合成生物学 ›› 2024, Vol. 5 ›› Issue (1): 202-216.DOI: 10.12211/2096-8280.2022-068
• 特约评述 • 上一篇
CRISPR-Cas系统在病原核酸检测中的研究进展
杜瑶1,2, 高宏丹1,2, 刘家坤3,4, 刘孝荣2, 邢志浩2, 张涛5, 马东礼2
- 1.蚌埠医学院检验医学院,安徽 蚌埠 233030
2.深圳市儿童医院儿科研究所,广东 深圳 518034
3.中国科学院深圳先进技术研究所深圳合成生物研究所,广东 深圳 518055
4.深圳市海微生物科技有限公司,广东 深圳 518057
5.蚌埠医学院病原生物学教研室,安徽 蚌埠 233030
-
收稿日期:2022-11-26修回日期:2023-02-15出版日期:2024-02-29发布日期:2024-03-20 -
通讯作者:马东礼 -
作者简介:杜瑶 (1999—),女,硕士研究生。研究方向为病原生物学及分子生物学。 E-mail:dy852578@163.com马东礼 (1967—),男,硕士生导师,主任技师。研究方向为病原微生物致病机制及基因精准诊断。 E-mail:madl1234@126.com -
基金资助:广东省高水平医院项目(SEY-GSP-YXPT-A02);深圳市发展和改革委员会赠款(2019)986
Research progress of the CRISPR-Cas system in the detecting pathogen nucleic acids
DU Yao1,2, GAO Hongdan1,2, LIU Jiakun3,4, LIU Xiaorong2, XING Zhihao2, ZHANG Tao5, MA Dongli2
- 1.School of Laboratory Medicine,Bengbu Medical College,Bengbu 233030,Anhui,China
2.Pediatric Research Institute,Shenzhen Children’s Hospital,Shenzhen 518034,Guangdong,China
3.Shenzhen Institute of Synthetic Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
4.Shenzhen Hapmic Biotechnology Company limited,Shenzhen 518057,Guangdong,China
5.Department of Pathogenic Biology,Bengbu Medical College,Bengbu 233030,Anhui,China
-
Received:2022-11-26Revised:2023-02-15Online:2024-02-29Published:2024-03-20 -
Contact:MA Dongli
摘要:
CRISPR-Cas系统作为原核生物获得性免疫系统,由簇状规则间隔短回文重复序列(clustered regularly interspaced short palindromic repeats, CRISPR)和CRISPR相关蛋白(CRISPR-associated proteins, Cas)构成,因其识别和切割特定DNA或RNA序列,而成为分子诊断领域研究的热点。研究人员利用Cas蛋白(Cas12、Cas13、Cas14、Cas3等)结合信号放大和转化技术(荧光法、电位法、比色法、侧向流动技术等),开发了许多高灵敏度、高特异性、低成本的诊断平台,为病原核酸检测提供了新途径。本文介绍了CRISPR-Cas系统的生物学机制及分类,总结现有的基于Cas蛋白反式切割活性开发的病原核酸检测技术,描述其特点、功能和应用场景,并对该系统的未来应用前景进行展望,期望CRISPR-Cas系统成为包括核酸检测在内的多靶标的理想检测平台。
中图分类号:
引用本文
杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216.
DU Yao, GAO Hongdan, LIU Jiakun, LIU Xiaorong, XING Zhihao, ZHANG Tao, MA Dongli. Research progress of the CRISPR-Cas system in the detecting pathogen nucleic acids[J]. Synthetic Biology Journal, 2024, 5(1): 202-216.
| CRISPR类型 | 技术名称 | 效应 蛋白 | 目标 分子 | 扩增 方式 | 检测线 | 病原体 | 检测技术 |
|---|---|---|---|---|---|---|---|
| Type Ⅰ-E | CONAN[ | Cas3 | DNA | RT-LAMP | 1 copy | SARS-CoV-2 IAV | 侧向流动分析 |
| Type Ⅴ | DETECTR[ | Cas12a | DNA | RPA | amol/L | HPV16/18 SARS-CoV-2 | 荧光信号 |
| OR-DETECTR[ | Cas12a | RNA | RT-RPA | 1~2,5 copies/μL | SARS-CoV-2 H1N1 | 荧光信号、 侧向流动分析 | |
| HOLMES[ | Cas12a | DNA RNA | PCR | amol/L | 日本脑炎病毒 伪狂犬病毒 | 荧光信号 | |
| HOLMESv2[ | Cas12b | DNA RNA | LAMP | amol/L | 日本脑炎病毒 | 荧光信号 | |
| CDetection[ | AaCas12b | DNA | RPA | nmol/L | HPV16/18 | 荧光信号 | |
| E-CRISPR[ | Cas12a | DNA | — | pmol/L | HPV1、B19 | 电化学 | |
| MoECS[ | Cas12 | DNA | — | fmol/L | SARS-CoV-2 Delta 变异株 | 电化学 | |
| CRISPR-ENHANCE[ | LbCas12a | RNA | RT-LAMP | SARS-CoV-2、HIV、HCV | 侧向流动分析 | ||
| AIOD-CRISPR[ | LbaCas12a | RNA | RPA | 11 copies | SARS-CoV-2 HIV-1 | 荧光信号、 视觉法 | |
| SCAN[ | Cas12a | DNA RNA | RT-PCR | 13.5 copies/μL | SARS-CoV-2 HIV-1 | 纳米孔传感器 | |
| TB-QUICK[ | Cas12b | DNA | LAMP | 1.3 copies/μL | 结核分枝杆菌 | 荧光信号 | |
| STOPCovid[ | Cas12 | DNA RNA | LAMP | 100 copies | SARS-CoV-2 | 荧光信号 | |
| STOPCovid.v2[ | Cas12 | DNA RNA | LAMP | 2000 copies/mL | SARS-CoV-2 | 荧光信号 | |
| sPAMC[ | LbCas12a | DNA RNA | RPA | 1 copy/μL | SARS-CoV-2 | 荧光信号、 侧向流动分析 | |
| Cas14-DETECTR[ | Cas14a | DNA | RPA | fmol/L | HBoV-1 | 荧光信号 | |
| ACasB[ | Cas14a1 | DNA | — | 400 CFU/mL | 金黄色葡萄球菌 | 荧光信号 | |
| Type Ⅵ | SHERLOCK[ | Cas13a | DNA RNA | RPA | amol/L | 病毒、细菌 | 荧光信号 |
| HUDSON[ | Cas13a | RNA | RT-RPA | 1copy/μL | 寨卡病毒、登革病毒 | 荧光信号、 侧向流动分析 | |
| OR-SHERLOCK[ | Cas13a | RNA | RT-RPA | 1~2 copies/μL, 5 copies/μL | SARS-CoV-2 H1N1 | 荧光信号、 侧向流动分析 | |
| SHINE[ | Cas13 | RNA | RT-RPA | 10 copies/μL | SARS-CoV-2 | 智能手机(管内荧光读数或侧向流动分析) | |
| FIND-IT[ | Lbu Cas13a | RNA | — | 31 copies/μL | SARS-CoV-2 | 荧光信号 (集成检测器) | |
| CARMEN[ | LwCas13a | DNA RNA | PCR或RPA | amol/L | 169种人类感染病毒 | 荧光信号 | |
| mCARMEN[ | LwCas13a | DNA RNA | PCR或RPA | 102 copies/μL | 21种人类呼吸道病毒 | 荧光信号 | |
Type Ⅴ Type Ⅵ Type Ⅲ | SHERLOCKv2[ | Cas13 Cas12a Csm6 | DNA RNA | RPA | zmol/L | 病毒、细菌 | 荧光信号、 侧向流动分析 |
表1 基于CRISPR-Cas系统开发的部分诊断技术
Table 1 Technologies for detecting pathogens based on the CRISPR-Cas system
| CRISPR类型 | 技术名称 | 效应 蛋白 | 目标 分子 | 扩增 方式 | 检测线 | 病原体 | 检测技术 |
|---|---|---|---|---|---|---|---|
| Type Ⅰ-E | CONAN[ | Cas3 | DNA | RT-LAMP | 1 copy | SARS-CoV-2 IAV | 侧向流动分析 |
| Type Ⅴ | DETECTR[ | Cas12a | DNA | RPA | amol/L | HPV16/18 SARS-CoV-2 | 荧光信号 |
| OR-DETECTR[ | Cas12a | RNA | RT-RPA | 1~2,5 copies/μL | SARS-CoV-2 H1N1 | 荧光信号、 侧向流动分析 | |
| HOLMES[ | Cas12a | DNA RNA | PCR | amol/L | 日本脑炎病毒 伪狂犬病毒 | 荧光信号 | |
| HOLMESv2[ | Cas12b | DNA RNA | LAMP | amol/L | 日本脑炎病毒 | 荧光信号 | |
| CDetection[ | AaCas12b | DNA | RPA | nmol/L | HPV16/18 | 荧光信号 | |
| E-CRISPR[ | Cas12a | DNA | — | pmol/L | HPV1、B19 | 电化学 | |
| MoECS[ | Cas12 | DNA | — | fmol/L | SARS-CoV-2 Delta 变异株 | 电化学 | |
| CRISPR-ENHANCE[ | LbCas12a | RNA | RT-LAMP | SARS-CoV-2、HIV、HCV | 侧向流动分析 | ||
| AIOD-CRISPR[ | LbaCas12a | RNA | RPA | 11 copies | SARS-CoV-2 HIV-1 | 荧光信号、 视觉法 | |
| SCAN[ | Cas12a | DNA RNA | RT-PCR | 13.5 copies/μL | SARS-CoV-2 HIV-1 | 纳米孔传感器 | |
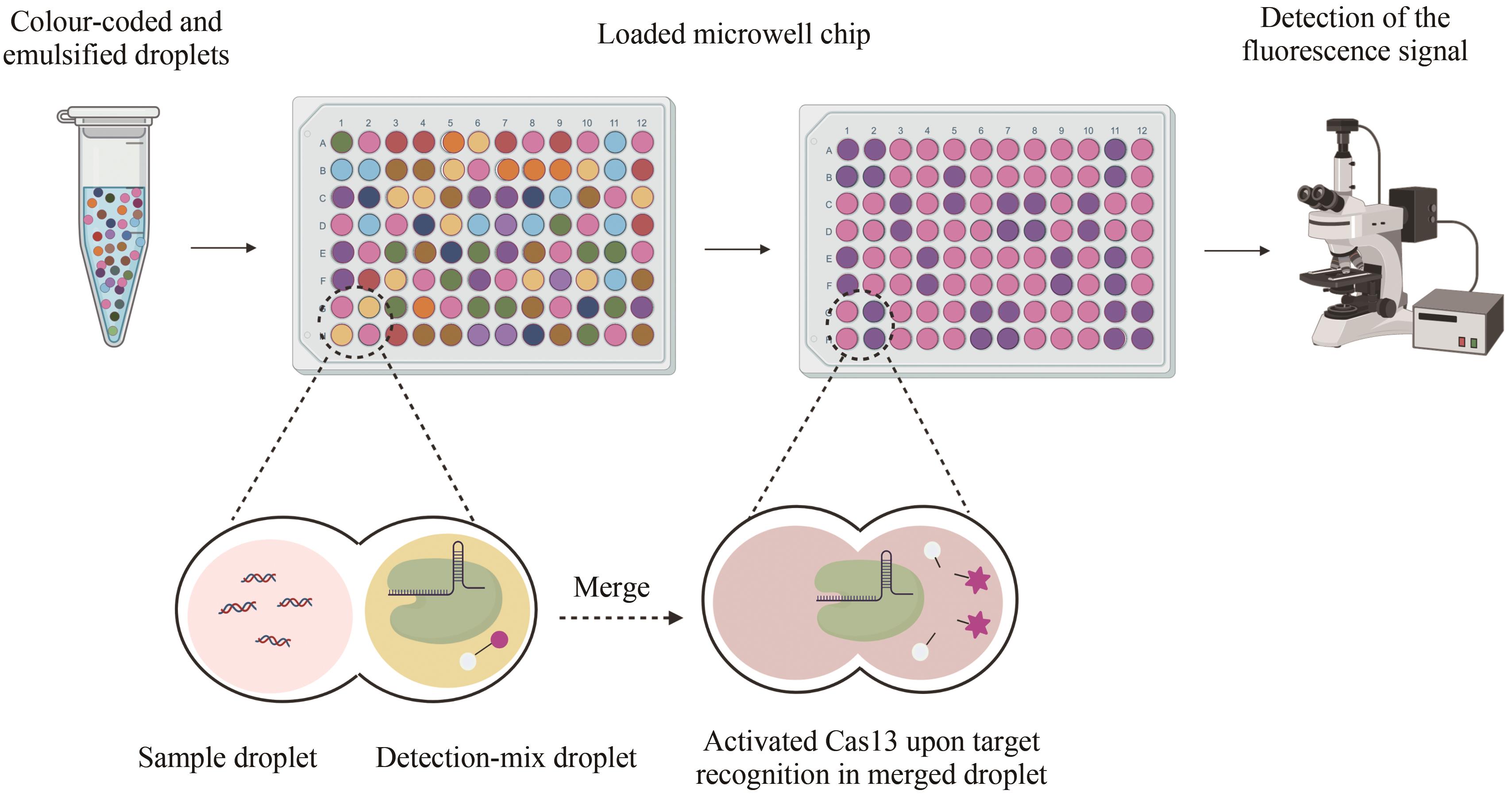
| TB-QUICK[ | Cas12b | DNA | LAMP | 1.3 copies/μL | 结核分枝杆菌 | 荧光信号 | |
| STOPCovid[ | Cas12 | DNA RNA | LAMP | 100 copies | SARS-CoV-2 | 荧光信号 | |
| STOPCovid.v2[ | Cas12 | DNA RNA | LAMP | 2000 copies/mL | SARS-CoV-2 | 荧光信号 | |
| sPAMC[ | LbCas12a | DNA RNA | RPA | 1 copy/μL | SARS-CoV-2 | 荧光信号、 侧向流动分析 | |
| Cas14-DETECTR[ | Cas14a | DNA | RPA | fmol/L | HBoV-1 | 荧光信号 | |
| ACasB[ | Cas14a1 | DNA | — | 400 CFU/mL | 金黄色葡萄球菌 | 荧光信号 | |
| Type Ⅵ | SHERLOCK[ | Cas13a | DNA RNA | RPA | amol/L | 病毒、细菌 | 荧光信号 |
| HUDSON[ | Cas13a | RNA | RT-RPA | 1copy/μL | 寨卡病毒、登革病毒 | 荧光信号、 侧向流动分析 | |
| OR-SHERLOCK[ | Cas13a | RNA | RT-RPA | 1~2 copies/μL, 5 copies/μL | SARS-CoV-2 H1N1 | 荧光信号、 侧向流动分析 | |
| SHINE[ | Cas13 | RNA | RT-RPA | 10 copies/μL | SARS-CoV-2 | 智能手机(管内荧光读数或侧向流动分析) | |
| FIND-IT[ | Lbu Cas13a | RNA | — | 31 copies/μL | SARS-CoV-2 | 荧光信号 (集成检测器) | |
| CARMEN[ | LwCas13a | DNA RNA | PCR或RPA | amol/L | 169种人类感染病毒 | 荧光信号 | |
| mCARMEN[ | LwCas13a | DNA RNA | PCR或RPA | 102 copies/μL | 21种人类呼吸道病毒 | 荧光信号 | |
Type Ⅴ Type Ⅵ Type Ⅲ | SHERLOCKv2[ | Cas13 Cas12a Csm6 | DNA RNA | RPA | zmol/L | 病毒、细菌 | 荧光信号、 侧向流动分析 |
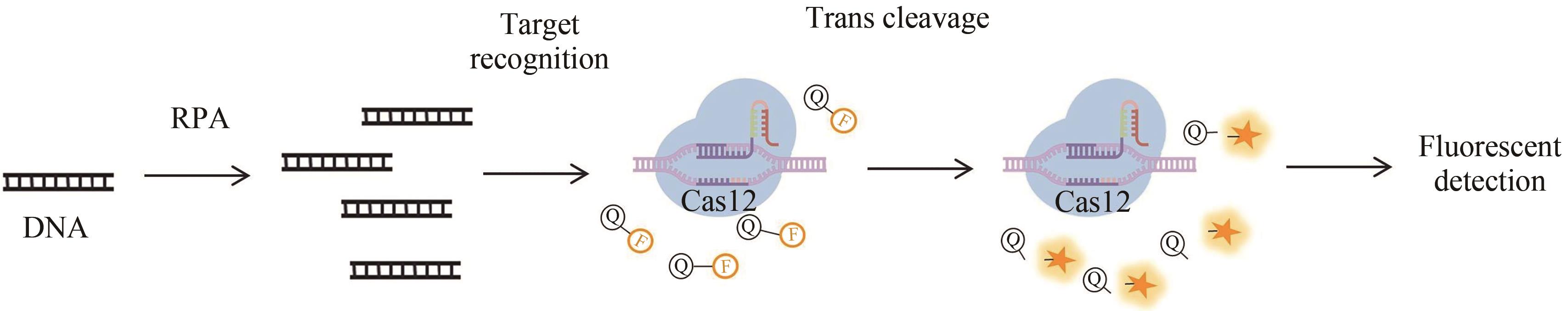
图1 基于CRISPR-Cas12a的DETECTR检测方法原理图(部分绘图素材来自Figdraw)
Fig. 1 Schematics of the CRISPR-Cas12a-based CRISPR-diagnostic method DETECTR(Some drawing elements are from Figdraw)

图2 基于CRISPR-Cas12a的E-CRISPR检测方法原理图(图片绘制来自BioRender)
Fig. 2 Schematics of the CRISPR-Cas12a-based diagnostic method E-CRISPR(The picture was created in BioRender)
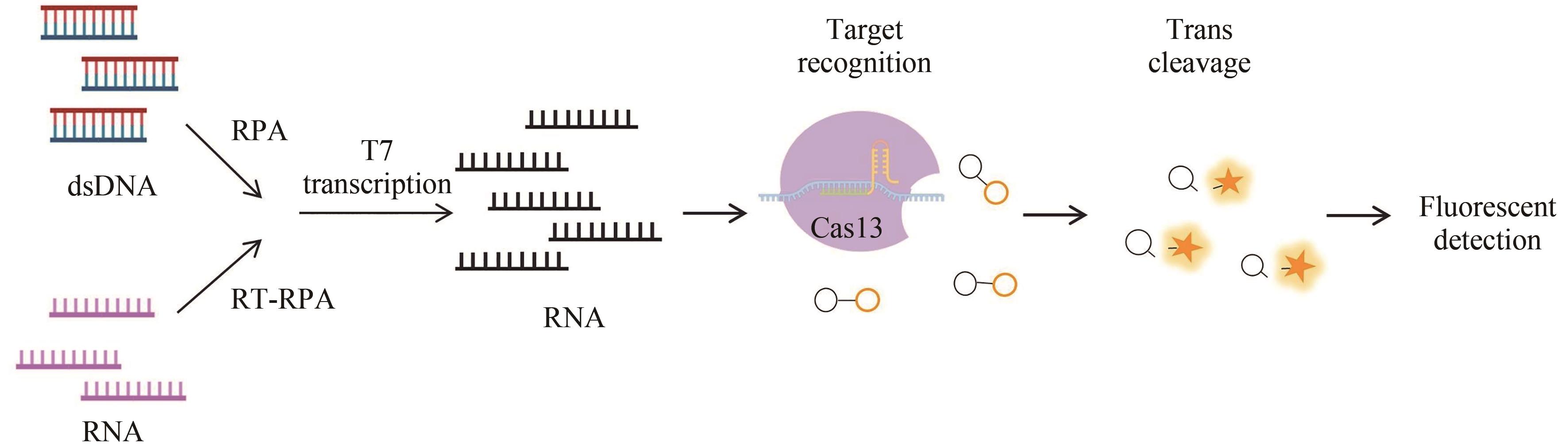
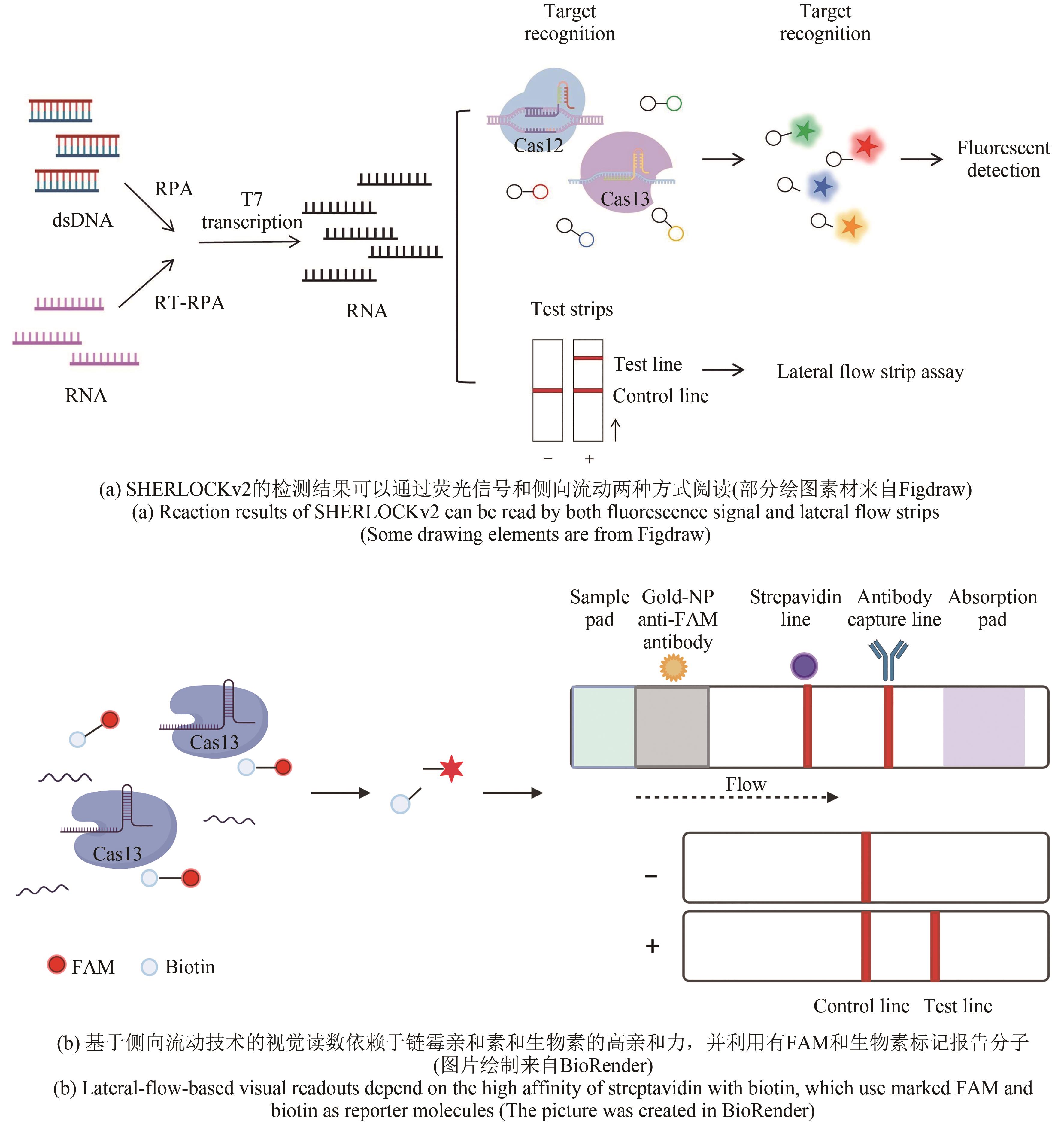
图3 基于CRISPR-Cas13a的SHERLOCK检测方法原理图(部分绘图素材来自Figdraw)
Fig. 3 Schematics of the CRISPR-Cas13-based diagnostic method SHERLOCK(Some drawing elements are from Figdraw)
| 1 | HWANG H, HWANG B Y, BUENO J. Biomarkers in infectious diseases[J]. Disease Markers, 2018, 2018: 8509127. |
| 2 | 库婷婷, 刘倩, 桑楠, 等. 几种典型呼吸道病毒的病原学特征及其检测方法[J]. 环境化学, 2020, 39(4): 841-851. |
| KU T T, LIU Q, SANG N, et al. Pathogenic characteristics and detection methods of several typical respiratory viruses[J]. Environmental Chemistry, 2020, 39(4): 841-851. | |
| 3 | MATTHIJS G, SOUCHE E, ALDERS M, et al. Guidelines for diagnostic next-generation sequencing[J]. European Journal of Human Genetics, 2016, 24(1): 2-5. |
| 4 | MORENS D M, FAUCI A S. Emerging pandemic diseases: how we got to COVID-19[J]. Cell, 2020, 182(5): 1077-1092. |
| 5 | 张礼堃, 邹秉杰, 宋沁馨, 等. PCR技术在新冠病毒核酸检测中的应用[J]. 医学研究生学报, 2021, 34(5): 539-544. |
| ZHANG L K, ZOU B J, SONG Q X, et al. Application of PCR in novel coronavirus nucleic acid detection[J]. Journal of Medical Postgraduates, 2021, 34(5): 539-544. | |
| 6 | PICKAR-OLIVER A, GERSBACH C A. The next generation of CRISPR-Cas technologies and applications[J]. Nature Reviews Molecular Cell Biology, 2019, 20(8): 490-507. |
| 7 | BHATTACHARYYA R P, THAKKU S G, HUNG D T. Harnessing CRISPR effectors for infectious disease diagnostics[J]. ACS Infectious Diseases, 2018, 4(9): 1278-1282. |
| 8 | ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. Journal of Bacteriology, 1987, 169(12): 5429-5433. |
| 9 | JANSEN R, EMBDEN J D, GAASTRA W, et al. Identification of genes that are associated with DNA repeats in prokaryotes[J]. Molecular Microbiology, 2002, 43(6): 1565-1575. |
| 10 | BARRANGOU R. CRISPR-Cas systems and RNA-guided interference[J]. Wiley Interdisciplinary Reviews RNA, 2013, 4(3): 267-278. |
| 11 | GARNEAU J E, DUPUIS M È, VILLION M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA[J]. Nature, 2010, 468(7320): 67-71. |
| 12 | YAO R L, LIU D, JIA X, et al. CRISPR-Cas9/Cas12a biotechnology and application in bacteria[J]. Synthetic and Systems Biotechnology, 2018, 3(3): 135-149. |
| 13 | JACKSON S A, MCKENZIE R E, FAGERLUND R D, et al. CRISPR-Cas: adapting to change[J]. Science, 2017, 356(6333): eaal5056. |
| 14 | HILLE F, RICHTER H, WONG S P, et al. The biology of CRISPR-Cas: backward and forward[J]. Cell, 2018, 172(6): 1239-1259. |
| 15 | GASIUNAS G, SINKUNAS T, SIKSNYS V. Molecular mechanisms of CRISPR-mediated microbial immunity[J]. Cellular and Molecular Life Sciences, 2014, 71(3): 449-465. |
| 16 | MAKAROVA K S, WOLF Y I, ALKHNBASHI O S, et al. An updated evolutionary classification of CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2015, 13(11): 722-736. |
| 17 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 18 | MAKAROVA K S, ZHANG F, KOONIN E V. SnapShot: class 1 CRISPR-cas systems[J]. Cell, 2017, 168(5): 946-946.e1. |
| 19 | SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems[J]. Molecular Cell, 2015, 60(3): 385-397. |
| 20 | LI P P, WANG L, YANG J N, et al. Applications of the CRISPR-Cas system for infectious disease diagnostics[J]. Expert Review of Molecular Diagnostics, 2021, 21(7): 723-732. |
| 21 | BROUNS S J J, JORE M M, LUNDGREN M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes[J]. Science, 2008, 321(5891): 960-964. |
| 22 | HAYES R P, XIAO Y B, DING F, et al. Structural basis for promiscuous PAM recognition in type Ⅰ-E Cascade from E. coli [J]. Nature, 2016, 530(7591): 499-503. |
| 23 | ZHENG Y L, LI J, WANG B Y, et al. Endogenous typeⅠCRISPR-Cas: from foreign DNA defense to prokaryotic engineering[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 62. |
| 24 | YOSHIMI K, TAKESHITA K, YAMAYOSHI S, et al. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus[J]. iScience, 2022, 25(2): 103830. |
| 25 | HOCHSTRASSER M L, TAYLOR D W, BHAT P, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(18): 6618-6623. |
| 26 | ZHAO H T, SHENG G, WANG J Y, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli [J]. Nature, 2014, 515(7525): 147-150. |
| 27 | KAZLAUSKIENE M, KOSTIUK G, VENCLOVAS Č, et al. A cyclic oligonucleotide signaling pathway in typeⅢCRISPR-Cas systems[J]. Science, 2017, 357(6351): 605-609. |
| 28 | PINILLA-REDONDO R, MAYO-MUÑOZ D, RUSSEL J, et al. Type Ⅳ CRISPR-Cas systems are highly diverse and involved in competition between plasmids[J]. Nucleic Acids Research, 2020, 48(4): 2000-2012. |
| 29 | SHMAKOV S, SMARGON A, SCOTT D, et al. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2017, 15(3): 169-182. |
| 30 | JIANG W Y, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| 31 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 32 | SWARTS D C, JINEK M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a[J]. Molecular Cell, 2019, 73(3): 589-600.e4. |
| 33 | ZENG R J, XU J H, LU L L, et al. Photoelectrochemical bioanalysis of microRNA on yolk-in-shell Au@CdS based on the catalytic hairpin assembly-mediated CRISPR-Cas12a system[J]. Chemical Communications, 2022, 58(54): 7562-7565. |
| 34 | CHEN H, LI Z Y, CHEN J S, et al. CRISPR/Cas12a-based electrochemical biosensor for highly sensitive detection of cTnI[J]. Bioelectrochemistry, 2022, 146: 108167. |
| 35 | KIM D, KIM J, HUR J K, et al. Erratum: Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells[J]. Nature Biotechnology, 2016, 34(8): 888. |
| 36 | XIN C C, YIN J H, YUAN S P, et al. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption[J]. Nature Communications, 2022, 13: 5623. |
| 37 | CASTELLE C J, WRIGHTON K C, THOMAS B C, et al. Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling[J]. Current Biology, 2015, 25(6): 690-701. |
| 38 | HARRINGTON L B, BURSTEIN D, CHEN J S, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes[J]. Science, 2018, 362(6416): 839-842. |
| 39 | KHAN M Z, HAIDER S, MANSOOR S, et al. Targeting plant ssDNA viruses with engineered miniature CRISPR-Cas14a[J]. Trends in Biotechnology, 2019, 37(8): 800-804. |
| 40 | ANANTHARAMAN V, MAKAROVA K S, BURROUGHS A M, et al. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing[J]. Biology Direct, 2013, 8: 15. |
| 41 | ABUDAYYEH O O, GOOTENBERG J S, ESSLETZBICHLER P, et al. RNA targeting with CRISPR-Cas13[J]. Nature, 2017, 550(7675): 280-284. |
| 42 | COX D B T, GOOTENBERG J S, ABUDAYYEH O O, et al. RNA editing with CRISPR-Cas13[J]. Science, 2017, 358(6366): 1019-1027. |
| 43 | MAKAROVA K S, WOLF Y I, KOONIN E V. Classification and nomenclature of CRISPR-Cas systems: where from here?[J]. The CRISPR Journal, 2018, 1(5): 325-336. |
| 44 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 45 | 李洋, 申晓林, 孙新晓, 等. CRISPR基因编辑技术在微生物合成生物学领域的研究进展[J]. 合成生物学, 2021, 2(1): 106-120. |
| LI Y, SHEN X L, SUN X X, et al. Advances of CRISPR gene editing in microbial synthetic biology[J]. Synthetic Biology Journal, 2021, 2(1): 106-120. | |
| 46 | PARDEE K, GREEN A A, TAKAHASHI M K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components[J]. Cell, 2016, 165(5): 1255-1266. |
| 47 | HILTON I B, D’IPPOLITO A M, VOCKLEY C M, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers[J]. Nature Biotechnology, 2015, 33(5): 510-517. |
| 48 | SLOMOVICS, PARDEEK, COLLINSJ J. Synthetic biology devices for in vitro and in vivo diagnostics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(47): 14429-14435. |
| 49 | LU T K, BOWERS J, KOERIS M S. Advancing bacteriophage-based microbial diagnostics with synthetic biology[J]. Trends in Biotechnology, 2013, 31(6): 325-327. |
| 50 | QUAN J, LANGELIER C, KUCHTA A, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences[J]. Nucleic Acids Research, 2019, 47(14): e83. |
| 51 | ZHOU W H, HU L, YING L M, et al. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection[J]. Nature Communications, 2018, 9: 5012. |
| 52 | TSOU J H, LENG Q X, JIANG F. A CRISPR test for detection of circulating nuclei acids[J]. Translational Oncology, 2019, 12(12): 1566-1573. |
| 53 | SUN Y Y, YU L, LIU C X, et al. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a[J]. Journal of Translational Medicine, 2021, 19(1): 74. |
| 54 | LI S Y, CHENG Q X, WANG J M, et al. CRISPR-Cas12a-assisted nucleic acid detection[J]. Cell Discovery, 2018, 4: 20. |
| 55 | LI L X, LI S Y, WU N, et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation[J]. ACS Synthetic Biology, 2019, 8(10): 2228-2237. |
| 56 | TENG F, GUO L, CUI T T, et al. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity[J]. Genome Biology, 2019, 20(1): 132. |
| 57 | DAI Y F, SOMOZA R A, WANG L, et al. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor[J]. Angewandte Chemie (International Ed in English), 2019, 58(48): 17399-17405. |
| 58 | WU C S, CHEN Z, LI C Z, et al. CRISPR-Cas12a-empowered electrochemical biosensor for rapid and ultrasensitive detection of SARS-CoV-2 delta variant[J]. Nano-Micro Letters, 2022, 14(1): 159. |
| 59 | NGUYEN L T, GURIJALA J, RANANAWARE S R, et al. CRISPR-ENHANCE: an enhanced nucleic acid detection platform using Cas12a[J]. Methods, 2022, 203: 116-124. |
| 60 | DING X, YIN K, LI Z Y, et al. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus[EB/OL]. BioRxiv. (2020-03-21) [2022-11-26]. . |
| 61 | NOURI R, JIANG Y Q, TANG Z F, et al. Detection of SARS-CoV-2 with solid-state CRISPR-Cas12a-assisted nanopores[J]. Nano Letters, 2021, 21(19): 8393-8400. |
| 62 | SAM I K, CHEN Y Y, MA J, et al. TB-QUICK: CRISPR-Cas12b-assisted rapid and sensitive detection of Mycobacterium tuberculosis [J]. Journal of Infection, 2021, 83(1): 54-60. |
| 63 | JOUNG J, LADHA A, SAITO M, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics[EB/OL]. MedRxiv. (2020-05-08) [2022-11-26]. . |
| 64 | JOUNG J, LADHA A, SAITO M, et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing[J]. The New England Journal of Medicine, 2020, 383(15): 1492-1494. |
| 65 | LU S H, TONG X H, HAN Y, et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a[J]. Nature Biomedical Engineering, 2022, 6(3): 286-297. |
| 66 | AQUINO-JARQUIN G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic[J]. Nanomedicine: Nanotechnology, Biology, and Medicine, 2019, 18: 428-431. |
| 67 | WEI Y D, TAO Z Z, WAN L, et al. Aptamer-based Cas14a1 biosensor for amplification-free live pathogenic detection[J]. Biosensors & Bioelectronics, 2022, 211: 114282. |
| 68 | KELLNER M J, KOOB J G, GOOTENBERG J S, et al. SHERLOCK: nucleic acid detection with CRISPR nucleases[J]. Nature Protocols, 2019, 14(10): 2986-3012. |
| 69 | ARIZTI-SANZ J, FREIJE C A, STANTON A C, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2[J]. Nature Communications, 2020, 11: 5921. |
| 70 | LIU T Y, KNOTT G J, SMOCK D C J, et al. Accelerated RNA detection using tandem CRISPR nucleases[J]. Nature Chemical Biology, 2021, 17(9): 982-988. |
| 71 | ACKERMAN C M, MYHRVOLD C, THAKKU S G, et al. Massively multiplexed nucleic acid detection with Cas13[J]. Nature, 2020, 582(7811): 277-282. |
| 72 | WELCH N L, ZHU M L, HUA C, et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants[J]. Nature Medicine, 2022, 28(5): 1083-1094. |
| 73 | GOOTENBERG J S, ABUDAYYEH O O, KELLNER M J, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6[J]. Science, 2018, 360(6387): 439-444. |
| 74 | YAN W X, HUNNEWELL P, ALFONSE L E, et al. Functionally diverse typeⅤCRISPR-Cas systems[J]. Science, 2019, 363(6422): 88-91. |
| 75 | LEUNG R K K, CHENG Q X, WU Z L, et al. CRISPR-Cas12-based nucleic acids detection systems[J]. Methods, 2022, 203: 276-281. |
| 76 | NGUYEN L T, SMITH B M, JAIN P K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection[J]. Nature Communications, 2020, 11: 4906. |
| 77 | CHEN J S, MA E B, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360(6387): 436-439. |
| 78 | BROUGHTON J P, DENG X D, YU G X, et al. CRISPR-Cas12-based detection of SARS-CoV-2[J]. Nature Biotechnology, 2020, 38(7): 870-874. |
| 79 | STRECKER J, JONES S, KOOPAL B, et al. Engineering of CRISPR-Cas12b for human genome editing[J]. Nature Communications, 2019, 10: 212. |
| 80 | ZHANG M Y, LIU C Z, SHI Y, et al. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application[J]. Talanta, 2020, 214: 120818. |
| 81 | MUKAMA O, WU J H, LI, Z Y, et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids[J]. Biosensors & Bioelectronics, 2020, 159: 112143. |
| 82 | XIAO X X, LIN Z Q, HUANG X H, et al. Rapid and sensitive detection of Vibrio vulnificus using CRISPR/Cas12a combined with a recombinase-aided amplification assay[J]. Frontiers in Microbiology, 2021, 12: 767315. |
| 83 | TENG F, CUI T T, FENG G H, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering[J]. Cell Discovery, 2018, 4: 63. |
| 84 | LIU X, BU S J, FENG J Q, et al. Electrochemical biosensor for detecting pathogenic bacteria based on a hybridization chain reaction and CRISPR-Cas12a[J]. Analytical and Bioanalytical Chemistry, 2022, 414(2): 1073-1080. |
| 85 | LI F, YE Q H, CHEN M T, et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection[J]. Biosensors & Bioelectronics, 2021, 179: 113073. |
| 86 | NOURI R, JIANG Y Q, LIAN X L, et al. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-Cas12a-assisted nanopores (SCAN)[J]. ACS Sensors, 2020, 5(5): 1273-1280. |
| 87 | GORBALENYA A E, BAKER S C, BARIC R S, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2[J]. Nature Microbiology, 2020, 5(4): 536-544. |
| 88 | RHEE S Y, CLUTTER D, FESSEL W J, et al. Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population[J]. Clinical Infectious Diseases, 2019, 68(2): 213-221. |
| 89 | ABUDAYYEH O O, GOOTENBERG J S, KONERMANN S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector[J]. Science, 2016, 353(6299): aaf5573. |
| 90 | HADIDI A. Next-generation sequencing and CRISPR/Cas13 editing in viroid research and molecular diagnostics[J]. Viruses, 2019, 11(2): 120. |
| 91 | GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442. |
| 92 | LEE R A, PUIG H, NGUYEN P Q, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(41): 25722-25731. |
| 93 | MYHRVOLD C, FREIJE C A, GOOTENBERG J S, et al. Field-deployable viral diagnostics using CRISPR-Cas13[J]. Science, 2018, 360(6387): 444-448. |
| 94 | BARNES K G, LACHENAUER A E, NITIDO A, et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time[J]. Nature Communications, 2020, 11: 4131. |
| 95 | GACH P C, IWAI K, KIM P W, et al. Droplet microfluidics for synthetic biology[J]. Lab on a Chip, 2017, 17(20): 3388-3400. |
| 96 | IWAI K, WEHRS M, GARBER M, et al. Scalable and automated CRISPR-based strain engineering using droplet microfluidics[J]. Microsystems & Nanoengineering, 2022, 8: 31. |
| 97 | CHEN Y, MEI Y X, ZHAO X H, et al. Reagents-loaded, automated assay that integrates recombinase-aided amplification and Cas12a nucleic acid detection for a point-of-care test[J]. Analytical Chemistry, 2020, 92(21): 14846-14852. |
| 98 | PATCHSUNG M, JANTARUG K, PATTAMA A, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA[J]. Nature Biomedical Engineering, 2020, 4(12): 1140-1149. |
| 99 | EIBERG H, TROELSEN J, NIELSEN M, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression[J]. Human Genetics, 2008, 123(2): 177-187. |
| 100 | KOONIN E V, MAKAROVA K S, ZHANG F. Diversity, classification and evolution of CRISPR-Cas systems[J]. Current Opinion in Microbiology, 2017, 37: 67-78. |
| 101 | XIAO Y B, LUO M, DOLAN A E, et al. Structure basis for RNA-guided DNA degradation by Cascade and Cas3[J]. Science, 2018, 361(6397): eaat0839. |
| 102 | KHAN W A, BARNEY R E, TSONGALIS G J. CRISPR-Cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting[J]. Journal of Clinical Virology, 2021, 145: 105019. |
| 103 | LIANG M D, LI Z L, WANG W S, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules[J]. Nature Communications, 2019, 10: 3672. |
| 104 | ARCHAKOV A I, ASEEV A L, BYKOV V A, et al. Challenges of the human proteome project: 10-year experience of the Russian consortium[J]. Journal of Proteome Research, 2019, 18(12): 4206-4214. |
| [1] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [2] | 赵静宇, 张健, 祁庆生, 王倩. 基于细菌双组分系统的生物传感器的研究进展[J]. 合成生物学, 2024, 5(1): 38-52. |
| [3] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [4] | 杨璐, 吴楠, 白茸茸, 董维亮, 周杰, 姜岷. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080. |
| [5] | 梁晓声, 郭永超, 门冬, 张先恩. 病毒-纳米金杂合导电网络结构在电化学分析的应用[J]. 合成生物学, 2022, 3(2): 415-427. |
| [6] | 张萍, 魏文平, 周英, 叶邦策. 解脂耶氏酵母中光控表达系统的构建及其应用研究[J]. 合成生物学, 2021, 2(5): 778-791. |
| [7] | 刘倩, 李金根, 张晨阳, 李芳雅, 田朝光. 工业丝状真菌基因组编辑技术研究进展[J]. 合成生物学, 2021, 2(2): 256-273. |
| [8] | 张博, 马永硕, 尚轶, 黄三文. 植物合成生物学研究进展[J]. 合成生物学, 2020, 1(2): 121-140. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||