合成生物学 ›› 2024, Vol. 5 ›› Issue (4): 782-794.DOI: 10.12211/2096-8280.2023-101
合成生物学在干细胞工程化改造中的研究进展
蔡冰玉1,2, 谭象天1,2, 李伟1,3,4
- 1.中国科学院动物研究所,干细胞与生殖生物学国家重点实验室,北京 100101
2.中国科学院大学,北京 100049
3.中国科学院干细胞与再生医学创新研究院,北京 100101
4.北京干细胞与再生医学研究院,北京 100101
-
收稿日期:2023-12-01修回日期:2024-03-12出版日期:2024-08-31发布日期:2024-09-19 -
通讯作者:李伟 -
作者简介:蔡冰玉 (1997—),女,博士研究生。研究方向为再生医学,合成生物学。E-mail:m18739087500@163.com李伟 (1982—),男,研究员,博士生导师。研究方向为结合基因工程、细胞工程和合成生物学等手段建立新的基因工程技术和细胞/动物模型。E-mail:liwei@ioz.ac.cn -
基金资助:国家重点研发计划(2019YFA0903800)
Advances in synthetic biology for engineering stem cell
CAI Bingyu1,2, TAN Xiangtian1,2, LI Wei1,3,4
- 1.State Key Laboratory of Stem Cell and Reproductive Biology,Institute of Zoology,Chinese Academy of Sciences,Beijing 100101,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
3.Institute for Stem Cell and Regenerative Medicine,Chinese Academy of Sciences,Beijing 100101,China
4.Beijing Institute for Stem Cell and Regenerative Medicine,Beijing 100101,China
-
Received:2023-12-01Revised:2024-03-12Online:2024-08-31Published:2024-09-19 -
Contact:LI Wei
摘要:
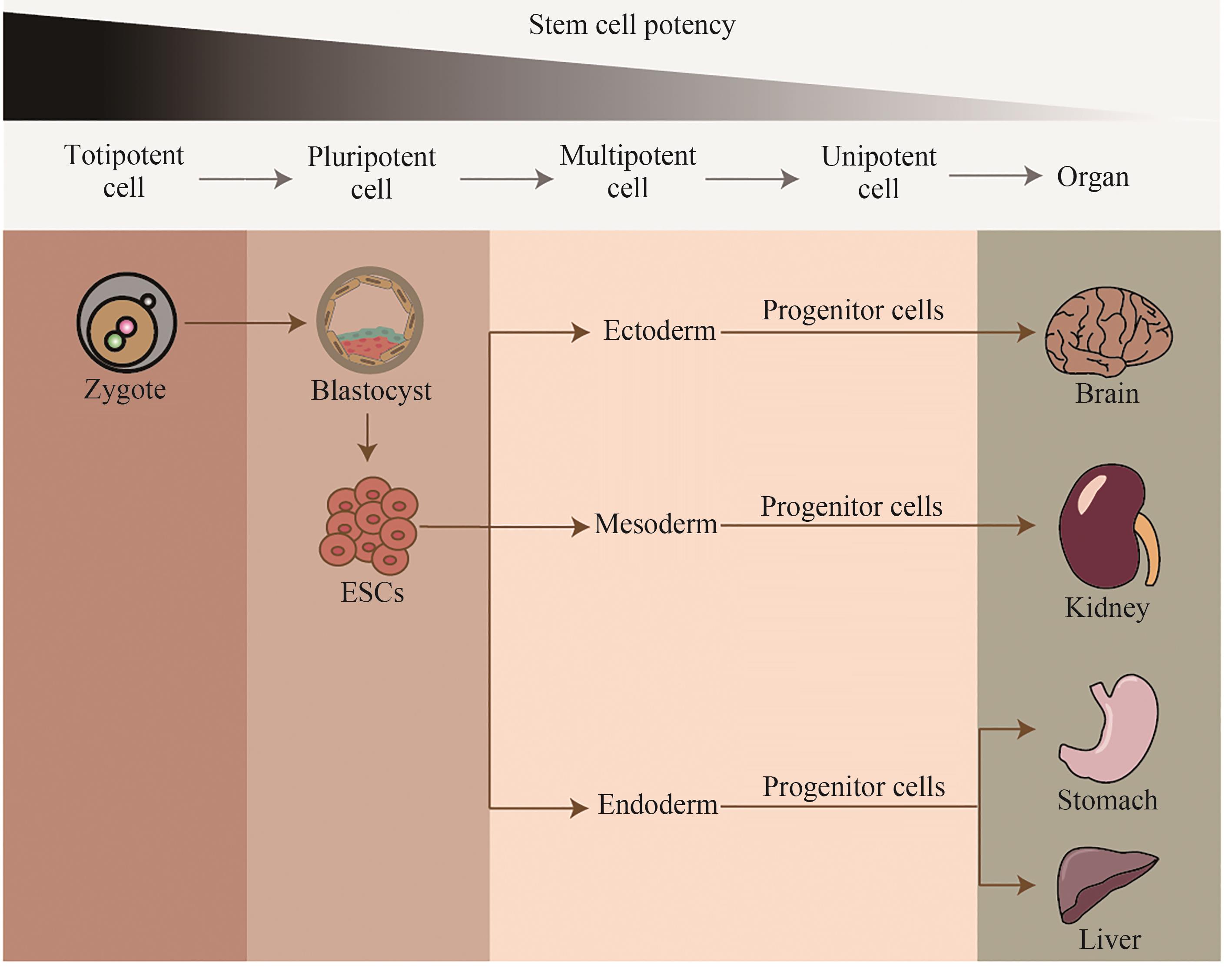
多能干细胞具备自我更新能力和多向分化潜能,其分化衍生的细胞及类器官在再生医学中具有巨大的应用潜力。但是干细胞临床转化仍然存在许多挑战。合成生物学“自上而下”的设计理念,结合基因编辑以及合成受体在内的强大工具库,能够赋予细胞新的功能,实现干细胞工程化改造。在此,本文总结了多能干细胞的临床应用和干细胞临床转化面临的主要挑战(干细胞分化衍生物的致瘤性、异质性、免疫原性),以及合成生物学在干细胞工程化改造中的应用(精确控制细胞命运、调控细胞通信、优化类器官结构功能、监测并清除致瘤细胞)。这些合成生物学工具为干细胞工程化改造提供了新的策略和平台,有望解决干细胞临床应用现存的诸多挑战,推动再生医学的进一步发展,实现“器官再生”这一核心目标。
中图分类号:
引用本文
蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794.
CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell[J]. Synthetic Biology Journal, 2024, 5(4): 782-794.
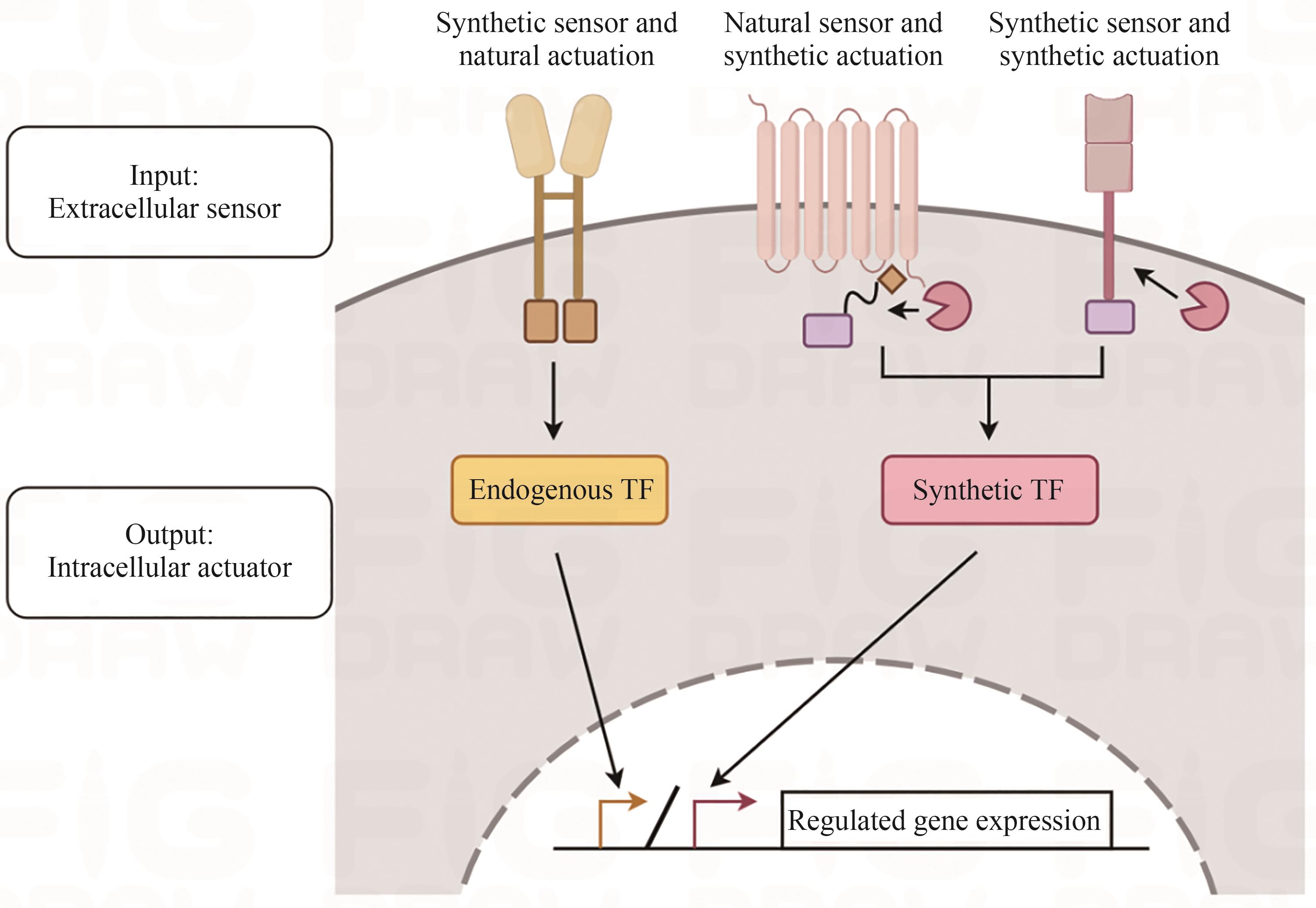
| 工程化干细胞改造技术 | ||||||
|---|---|---|---|---|---|---|
| 基因编辑技术 | 合成受体 | |||||
| 条件性基因敲除系统 | ZFN系统 | TALEN系统 | CRISPR/Cas系统 | 合成传感器 + 天然致动器 | 天然传感器 + 合成致动器 | 合成传感器 + 合成致动器 |
| CARs、SyCyR | Tango、ChaCha | synNotch、SNIPR、MESA | ||||
| 工程化干细胞改造策略 | ||||||
| 细胞命运决定 | 细胞通信 | 类器官结构功能优化 | 强化细胞治疗功能 | 监测并消除致瘤细胞 | ||
表1 合成生物学在干细胞中的应用
Table 1 Applications of synthetic biology in stem cells
| 工程化干细胞改造技术 | ||||||
|---|---|---|---|---|---|---|
| 基因编辑技术 | 合成受体 | |||||
| 条件性基因敲除系统 | ZFN系统 | TALEN系统 | CRISPR/Cas系统 | 合成传感器 + 天然致动器 | 天然传感器 + 合成致动器 | 合成传感器 + 合成致动器 |
| CARs、SyCyR | Tango、ChaCha | synNotch、SNIPR、MESA | ||||
| 工程化干细胞改造策略 | ||||||
| 细胞命运决定 | 细胞通信 | 类器官结构功能优化 | 强化细胞治疗功能 | 监测并消除致瘤细胞 | ||
| 1 | JACOBSON L O, SIMMONS E L, MARKS E K, et al. Recovery from radiation injury[J]. Science, 1951, 113(2940): 510-511. |
| 2 | TILL J E, MCCULLOCH E A. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells[J]. Radiation Research, 1961, 14: 213-222. |
| 3 | EVANS M J, KAUFMAN M H. Establishment in culture of pluripotential cells from mouse embryos[J]. Nature, 1981, 292(5819): 154-156. |
| 4 | THOMSON J A, ITSKOVITZ-ELDOR J, SHAPIRO S S, et al. Embryonic stem cell lines derived from human blastocysts[J]. Science, 1998, 282(5391): 1145-1147. |
| 5 | TAKAHASHI K, TANABE K, OHNUKI M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors[J]. Cell, 2007, 131(5): 861-872. |
| 6 | GUAN J Y, WANG G, WANG J L, et al. Chemical reprogramming of human somatic cells to pluripotent stem cells[J]. Nature, 2022, 605(7909): 325-331. |
| 7 | SCHWARTZ S D, REGILLO C D, LAM B L, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies[J]. Lancet, 2015, 385(9967): 509-516. |
| 8 | KAMAO H, MANDAI M, OKAMOTO S, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application[J]. Stem Cell Reports, 2014, 2(2): 205-218. |
| 9 | PAGLIUCA F W, MILLMAN J R, GÜRTLER M, et al. Generation of functional human pancreatic β cells in vitro [J]. Cell, 2014, 159(2): 428-439. |
| 10 | SATO T, VRIES R G, SNIPPERT H J, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche[J]. Nature, 2009, 459(7244): 262-265. |
| 11 | SPENCE J R, MAYHEW C N, RANKIN S A, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro [J]. Nature, 2011, 470(7332): 105-109. |
| 12 | MCCRACKEN K W, CATÁ E M, CRAWFORD C M, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids[J]. Nature, 2014, 516(7531): 400-404. |
| 13 | MONZEL A S, SMITS L M, HEMMER K, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells[J]. Stem Cell Reports, 2017, 8(5): 1144-1154. |
| 14 | MUGURUMA K, NISHIYAMA A, KAWAKAMI H, et al. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells[J]. Cell Reports, 2015, 10(4): 537-550. |
| 15 | DYE B R, HILL D R, FERGUSON M A, et al. In vitro generation of human pluripotent stem cell derived lung organoids[J]. eLife, 2015, 4: e05098. |
| 16 | CAMP J G, SEKINE K, GERBER T, et al. Multilineage communication regulates human liver bud development from pluripotency[J]. Nature, 2017, 546(7659): 533-538. |
| 17 | TAKEBE T, SEKINE K, ENOMURA M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant[J]. Nature, 2013, 499(7459): 481-484. |
| 18 | TAKASATO M, ER P X, BECROFT M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney[J]. Nature Cell Biology, 2014, 16(1): 118-126. |
| 19 | TAKEBE T, ENOMURA M, YOSHIZAWA E, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation[J]. Cell Stem Cell, 2015, 16(5): 556-565. |
| 20 | KOIKE H, IWASAWA K, OUCHI R, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary[J]. Nature, 2019, 574(7776): 112-116. |
| 21 | MIURA K, OKADA Y, AOI T, et al. Variation in the safety of induced pluripotent stem cell lines[J]. Nature Biotechnology, 2009, 27(8): 743-745. |
| 22 | The International Stem Cell Initiative. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage[J]. Nature Biotechnology, 2011, 29(12): 1132-1144. |
| 23 | OSAFUNE K, CARON L, BOROWIAK M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines[J]. Nature Biotechnology, 2008, 26(3): 313-315. |
| 24 | MACFARLAN T S, GIFFORD W D, DRISCOLL S, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity[J]. Nature, 2012, 487(7405): 57-63. |
| 25 | ZHAO T B, ZHANG Z N, RONG Z L, et al. Immunogenicity of induced pluripotent stem cells[J]. Nature, 2011, 474(7350): 212-215. |
| 26 | DEUSE T, HU X M, AGBOR-ENOH S, et al. De novo mutations in mitochondrial DNA of iPSCs produce immunogenic neoepitopes in mice and humans[J]. Nature Biotechnology, 2019, 37(10): 1137-1144. |
| 27 | CAMP J G, BADSHA F, FLORIO M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(51): 15672-15677. |
| 28 | BAXTER M, WITHEY S, HARRISON S, et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes[J]. Journal of Hepatology, 2015, 62(3): 581-589. |
| 29 | GORDON A, YOON S J, TRAN S S, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions[J]. Nature Neuroscience, 2021, 24(3): 331-342. |
| 30 | KITADA T, DIANDRETH B, TEAGUE B, et al. Programming gene and engineered-cell therapies with synthetic biology[J]. Science, 2018, 359(6376): eaad1067. |
| 31 | TOLLE F, STÜCHELI P, FUSSENEGGER M. Genetic circuitry for personalized human cell therapy[J]. Current Opinion in Biotechnology, 2019, 59: 31-38. |
| 32 | MANSOURI M, FUSSENEGGER M. Therapeutic cell engineering: designing programmable synthetic genetic circuits in mammalian cells[J]. Protein & Cell, 2022, 13(7): 476-489. |
| 33 | KIM Y G, CHA J, CHANDRASEGARAN S. Hybrid restriction enzymes: zinc finger fusions to Fok Ⅰ cleavage domain[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(3): 1156-1160. |
| 34 | CHRISTIAN M, CERMAK T, DOYLE E L, et al. Targeting DNA double-strand breaks with TAL effector nucleases[J]. Genetics, 2010, 186(2): 757-761. |
| 35 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 36 | STRICKLETT P K, NELSON R D, KOHAN D E. Site-specific recombination using an epitope tagged bacteriophage P1 Cre recombinase[J]. Gene, 1998, 215(2): 415-423. |
| 37 | RAMÍREZ-SOLIS R, LIU P, BRADLEY A. Chromosome engineering in mice[J]. Nature, 1995, 378(6558): 720-724. |
| 38 | SU H, WANG X, BRADLEY A. Nested chromosomal deletions induced with retroviral vectors in mice[J]. Nature Genetics, 2000, 24(1): 92-95. |
| 39 | RIVENBARK A G, STOLZENBURG S, BELTRAN A S, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation[J]. Epigenetics, 2012, 7(4): 350-360. |
| 40 | KONERMANN S, BRIGHAM M D, TREVINO A, et al. Optical control of mammalian endogenous transcription and epigenetic states[J]. Nature, 2013, 500(7463): 472-476. |
| 41 | KEARNS N A, GENGA R M, ENUAMEH M S, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells[J]. Development, 2014, 141(1): 219-223. |
| 42 | GILBERT L A, LARSON M H, MORSUT L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes[J]. Cell, 2013, 154(2): 442-451. |
| 43 | KONERMANN S, BRIGHAM M D, TREVINO A E, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex[J]. Nature, 2015, 517(7536): 583-588. |
| 44 | LIU P, CHEN M, LIU Y X, et al. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency[J]. Cell Stem Cell, 2018, 22(2): 252-261. e4. |
| 45 | CHAVEZ A, SCHEIMAN J, VORA S, et al. Highly efficient Cas9-mediated transcriptional programming[J]. Nature Methods, 2015, 12(4): 326-328. |
| 46 | LIU X S, WU H, KRZISCH M, et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene[J]. Cell, 2018, 172(5): 979-992. e6. |
| 47 | MANHAS J, EDELSTEIN H I, LEONARD J N, et al. The evolution of synthetic receptor systems[J]. Nature Chemical Biology, 2022, 18(3): 244-255. |
| 48 | BRENNER J, CHO J H, WONG W W. Synthetic biology: sensing with modular receptors[J]. Nature Chemical Biology, 2017, 13(2): 131-132. |
| 49 | LABANIEH L, MACKALL C L. CAR immune cells: design principles, resistance and the next generation[J]. Nature, 2023, 614(7949): 635-648. |
| 50 | CAPPELL K M, KOCHENDERFER J N. Long-term outcomes following CAR T cell therapy: what we know so far[J]. Nature Reviews Clinical Oncology, 2023, 20(6): 359-371. |
| 51 | ENGELOWSKI E, SCHNEIDER A, FRANKE M, et al. Synthetic cytokine receptors transmit biological signals using artificial ligands[J]. Nature Communications, 2018, 9(1): 2034. |
| 52 | ISHIZUKA S, LAI C Y, OTSU M, et al. Designing motif-engineered receptors to elucidate signaling molecules important for proliferation of hematopoietic stem cells[J]. ACS Synthetic Biology, 2018, 7(7): 1709-1714. |
| 53 | MOSSNER S, KUCHNER M, FAZEL MODARES N, et al. Synthetic interleukin 22 (IL-22) signaling reveals biological activity of homodimeric IL-10 receptor 2 and functional cross-talk with the IL-6 receptor gp130[J]. The Journal of Biological Chemistry, 2020, 295(35): 12378-12397. |
| 54 | SCHELLER L, STRITTMATTER T, FUCHS D, et al. Generalized extracellular molecule sensor platform for programming cellular behavior[J]. Nature Chemical Biology, 2018, 14(7): 723-729. |
| 55 | KROEZE W K, SASSANO M F, HUANG X P, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome[J]. Nature Structural & Molecular Biology, 2015, 22(5): 362-369. |
| 56 | KIPNISS N H, DINGAL P C D P, ABBOTT T R, et al. Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system[J]. Nature Communications, 2017, 8(1): 2212. |
| 57 | MORSUT L, ROYBAL K T, XIONG X, et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors[J]. Cell, 2016, 164(4): 780-791. |
| 58 | ZHU I, LIU R, GARCIA J M, et al. Modular design of synthetic receptors for programmed gene regulation in cell therapies[J]. Cell, 2022, 185(8): 1431-1443. e16. |
| 59 | DARINGER N M, DUDEK R M, SCHWARZ K A, et al. Modular extracellular sensor architecture for engineering mammalian cell-based devices[J]. ACS Synthetic Biology, 2014, 3(12): 892-902. |
| 60 | MALAGUTI M, PORTERO MIGUELES R, ANNOH J, et al. SyNPL: Synthetic Notch pluripotent cell lines to monitor and manipulate cell interactions in vitro and in vivo [J]. Development, 2022, 149(12): dev200226. |
| 61 | SAXENA P, BOJAR D, ZULEWSKI H, et al. Generation of glucose-sensitive insulin-secreting beta-like cells from human embryonic stem cells by incorporating a synthetic lineage-control network[J]. Journal of Biotechnology, 2017, 259: 39-45. |
| 62 | CAHAN P, LI H, MORRIS S A, et al. CellNet: network biology applied to stem cell engineering[J]. Cell, 2014, 158(4): 903-915. |
| 63 | TODA S, BLAUCH L R, TANG S K Y, et al. Programming self-organizing multicellular structures with synthetic cell-cell signaling[J]. Science, 2018, 361(6398): 156-162. |
| 64 | ZHANG S H, ZHAO H, LIU Z X, et al. Monitoring of cell-cell communication and contact history in mammals[J]. Science, 2022, 378(6623): eabo5503. |
| 65 | MA Y T, BUDDE M W, MAYALU M N, et al. Synthetic mammalian signaling circuits for robust cell population control[J]. Cell, 2022, 185(6): 967-979. e12. |
| 66 | CHAO G, WANNIER T M, GUTIERREZ C, et al. helixCAM: a platform for programmable cellular assembly in bacteria and human cells[J]. Cell, 2022, 185(19): 3551-3567. e39. |
| 67 | STEVENS A J, HARRIS A R, GERDTS J, et al. Programming multicellular assembly with synthetic cell adhesion molecules[J]. Nature, 2023, 614(7946): 144-152. |
| 68 | MIKI K, ENDO K, TAKAHASHI S, et al. Efficient detection and purification of cell populations using synthetic microRNA switches[J]. Cell Stem Cell, 2015, 16(6): 699-711. |
| 69 | JUILLERAT A, TKACH D, BUSSER B W, et al. Modulation of chimeric antigen receptor surface expression by a small molecule switch[J]. BMC Biotechnology, 2019, 19(1): 44. |
| 70 | ZAH E, LIN M Y, SILVA-BENEDICT A, et al. T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells[J]. Cancer Immunology Research, 2016, 4(6): 498-508. |
| 71 | FRY T J, SHAH N N, ORENTAS R J, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy[J]. Nature Medicine, 2018, 24(1): 20-28. |
| 72 | STRATI P, BACHANOVA V, GOODMAN A, et al. Preliminary results of a phase Ⅰ trial of FT516, an off-the-shelf natural killer (NK) cell therapy derived from a clonal master induced pluripotent stem cell (iPSC) line expressing high-affinity, non-cleavable CD16 (hnCD16), in patients (pts) with relapsed/refractory (R/R) B-cell lymphoma (BCL)[J]. Journal of Clinical Oncology, 2021, 39(): 7541. |
| 73 | ITAKURA G, KAWABATA S, ANDO M, et al. Fail-safe system against potential tumorigenicity after transplantation of iPSC derivatives[J]. Stem Cell Reports, 2017, 8(3): 673-684. |
| 74 | KOJIMA K, MIYOSHI H, NAGOSHI N, et al. Selective ablation of tumorigenic cells following human induced pluripotent stem cell-derived neural stem/progenitor cell transplantation in spinal cord injury[J]. Stem Cells Translational Medicine, 2019, 8(3): 260-270. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 艾宗勇, 张成庭, 牛宝华, 尹宇, 杨洁, 李天晴. 人胚胎早期发育与干细胞[J]. 合成生物学, 2024, 5(4): 700-718. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 胡可儿, 王汉奇, 黄儒麒, 张灿阳, 邢新会, 马少华. 整合设计策略下的工程化类器官与类器官芯片技术[J]. 合成生物学, 2024, 5(4): 883-897. |
| [15] | 李石开, 曾东鳌, 杜方舟, 张京钟, 余爽. 血管化类器官的构建方法及生物材料[J]. 合成生物学, 2024, 5(4): 851-866. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||