 7565-7570. |
| 58 | JOST M, FERNÁNDEZ-ZAPATA J, POLANCO M C, et al. Structural basis for gene regulation by a B12-dependent photoreceptor[J]. Nature, 2015, 526(7574): 536-541. |
| 59 | KUTTA R J, HARDMAN S J O, JOHANNISSEN L O, et al. The photochemical mechanism of a B12-dependent photoreceptor protein[J]. Nature Communications, 2015, 6: 7907. |
| 60 | HULTSCHIG C, HECHT H J, FRANK R. Systematic delineation of a calmodulin peptide interaction[J]. Journal of Molecular Biology, 2004, 343(3): 559-568. |
| 61 | LUO J R, SUN F. Calcium-responsive hydrogels enabled by inducible protein-protein interactions[J]. Polymer Chemistry, 2020, 11(31): 4973-4977. |
| 62 | KUBONIWA H, TJANDRA N, GRZESIEK S, et al. Solution structure of calcium-free calmodulin[J]. Nature Structural Biology, 1995, 2(9): 768-776. |
| 63 | LIU X T, YANG X, YANG Z G, et al. Versatile engineered protein hydrogels enabling decoupled mechanical and biochemical tuning for cell adhesion and neurite growth[J]. ACS Applied Nano Materials, 2018, 1(4): 1579-1585. |
| 64 | UHRICH K E, CANNIZZARO S M, LANGER R S, et al. Polymeric systems for controlled drug release[J]. Chemical Reviews, 1999, 99(11): 3181-3198. |
| 65 | JIANG B J, LIU X T, YANG C, et al. Injectable, photoresponsive hydrogels for delivering neuroprotective proteins enabled by metal-directed protein assembly[J]. Science Advances, 2020, 6(41): eabc4824. |
| 66 | PENG H T, SHEK P N. Novel wound sealants: biomaterials and applications[J]. Expert Review of Medical Devices, 2010, 7(5): 639-659. |
| 67 | MEHDIZADEH M, YANG J. Design strategies and applications of tissue bioadhesives[J]. Macromolecular Bioscience, 2013, 13(3): 271-288. |
| 68 | TROTT A T. Cyanoacrylate tissue adhesives. An advance in wound care[J]. JAMA: the Journal of the American Medical Association, 1997, 277(19): 1559-1560. |
| 69 | MARTINOWITZ U, SALTZ R. Fibrin sealant[J]. Current Opinion in Hematology, 1996, 3(5): 395-402. |
| 70 | ALBES J M, KRETTEK C, HAUSEN B, et al. Biophysical properties of the gelatin-resorcin-formaldehyde/glutaraldehyde adhesive[J]. The Annals of Thoracic Surgery, 1993, 56(4): 910-915. |
| 71 | JAYAKUMAR R, PRABAHARAN M, SUDHEESH KUMAR P T, et al. Biomaterials based on chitin and chitosan in wound dressing applications[J]. Biotechnology Advances, 2011, 29(3): 322-337. |
| 72 | LEE H, DELLATORE S M, MILLER W M, et al. Mussel-inspired surface chemistry for multifunctional coatings[J]. Science, 2007, 318(5849): 426-430. |
| 73 | LEE B P, MESSERSMITH P B, ISRAELACHVILI J N, et al. Mussel-inspired adhesives and coatings[J]. Annual Review of Materials Research, 2011, 41: 99-132. |
| 74 | CHOI Y S, YANG Y J, YANG B, et al. In vivo modification of tyrosine residues in recombinant mussel adhesive protein by tyrosinase co-expression in Escherichia coli [J]. Microbial Cell Factories, 2012, 11: 139. |
| 75 | LUO J R, LIU X T, YANG Z G, et al. Synthesis of entirely protein-based hydrogels by enzymatic oxidation enabling water-resistant bioadhesion and stem cell encapsulation[J]. ACS Applied Bio Materials, 2018, 1(5): 1735-1740. |
| 76 | PARK B M, LUO J R, SUN F. Enzymatic assembly of adhesive molecular networks with sequence-dependent mechanical properties inspired by mussel foot proteins[J]. Polymer Chemistry, 2019, 10(7): 823-826. |
| 77 | LEGGETT C J, ENDRIZZI F, RAO L F. Scientific basis for efficient extraction of uranium from seawater (II): Fundamental thermodynamic and structural studies[J]. Industrial & Engineering Chemistry Research, 2016, 55(15): 4257-4263. |
| 78 | MEJÁRE M, BÜLOW L. Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals[J]. Trends in Biotechnology, 2001, 19(2): 67-73. |
| 79 | ZHOU L, BOSSCHER M, ZHANG C S, et al. A protein engineered to bind uranyl selectively and with femtomolar affinity[J]. Nature Chemistry, 2014, 6(3): 236-241. |
| 80 | AKPOR O, MUCHIE M. Remediation of heavy metals in drinking water and wastewater treatment systems: Processes and applications[J]. International Journal of the Physical Sciences, 2010, 5(12): 1807-1817. |
| 81 | LI Y Y, LENEGHAN D B, MIURA K, et al. Enhancing immunogenicity and transmission-blocking activity of malaria vaccines by fusing Pfs25 to IMX313 multimerization technology[J]. Scientific Reports, 2016, 6: 18848. |
| 82 | POTEET E, LEWIS P, CHEN C Y, et al. Toll-like receptor 3 adjuvant in combination with virus-like particles elicit a humoral response against HIV[J]. Vaccine, 2016, 34(48): 5886-5894. |
| 83 | ASADI K, GHOLAMI A. Virosome-based nanovaccines; a promising bioinspiration and biomimetic approach for preventing viral diseases: a review[J]. International Journal of Biological Macromolecules, 2021, 182: 648-658. |
| 84 | PAN J D, CUI Z Q. Self-assembled nanoparticles: Exciting platforms for vaccination[J]. Biotechnology Journal, 2020, 15(12): e2000087. |
| 85 | JANSSENS M E, GEYSEN D, BROOS K, et al. Folding properties of the hepatitis B core as a carrier protein for vaccination research[J]. Amino Acids, 2010, 38(5): 1617-1626. |
| 86 | BRUNE K D, LENEGHAN D B, BRIAN I J, et al. Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization[J]. Scientific Reports, 2016, 6: 19234. |
| 87 | THRANE S, JANITZEK C M, MATONDO S, et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines[J]. Journal of Nanobiotechnology, 2016, 14: 30. |
| 88 | JANITZEK C M, MATONDO S, THRANE S, et al. Bacterial superglue generates a full-length circumsporozoite protein virus-like particle vaccine capable of inducing high and durable antibody responses[J]. Malaria Journal, 2016, 15(1): 545. |
| 89 | SINGH S K, THRANE S, JANITZEK C M, et al. Improving the malaria transmission-blocking activity of a Plasmodium falciparum 48/45 based vaccine antigen by SpyTag/SpyCatcher mediated virus-like display[J]. Vaccine, 2017, 35(30): 3726-3732. |
| 90 | LENEGHAN D B, MIURA K, TAYLOR I J, et al. Nanoassembly routes stimulate conflicting antibody quantity and quality for transmission-blocking malaria vaccines[J]. Scientific Reports, 2017, 7: 3811. |
| 91 | BRUNE K D, BULDUN C M, LI Y Y, et al. Dual plug-and-display synthetic assembly using orthogonal reactive proteins for twin antigen immunization[J]. Bioconjugate Chemistry, 2017, 28(5): 1544-1551. |
| 92 | BRUUN T U J, ANDERSSON A M C, DRAPER S J, et al. Engineering a rugged nanoscaffold to enhance plug-and-display vaccination[J]. ACS Nano, 2018, 12(9): 8855-8866. |
| 93 | URA T, OKUDA K, SHIMADA M. Developments in viral vector-based vaccines[J]. Vaccines, 2014, 2(3): 624-641. |
| 94 | WANG W J, LIU Z D, ZHOU X X, et al. Ferritin nanoparticle-based SpyTag/SpyCatcher-enabled click vaccine for tumor immunotherapy[J]. Nanomedicine: Nanotechnology, Biology and Medicine, 2019, 16: 69-78. |
| 95 | ZHANG B S, CHAO C W, TSYBOVSKY Y, et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone[J]. Scientific Reports, 2020, 10: 18149. |
| 96 | KANG Y F, SUN C, ZHUANG Z, et al. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates[J]. ACS Nano, 2021, 15(2): 2738-2752. |
| 97 | MA X C, ZOU F, YU F, et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses[J]. Immunity, 2020, 53(6): 1315-1330.e9. |
| 98 | COHEN A A, GNANAPRAGASAM P N P, LEE Y E, et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice[J]. Science, 2021, 371(6530): 735-741. |
| 99 | COHEN A A, YANG Z, GNANAPRAGASAM P N P, et al. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers[J]. PLoS One, 2021, 16(3): e0247963. |
| 100 | TAN T K, RIJAL P, RAHIKAINEN R, et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses[J]. Nature Communications, 2021, 12: 542. |
| 101 | RAHIKAINEN R, RIJAL P, TAN T K, et al. Overcoming symmetry mismatch in vaccine nanoassembly through spontaneous amidation[J]. Angewandte Chemie International Edition, 2021, 60(1): 321-330. |
| 102 | NGUYEN P Q, COURCHESNE N M D, DURAJ-THATTE A, et al. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials[J]. Advanced Materials, 2018, 30(19): e1704847. |
| 103 | XU L J, WANG X Y, SUN F, et al. Harnessing proteins for engineered living materials[J]. Current Opinion in Solid State and Materials Science, 2021, 25(1): 100896. |
| 104 | NAGAHAMA K, KIMURA Y, TAKEMOTO A. Living functional hydrogels generated by bioorthogonal cross-linking reactions of azide-modified cells with alkyne-modified polymers[J]. Nature Communications, 2018, 9: 2195. |
| 105 | WANG X Y, PU J H, AN B L, et al. Programming cells for dynamic assembly of inorganic nano-objects with spatiotemporal control[J]. Advanced Materials, 2018, 30(16): e1705968. |
| 106 | CHEN A Y, ZHONG C, LU T K. Engineering living functional materials[J]. ACS Synthetic Biology, 2015, 4(1): 8-11. |
| 107 | LEE S Y, CHOI J H, XU Z H. Microbial cell-surface display[J]. Trends in Biotechnology, 2003, 21(1): 45-52. |
| 108 | LINK A J, TIRRELL D A. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2]cycloaddition[J]. Journal of the American Chemical Society, 2003, 125(37): 11164-11165. |
| 109 | ISODA R, SIMANSKI S P, PATHANGEY L, et al. Expression of a Porphyromonas gingivalis hemagglutinin on the surface of a Salmonella vaccine vector[J]. Vaccine, 2007, 25(1): 117-126. |
| 110 | GILBERT C, ELLIS T. Biological engineered living materials: growing functional materials with genetically programmable properties[J]. ACS Synthetic Biology, 2019, 8(1): 1-15. |
| 111 | NGUYEN P Q, BOTYANSZKI Z, TAY P K R, et al. Programmable biofilm-based materials from engineered curli nanofibres[J]. Nature Communications, 2014, 5: 4945. |
| 112 | BOTYANSZKI Z, TAY P K R, NGUYEN P Q, et al. Engineered catalytic biofilms: site-specific enzyme immobilization onto E. coli curli nanofibers[J]. Biotechnology and Bioengineering, 2015, 112(10): 2016-2024. |
| 113 | CHEN A Y, DENG Z T, BILLINGS A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature Materials, 2014, 13(5): 515-523. |
| 114 | CHARRIER M, LI D, MANN V R, et al. Engineering the S-layer of Caulobacter crescentus as a foundation for stable, high-density, 2D living materials[J]. ACS Synthetic Biology, 2019, 8(1): 181-190. |
| 115 | OROZCO-HIDALGO M T, CHARRIER M, TJAHJONO N, et al. Engineering high-yield biopolymer secretion creates an extracellular protein matrix for living materials[J]. mSystems, 2021, 6(2): e00903-e00920. |
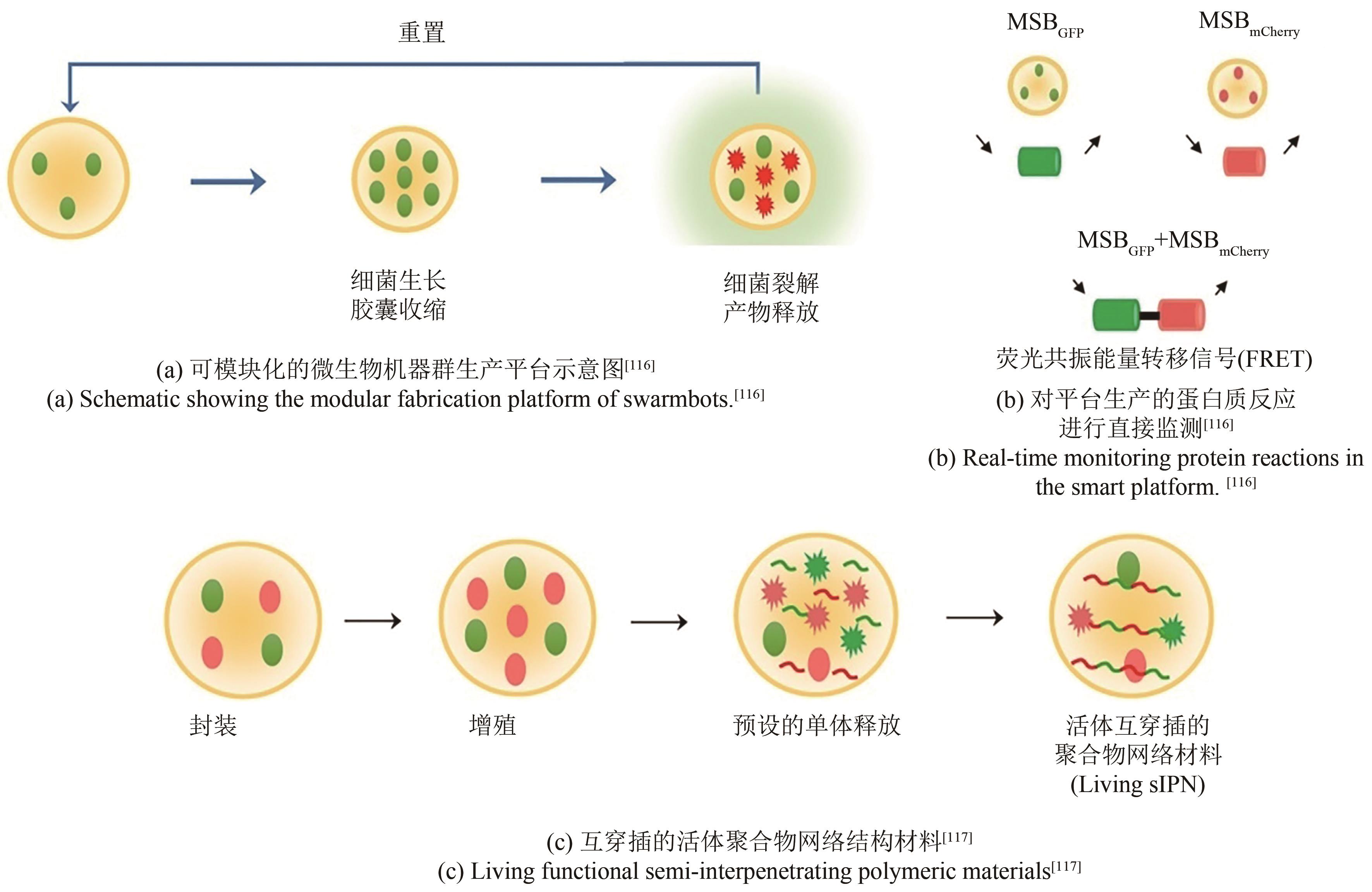
| 116 | DAI Z J, LEE A J, ROBERTS S, et al. Versatile biomanufacturing through stimulus-responsive cell-material feedback[J]. Nature Chemical Biology, 2019, 15(10): 1017-1024. |
| 117 | DAI Z J, YANG X Y, WU F L, et al. Living fabrication of functional semi-interpenetrating polymeric materials[J]. Nature Communications, 2021, 12: 3422. |
| 118 | KOZLOWSKI M T, SILVERMAN B R, JOHNSTONE C P, et al. Genetically programmable microbial assembly[J]. ACS Synthetic Biology, 2021, 10(6): 1351-1359. |
| 119 | LIU Z J, CAO S, LIU M, et al. Self-assembled multienzyme nanostructures on synthetic protein scaffolds[J]. ACS Nano, 2019, 13(10): 11343-11352. |
| 120 | YANG Z G, FOK H K F, LUO J R, et al. B12-induced reassembly of split photoreceptor protein enables photoresponsive hydrogels with tunable mechanics[J]. Science Advances, 2022, 8(13): abm5482. |
| [1] | 袁为锋, 赵永亮, 吴芷萱, 徐可. 合成生物学在新冠病毒广谱疫苗研发中的应用[J]. 合成生物学, 2024, 5(2): 369-384. |
| [2] | 章金勇, 顾江, 关山, 李海波, 曾浩, 邹全明. 合成生物学助力细菌疫苗研发[J]. 合成生物学, 2024, 5(2): 321-337. |
| [3] | 叶青, 秦成峰. “国际公共卫生紧急事件”下的mRNA疫苗研发[J]. 合成生物学, 2024, 5(2): 310-320. |
| [4] | 江莎莎, 王晨, 路冉, 刘俸君, 李俊, 王斌. T细胞免疫反应载体疫苗在人类疾病预防和治疗中的应用[J]. 合成生物学, 2024, 5(2): 294-309. |
| [5] | 王步森, 徐婧含, 高智强, 侯利华. 病毒载体疫苗研究进展[J]. 合成生物学, 2024, 5(2): 281-293. |
| [6] | 郭茜亚, 陈积, 董铭心. 流感病毒改造新策略及其应用[J]. 合成生物学, 2024, 5(2): 267-280. |
| [7] | 方超, 黄卫人. 合成生物学在肿瘤疫苗设计中的应用进展[J]. 合成生物学, 2024, 5(2): 239-253. |
| [8] | 谭子斌, 梁康, 陈有海. 合成生物学在基于微生物载体肿瘤疫苗设计中的应用[J]. 合成生物学, 2024, 5(2): 221-238. |
| [9] | 涂辉阳, 韩为东, 张斌. 肿瘤新抗原疫苗的设计与优化策略[J]. 合成生物学, 2024, 5(2): 254-266. |
| [10] | 刘泽众, 周洁, 朱赟, 陆路, 姜世勃. 基于重组人Ⅲ型胶原蛋白的三聚体抗原疫苗策略在新冠和流感疫苗中的应用[J]. 合成生物学, 2024, 5(2): 385-395. |
| [11] | 马雪璟, 郭畅, 华兆琳, 侯百东. 合成生物技术助力纳米颗粒疫苗理性设计时代的到来[J]. 合成生物学, 2024, 5(2): 353-368. |
| [12] | 叶精勤, 黄文华, 潘超, 朱力, 王恒樑. 合成生物学在多糖结合疫苗研发中的应用[J]. 合成生物学, 2024, 5(2): 338-352. |
| [13] | 余茜, 刘建英, 程功. 蚊媒黄病毒传播机制及疫苗与药物研发进展[J]. 合成生物学, 2023, 4(2): 347-372. |
| [14] | 申赵铃, 吴艳玲, 应天雷. 合成生物学与病毒疫苗研发[J]. 合成生物学, 2023, 4(2): 333-346. |
| [15] | 林思思, 潘超, 张一帆, 刘尽尧. 基于表面涂层益生菌的肿瘤抗原口服递送系统[J]. 合成生物学, 2022, 3(4): 810-820. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||