Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (5): 941-959.DOI: 10.12211/2096-8280.2024-002
• Invited Review • Previous Articles Next Articles
Recent advances in chemoenzymatic synthesis of important steroids
ZHENG Mengmeng1,2, LIU Benben1,2, LIN Zhi1,2, QU Xudong1,2
- 1.State Key Laboratory of Microbial Metabolism,School of Life Sciences and Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
2.Zhangjiang Institute for Advanced Study,Shanghai Jiao Tong University,Shanghai 201203,China
-
Received:2024-01-02Revised:2024-03-20Online:2024-11-20Published:2024-10-31 -
Contact:QU Xudong
重要甾体化合物的化学酶法合成研究进展
郑梦梦1,2, 刘犇犇1,2, 林芝1,2, 瞿旭东1,2
- 1.上海交通大学生命科学技术学院,微生物代谢国家重点实验室,上海 200240
2.上海交通大学张江高等研究院,上海 201203
-
通讯作者:瞿旭东 -
作者简介:郑梦梦 (1993—),女,博士后。研究方向为天然产物生物合成。 E-mail:sjtu7126164@sjtu.edu.cn瞿旭东 (1980—),男,博士,教授,博士生导师。研究方向为天然骨架的定向生物合成。 E-mail:quxd@sjtu.edu.cn -
基金资助:国家自然科学基金青年科学基金(32301216)
CLC Number:
Cite this article
ZHENG Mengmeng, LIU Benben, LIN Zhi, QU Xudong. Recent advances in chemoenzymatic synthesis of important steroids[J]. Synthetic Biology Journal, 2024, 5(5): 941-959.
郑梦梦, 刘犇犇, 林芝, 瞿旭东. 重要甾体化合物的化学酶法合成研究进展[J]. 合成生物学, 2024, 5(5): 941-959.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-002
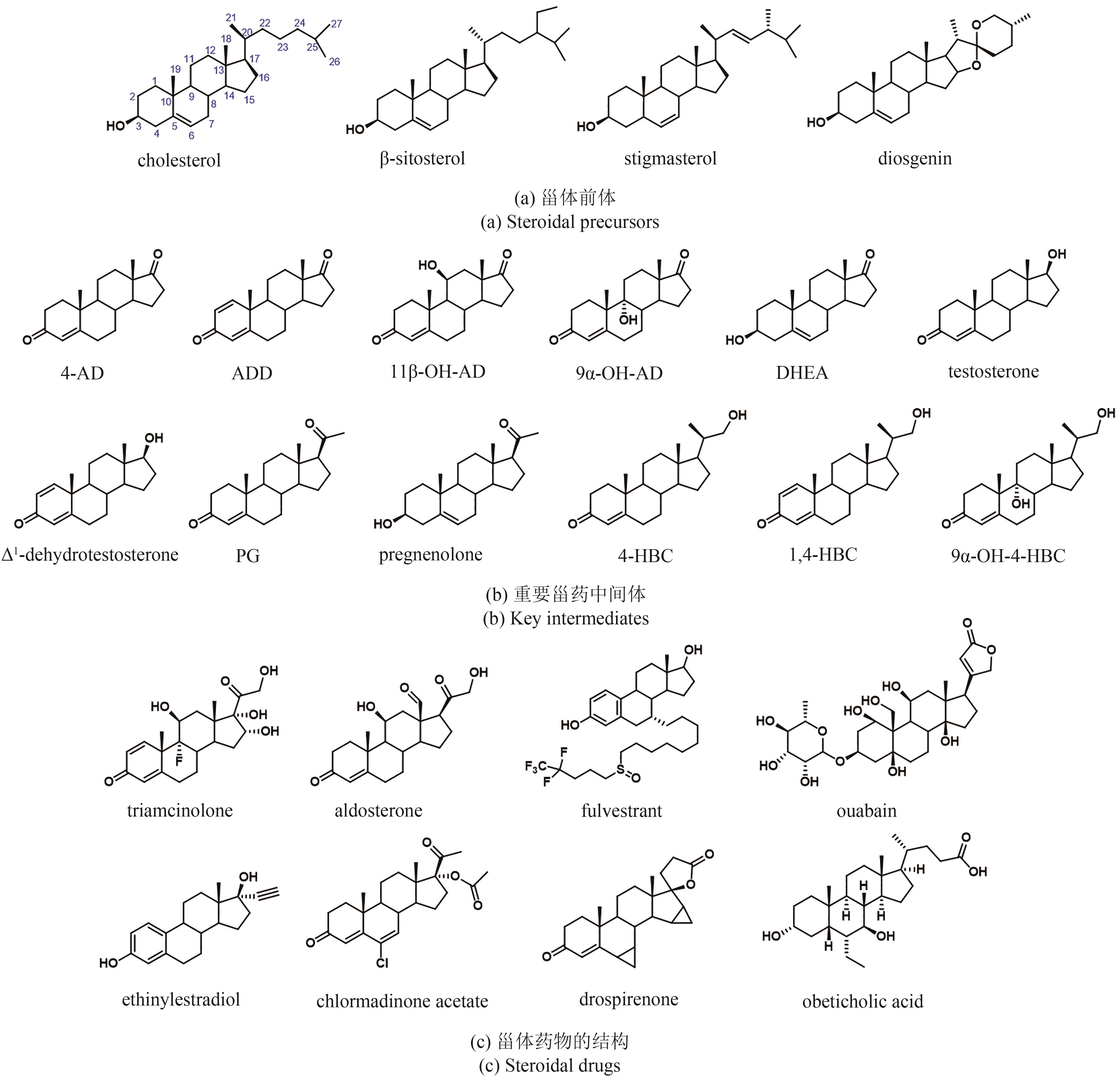
| 1 | WANG F Q, YAO K, WEI D Z. From soybean phytosterols to steroid hormones[M/OL]//Soybean and health. Rijeka, Croatia: InTech(2011-09-12)[2023-12-01]. . |
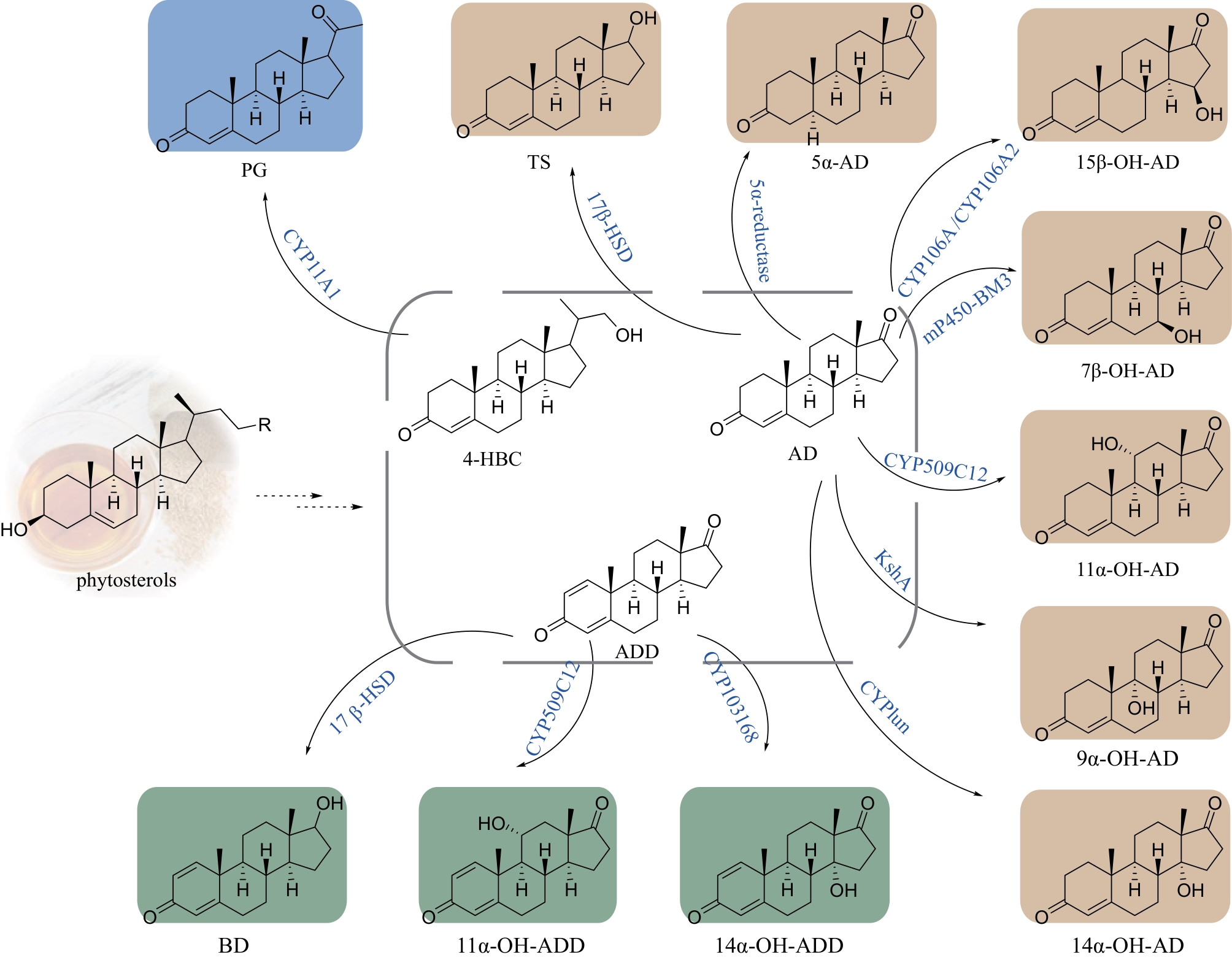
| 2 | PENG H D, WANG Y Y, JIANG K, et al. A dual role reductase from phytosterols catabolism enables the efficient production of valuable steroid precursors[J]. Angewandte Chemie International Edition, 2021, 60(10): 5414-5420. |
| 3 | DONOVA M V, EGOROVA O V. Microbial steroid transformations: current state and prospects[J]. Applied Microbiology and Biotechnology, 2012, 94(6): 1423-1447. |
| 4 | TONG W Y, DONG X. Microbial biotransformation: recent developments on steroid drugs[J]. Recent Patents on Biotechnology, 2009, 3(2): 141-153. |
| 5 | FENG J H, WU Q Q, ZHU D M, et al. Biotransformation enables innovations toward green synthesis of steroidal pharmaceuticals[J]. ChemSusChem, 2022, 15(9): e202102399. |
| 6 | 熊亮斌, 宋璐, 赵云秋, 等. 甾体化合物绿色生物制造:从生物转化到微生物从头合成[J]. 合成生物学, 2021, 2(6): 942-963. |
| XIONG L B, SONG L, ZHAO Y Q, et al. Green biomanufacturing of steroids: from biotransformation to de novo synthesis by microorganisms[J]. Synthetic Biology Journal, 2021, 2(6): 942-963. | |
| 7 | FERNÁNDEZ-CABEZÓN L, GALÁN B, GARCÍA J L. New insights on steroid biotechnology[J]. Frontiers in Microbiology, 2018, 9: 958. |
| 8 | ZHANG Y, XIAO P Y, PAN D L, et al. New insights into the modification of the non-core metabolic pathway of steroids in Mycolicibacterium and the application of fermentation biotechnology in C-19 steroid production[J]. International Journal of Molecular Sciences, 2023, 24(6): 5236. |
| 9 | SONG S K, HE J X, GAO M, et al. Loop pathways are responsible for tuning the accumulation of C19- and C22-sterol intermediates in the mycobacterial phytosterol degradation pathway[J]. Microbial Cell Factories, 2023, 22(1): 19. |
| 10 | FERNÁNDEZ-CABEZÓN L, GALÁN B, GARCÍA J L. Engineering Mycobacterium smegmatis for testosterone production[J]. Microbial Biotechnology, 2017, 10(1): 151-161. |
| 11 | ZHAO Y Q, SHEN Y B, MA S, et al. Production of 5α-androstene-3,17-dione from phytosterols by co-expression of 5α-reductase and glucose-6-phosphate dehydrogenase in engineered Mycobacterium neoaurum [J]. Green Chemistry, 2019, 21(7): 1809-1815. |
| 12 | TANG R, REN X X, XIA M L, et al. Efficient one-step biocatalytic multienzyme cascade strategy for direct conversion of phytosterol to C-17-hydroxylated steroids[J]. Applied and Environmental Microbiology, 2021, 87(24): e0032121. |
| 13 | FELPETO-SANTERO C, GALÁN B, GARCÍA J L. Production of 11α-hydroxysteroids from sterols in a single fermentation step by Mycolicibacterium smegmatis [J]. Microbial Biotechnology, 2021, 14(6): 2514-2524. |
| 14 | FELPETO-SANTERO C, GALÁN B, GARCÍA J L. Engineering the steroid hydroxylating system from Cochliobolus lunatus in Mycolicibacterium smegmatis [J]. Microorganisms, 2021, 9(7): 1499. |
| 15 | ZHANG C W, SHEN Y B, GAO Y Y, et al. Efficient production of 14α-OH-AD by engineered Mycolicibacterium neoaurum via coupled cofactor and reconstructed electron transport system[J]. Systems Microbiology and Biomanufacturing, 2023, 3(2): 358-369. |
| 16 | LI X, CHEN T, PENG F, et al. Efficient conversion of phytosterols into 4-androstene-3,17-dione and its C1, 2-dehydrogenized and 9α-hydroxylated derivatives by engineered Mycobacteria[J]. Microbial Cell Factories, 2021, 20(1): 158. |
| 17 | KARPOV M V, NIKOLAEVA V M, FOKINA V V, et al. Creation and functional analysis of Mycolicibacterium smegmatis recombinant strains carrying the bacillary cytochromes CYP106A1 and CYP106A2 genes[J]. Applied Biochemistry and Microbiology, 2022, 58(9): 947-957. |
| 18 | LIU K, WANG F Q, LIU K, et al. Light-driven progesterone production by InP-(M. neoaurum) biohybrid system[J]. Bioresources and Bioprocessing, 2022, 9(1): 93. |
| 19 | ZHAO Y Q, LIU Y J, JI W T, et al. One-pot biosynthesis of 7β-hydroxyandrost-4-ene-3,17-dione from phytosterols by cofactor regeneration system in engineered Mycolicibacterium neoaurum [J]. Microbial Cell Factories, 2022, 21(1): 59. |
| 20 | ABAS H, BLENCOWE P, BROOKFIELD J, et al. Selective hydroxylation of C(sp3)—H bonds in steroids[J]. Chemistry, 2023, 29(4): e202301066. |
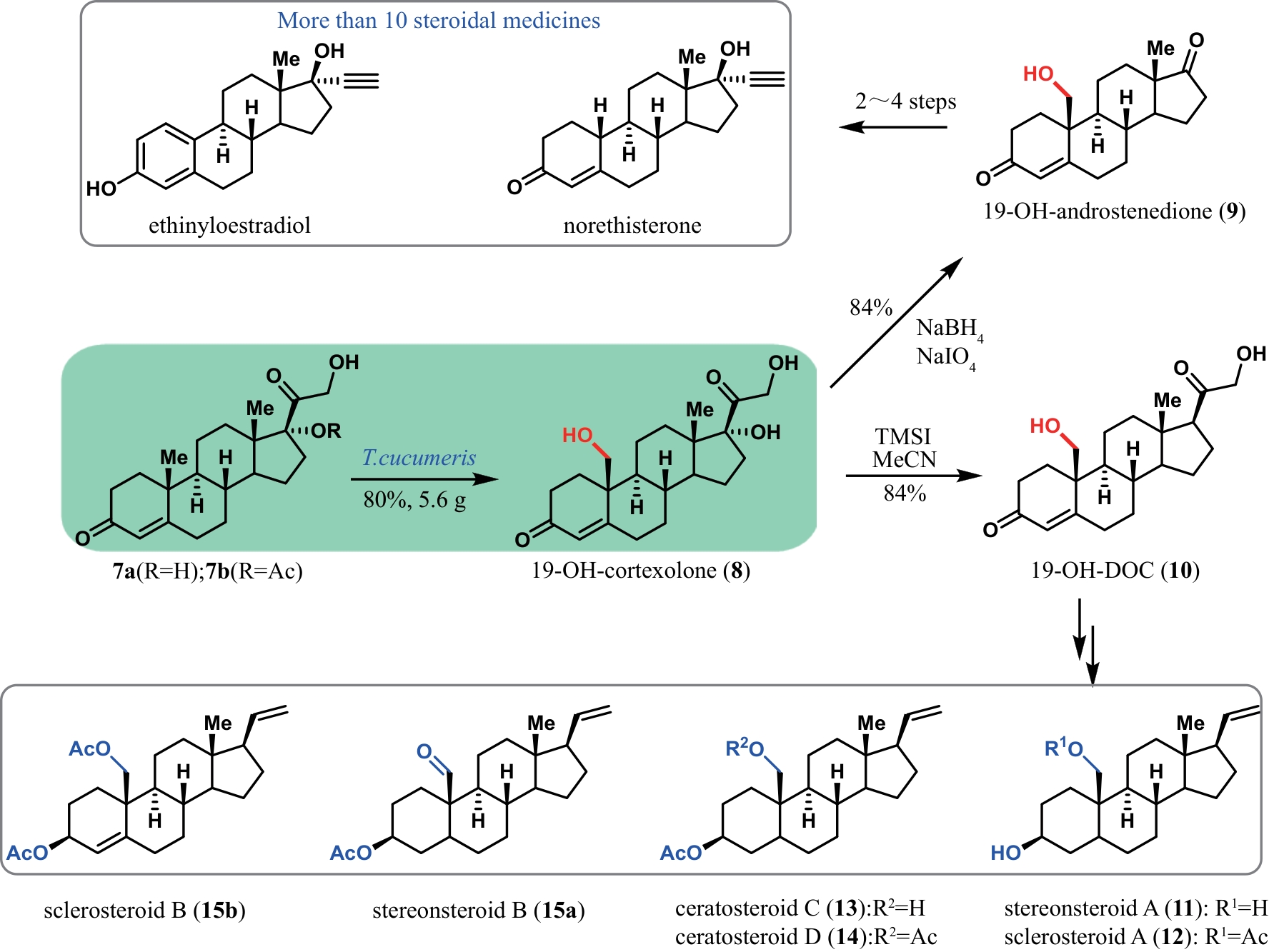
| 21 | RUDROFF F, MIHOVILOVIC M D, GRÖGER H, et al. Opportunities and challenges for combining chemo- and biocatalysis[J]. Nature Catalysis, 2018, 1: 12-22. |
| 22 | CHAKRABARTY S, ROMERO E O, PYSER J B, et al. Chemoenzymatic total synthesis of natural products[J]. Accounts of Chemical Research, 2021, 54(6): 1374-1384. |
| 23 | KILLE S, ZILLY F E, ACEVEDO J P, et al. Regio- and stereoselectivity of P450-catalysed hydroxylation of steroids controlled by laboratory evolution[J]. Nature Chemistry, 2011, 3(9): 738-743. |
| 24 | KASPAR F, SCHALLMEY A. Chemo-enzymatic synthesis of natural products and their analogs[J]. Current Opinion in Biotechnology, 2022, 77: 102759. |
| 25 | LI F Z, DENG H P, RENATA H. Chemoenzymatic approaches for exploring structure-activity relationship studies of bioactive natural products[J]. Nature Synthesis, 2023, 2: 708-718. |
| 26 | LI J, AMATUNI A, RENATA H. Recent advances in the chemoenzymatic synthesis of bioactive natural products[J]. Current Opinion in Chemical Biology, 2020, 55: 111-118. |
| 27 | PAULSEL T Q, WILLIAMS G J. Current state-of-the-art toward chemoenzymatic synthesis of polyketide natural products[J]. ChemBioChem, 2023, 24(21): e202300386. |
| 28 | RODDAN R, CARTER E M, THAIR B, et al. Chemoenzymatic approaches to plant natural product inspired compounds[J]. Natural Product Reports, 2022, 39(7): 1375-1382. |
| 29 | STOUT C N, WASFY N M, CHEN F, et al. Charting the evolution of chemoenzymatic strategies in the syntheses of complex natural products[J]. Journal of the American Chemical Society, 2023, 145(33): 18161-18181. |
| 30 | VANABLE E P, HABGOOD L G, PATRONE J D. Current progress in the chemoenzymatic synthesis of natural products[J]. Molecules, 2022, 27(19): 6373. |
| 31 | LATHE R. Steroid and sterol 7-hydroxylation: ancient pathways[J]. Steroids, 2002, 67(12): 967-977. |
| 32 | LI H P, LIU H M, GE W Z, et al. Synthesis of 7α-hydroxy-dehydroepiandrosterone and 7β-hydroxy-dehydroepiandrosterone[J]. Steroids, 2005, 70(14): 970-973. |
| 33 | ZHU R, LIU Y, YANG Y Y, et al. Cytochrome P450 monooxygenases catalyse steroid nucleus hydroxylation with regio- and stereo-selectivity[J]. Advanced Synthesis & Catalysis, 2022, 364(16): 2701-2719. |
| 34 | SZALENIEC M, WOJTKIEWICZ A M, BERNHARDT R, et al. Bacterial steroid hydroxylases: enzyme classes, their functions and comparison of their catalytic mechanisms[J]. Applied Microbiology and Biotechnology, 2018, 102(19): 8153-8171. |
| 35 | ZHANG X D, PENG Y Q, ZHAO J, et al. Bacterial cytochrome P450-catalyzed regio- and stereoselective steroid hydroxylation enabled by directed evolution and rational design[J]. Bioresources and Bioprocessing, 2020, 7(1): 2. |
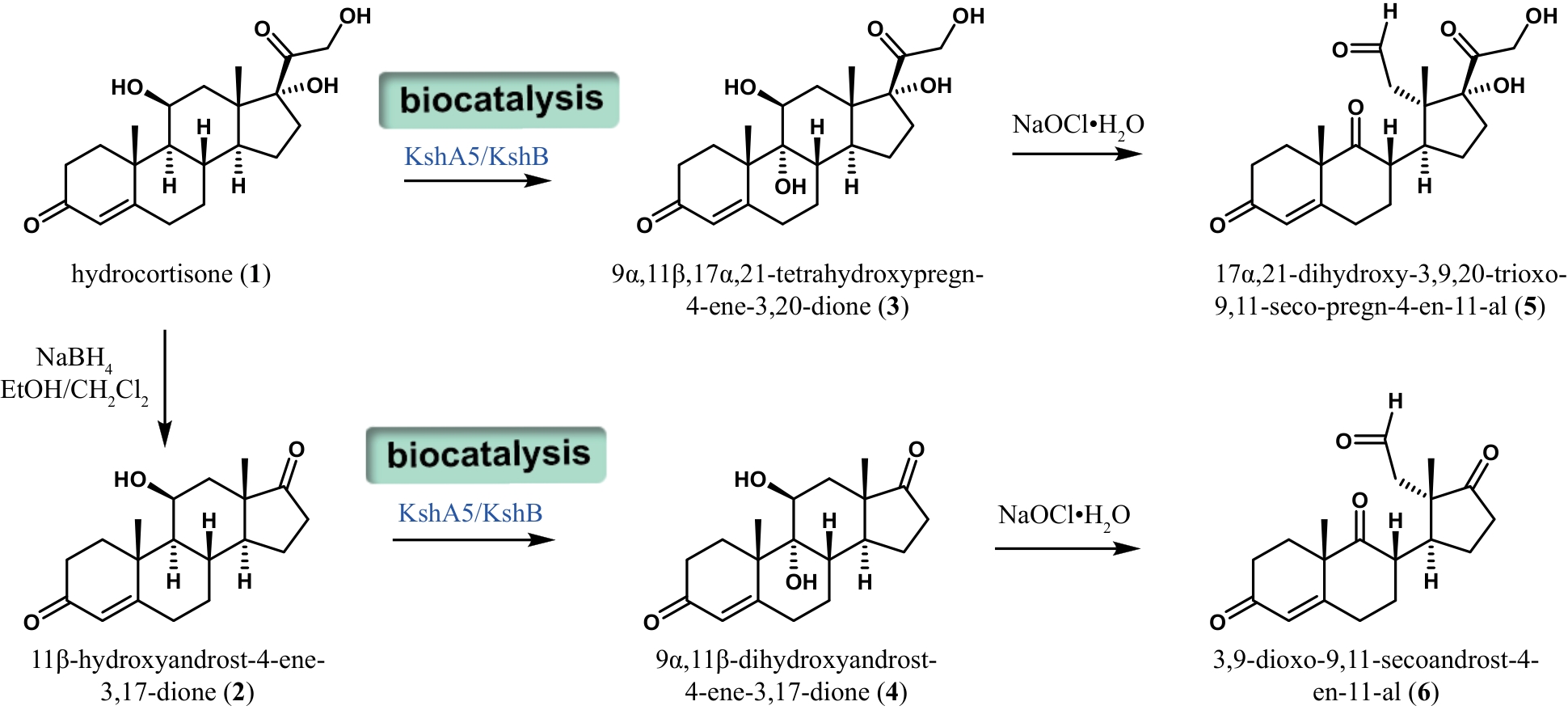
| 36 | WANG L, WU X W, GAO C H, et al. A fungal P450 enzyme from Fusarium graminearum with unique 12β-steroid hydroxylation activity[J]. Applied and Environmental Microbiology, 2023, 89(3): 22. |
| 37 | FERNANDES P, CRUZ A, ANGELOVA B, et al. Microbial conversion of steroid compounds: recent developments[J]. Enzyme and Microbial Technology, 2003, 32(6): 688-705. |
| 38 | CAPYK J K, D’ANGELO I, STRYNADKA N C, et al. Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis[J]. The Journal of Biological Chemistry, 2009, 284(15): 9937-9946. |
| 39 | LIU H H, XU L Q, YAO K, et al. Engineered 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum: an efficient platform for production of steroid drugs[J]. Applied and Environmental Microbiology, 2018, 84(14): e02777-17. |
| 40 | CHANG Y C, LAI K H, KUMAR S, et al. 1H NMR-based isolation of anti-inflammatory 9,11-secosteroids from the octocoral Sinularia leptoclados [J]. Marine Drugs, 2020, 18(5): 271. |
| 41 | HE Y Q, LEE CAPLAN S, SCESA P, et al. Cyclized 9,11-secosterol enol-ethers from the Gorgonian Pseudopterogorgia americana [J]. Steroids, 2017, 125: 47-53. |
| 42 | KÕLLO M, KASARI M, KASARI V, et al. Designed whole-cell-catalysis-assisted synthesis of 9,11-secosterols[J]. Beilstein Journal of Organic Chemistry, 2021, 17: 581-588. |
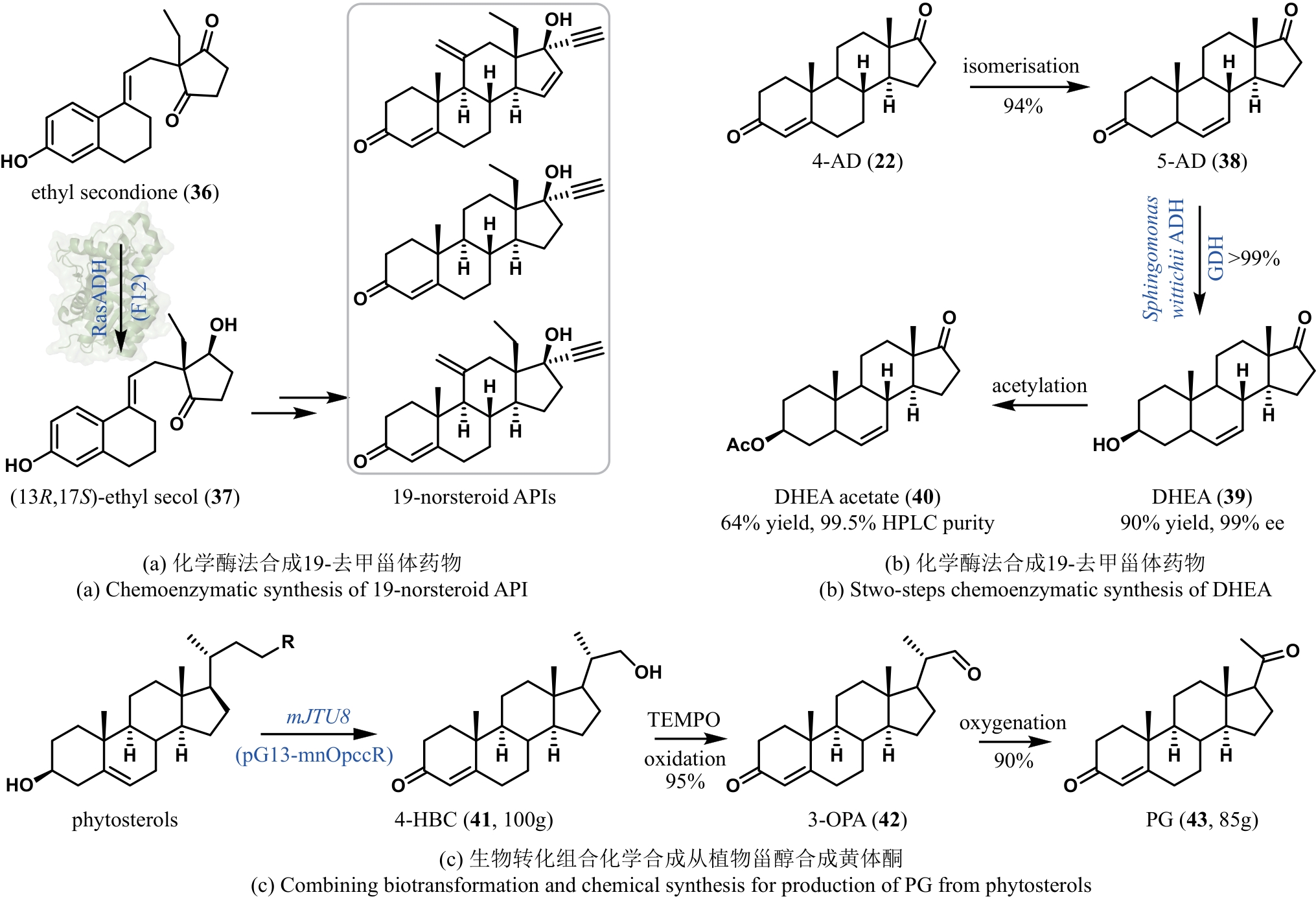
| 43 | RENATA H, ZHOU Q H, DÜNSTL G, et al. Development of a concise synthesis of ouabagenin and hydroxylated corticosteroid analogues[J]. Journal of the American Chemical Society, 2015, 137(3): 1330-1340. |
| 44 | KAPLAN W, KHATRI H R, NAGORNY P. Concise enantioselective total synthesis of cardiotonic steroids 19-hydroxysarmentogenin and trewianin aglycone[J]. Journal of the American Chemical Society, 2016, 138(22): 7194-7198. |
| 45 | LU W, CHEN X, FENG J H, et al. A fungal P450 enzyme from Thanatephorus cucumeris with steroid hydroxylation capabilities[J]. Applied and Environmental Microbiology, 2018, 84(13): e00503-18. |
| 46 | WANG J L, ZHANG Y N, LIU H H, et al. A biocatalytic hydroxylation-enabled unified approach to C19-hydroxylated steroids[J]. Nature Communications, 2019, 10(1): 3378. |
| 47 | EL-SEEDI H R, KHALIFA S A M, TAHER E A, et al. Cardenolides: insights from chemical structure and pharmacological utility[J]. Pharmacological Research, 2019, 141: 123-175. |
| 48 | ZHONG Y, ZHAO C, WU W Y, et al. Total synthesis, chemical modification and structure-activity relationship of bufadienolides[J]. European Journal of Medicinal Chemistry, 2020, 189: 112038. |
| 49 | CHEN J, TANG J L, XI Y Y, et al. Production of 14α-hydroxysteroids by a recombinant Saccharomyces cerevisiae biocatalyst expressing of a fungal steroid 14α-hydroxylation system[J]. Applied Microbiology and Biotechnology, 2019, 103(20): 8363-8374. |
| 50 | SONG F Z, ZHENG M M, WANG J L, et al. Chemoenzymatic synthesis of C14-functionalized steroids[J]. Nature Synthesis, 2023, 2: 729-739. |
| 51 | ZHAO Y, ZHANG B, SUN Z Q, et al. Biocatalytic C14-hydroxylation on androstenedione enabled modular synthesis of cardiotonic steroids[J]. ACS Catalysis, 2022, 12(16): 9839-9845. |
| 52 | SAMUEL S, NGUYEN T, CHOI H A. Pharmacologic characteristics of corticosteroids[J]. Journal of Neurocritical Care, 2017, 10(2): 53-59. |
| 53 | ZHANG B, ZHOU D F, LI M J, et al. Progress of 3-ketosteroid Δ1-dehydrogenases for steroid production[J]. Systems Microbiology and Biomanufacturing, 2024, 4(2): 631-660. |
| 54 | PETRUSMA M, VAN DER GEIZE R, DIJKHUIZEN L. 3-Ketosteroid 9α-hydroxylase enzymes: Rieske non-heme monooxygenases essential for bacterial steroid degradation[J]. Antonie Van Leeuwenhoek, 2014, 106(1): 157-172. |
| 55 | CHENG H J, SUN Y H, CHANG H W, et al. Compatible solutes adaptive alterations in Arthrobacter simplex during exposure to ethanol, and the effect of trehalose on the stress resistance and biotransformation performance[J]. Bioprocess and Biosystems Engineering, 2020, 43(5): 895-908. |
| 56 | LUO J M, ZHU W C, CAO S T, et al. Improving biotransformation efficiency of Arthrobacter simplex by enhancement of cell stress tolerance and enzyme activity[J]. Journal of Agricultural and Food Chemistry, 2021, 69(2): 704-716. |
| 57 | TANG J, ZENG C L, XIE L Y, et al. Improved synthesis of fluocinolone acetonide and process research of 6α,9α- fluorination[J]. Chemistry Letters, 2018, 47(1): 110-112. |
| 58 | ZHANG R J, XU X X, CAO H J, et al. Purification, characterization, and application of a high activity 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum DSM 1381[J]. Applied Microbiology and Biotechnology, 2019, 103(16): 6605-6616. |
| 59 | WÓJCIK P, GLANOWSKI M, WOJTKIEWICZ A M, et al. Universal capability of 3-ketosteroid Δ1-dehydrogenases to catalyze Δ1-dehydrogenation of C17-substituted steroids[J]. Microbial Cell Factories, 2021, 20(1): 119. |
| 60 | WU Y, MA J N, SHI J H, et al. iTRAQ-based quantitative proteomic analysis of Arthrobacter simplex in response to cortisone acetate and its mutants with improved Δ1-dehydrogenation efficiency[J]. Journal of Agricultural and Food Chemistry, 2023, 71(16): 6376-6388. |
| 61 | WANG Y, ZHANG R, FENG J H, et al. A new 3-ketosteroid-Δ1-dehydrogenase with high activity and broad substrate scope for efficient transformation of hydrocortisone at high substrate concentration[J]. Microorganisms, 2022, 10(3): 508. |
| 62 | ZHANG Y J, LIU M J, WANG H J, et al. Focused mutagenesis in non-catalytic cavity for improving catalytic efficiency of 3-ketosteroid-Δ1-dehydrogenase[J]. Molecular Catalysis, 2022, 531: 112661. |
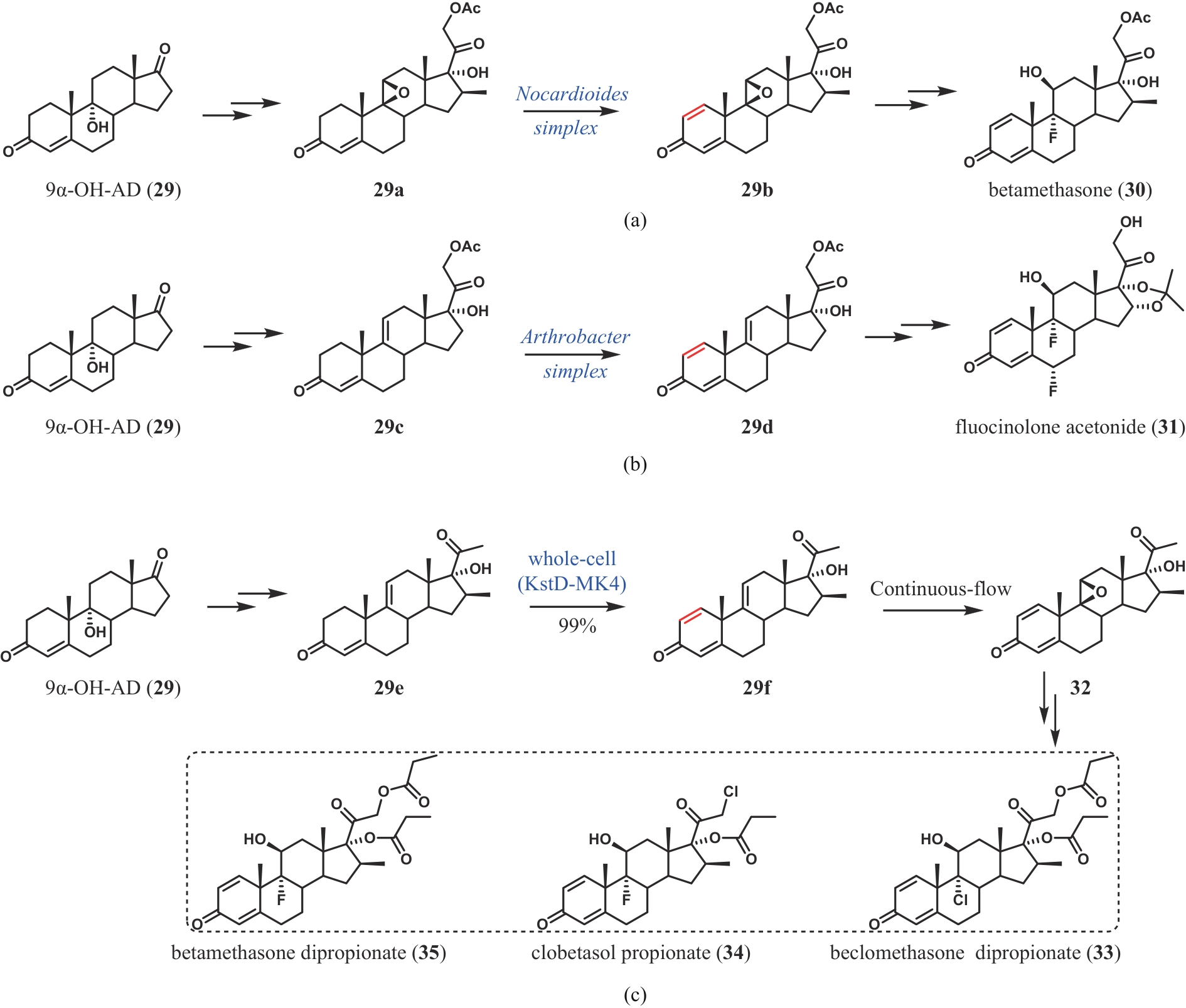
| 63 | WEI J H, ZHANG Y J, LIU M J, et al. Divergent chemo- and biocatalytic route to 16β-methylcorticoids: asymmetric synthesis of betamethasone dipropionate, clobetasol propionate, and beclomethasone dipropionate[J]. Angewandte Chemie International Edition, 2024, 63(4): e202313952. |
| 64 | CONTENTE M L, MOLINARI F, SERRA I, et al. Stereoselective enzymatic reduction of ethyl secodione: preparation of a key intermediate for the total synthesis of steroids[J]. European Journal of Organic Chemistry, 2016, 2016(7): 1260-1263. |
| 65 | CHEN X, ZHANG H L, MARIA-SOLANO M A, et al. Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase[J]. Nature Catalysis, 2019, 2: 931-941. |
| 66 | DING H X, LIU K K C, SAKYA S M, et al. Synthetic approaches to the 2011 new drugs[J]. Bioorganic & Medicinal Chemistry, 2013, 21(11): 2795-2825. |
| 67 | FRYSZKOWSKA A, PETERSON J, DAVIES N L, et al. Development of a chemoenzymatic process for dehydroepiandrosterone acetate synthesis[J]. Organic Process Research & Development, 2016, 20(8): 1520-1528. |
| 68 | MONTI D, FERRANDI E E, ZANELLATO I, et al. One-pot multienzymatic synthesis of 12-ketoursodeoxycholic acid: subtle cofactor specificities rule the reaction equilibria of five biocatalysts working in a row[J]. Advanced Synthesis & Catalysis, 2009, 351(9): 1303-1311. |
| 69 | SUN B Q, KANTZOW C, BRESCH S, et al. Multi-enzymatic one-pot reduction of dehydrocholic acid to 12-keto-ursodeoxycholic acid with whole-cell biocatalysts[J]. Biotechnology and Bioengineering, 2013, 110(1): 68-77. |
| 70 | TONIN F, ARENDS I W C E. Latest development in the synthesis of ursodeoxycholic acid (UDCA): a critical review[J]. Beilstein Journal of Organic Chemistry, 2018, 14: 470-483. |
| 71 | LIU L, BRAUN M, GEBHARDT G, et al. One-step synthesis of 12-ketoursodeoxycholic acid from dehydrocholic acid using a multienzymatic system[J]. Applied Microbiology and Biotechnology, 2013, 97(2): 633-639. |
| 72 | SHI S C, YOU Z N, ZHOU K, et al. Efficient synthesis of 12-oxochenodeoxycholic acid using a 12α-hydroxysteroid dehydrogenase from Rhodococcus ruber [J]. Advanced Synthesis & Catalysis, 2019, 361(20): 4661-4668. |
| 73 | GROBE S, BADENHORST C P S, BAYER T, et al. Engineering regioselectivity of a P450 monooxygenase enables the synthesis of ursodeoxycholic acid via 7β-hydroxylation of lithocholic acid[J]. Angewandte Chemie International Edition, 2021, 60(2): 753-757. |
| 74 | YOU Z N, CHEN Q, SHI S C, et al. Switching cofactor dependence of 7β-hydroxysteroid dehydrogenase for cost-effective production of ursodeoxycholic acid[J]. ACS Catalysis, 2019, 9(1): 466-473. |
| 75 | ZHENG M M, WANG R F, LI C X, et al. Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β- hydroxysteroid dehydrogenase from Ruminococcus torques [J]. Process Biochemistry, 2015, 50(4): 598-604. |
| 76 | ZHENG M M, CHEN F F, LI H, et al. Continuous production of ursodeoxycholic acid by using two cascade reactors with co-immobilized enzymes[J]. ChemBioChem, 2018, 19(4): 347-353. |
| 77 | LI H P, YOU Z N, LIU Y Y, et al. Continuous-flow microreactor-enhanced clean NAD+ regeneration for biosynthesis of 7-oxo-lithocholic acid[J]. ACS Sustainable Chemistry & Engineering, 2022, 10(1): 456-463. |
| 78 | ZHENG M M, CHEN K C, WANG R F, et al. Engineering 7β-hydroxysteroid dehydrogenase for enhanced ursodeoxycholic acid production by multiobjective directed evolution[J]. Journal of Agricultural and Food Chemistry, 2017, 65(6): 1178-1185. |
| 79 | JI Q Z, TAN J, ZHU L C, et al. Preparing tauroursodeoxycholic acid (TUDCA) using a double-enzyme-coupled system[J]. Biochemical Engineering Journal, 2016, 105: 1-9. |
| 80 | PENG Y Q, GAO C H, ZHANG Z L, et al. A chemoenzymatic strategy for the synthesis of steroid drugs enabled by P450 monooxygenase-mediated steroidal core modification[J]. ACS Catalysis, 2022, 12(5): 2907-2914. |
| 81 | ZHANG Z L, GAO C H, ZHAO J, et al. A designed chemoenzymatic route for efficient synthesis of 6-dehydronandrolone acetate: a key precursor in the synthesis of C7-functionalized steroidal drugs[J]. ACS Catalysis, 2023, 13(19): 13111-13116. |
| 82 | ZHANG Y J, LIU M J, YANG Z X, et al. Batch and continuous flow asymmetric synthesis of anabolic-androgenic steroids via a single-cell biocatalytic Δ1-dehydrogenation and C17β- carbonyl reduction cascade[J]. Green Chemistry, 2023, 25(8): 3223-3235. |
| 83 | SZCZEBARA F M, CHANDELIER C, VILLERET C, et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast[J]. Nature Biotechnology, 2003, 21(2): 143-149. |
| 84 | BROCARD-MASSON C, BONNIN I, DUMAS B. Process for preparing genetically transformed yeasts capable of producing a molecule of interest at a high titre: US20140186885[P]. 2014-07-03. |
| 85 | GU Y H, JIAO X, YE L D, et al. Metabolic engineering strategies for de novo biosynthesis of sterols and steroids in yeast[J]. Bioresources and Bioprocessing, 2021, 8(1): 110. |
| 86 | XU S H, LI Y R. Yeast as a promising heterologous host for steroid bioproduction[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47: 829-843. |
| 87 | JIANG Y Q, LIN J P. Recent progress in strategies for steroid production in yeasts[J]. World Journal of Microbiology & Biotechnology, 2022, 38(6): 93. |
| 88 | SU W, XIAO W H, WANG Y, et al. Alleviating redox imbalance enhances 7-dehydrocholesterol production in engineered Saccharomyces cerevisiae [J]. PLoS One, 2015, 10(6): e0130840. |
| 89 | GUO X J, XIAO W H, WANG Y, et al. Metabolic engineering of Saccharomyces cerevisiae for 7-dehydrocholesterol overproduction[J]. Biotechnology for Biofuels, 2018, 11: 192. |
| 90 | GUO X J, YAO M D, XIAO W H, et al. Compartmentalized reconstitution of post-squalene pathway for 7-dehydrocholesterol overproduction in Saccharomyces cerevisiae [J]. Frontiers in Microbiology, 2021, 12: 663973. |
| 91 | QU L S, XIU X, SUN G Y, et al. Engineered yeast for efficient de novo synthesis of 7-dehydrocholesterol[J]. Biotechnology and Bioengineering, 2022, 119(5): 1278-1289. |
| 92 | XIU X, SUN Y, WU Y K, et al. Modular remodeling of sterol metabolism for overproduction of 7-dehydrocholesterol in engineered yeast[J]. Bioresource Technology, 2022, 360: 127572. |
| 93 | WEI W Q, GAO S, YI Q, et al. Reengineering of 7-dehydrocholesterol biosynthesis in Saccharomyces cerevisiae using combined pathway and organelle strategies[J]. Frontiers in Microbiology, 2022, 13: 978074. |
| 94 | GU Y H, CHEN S H, JIAO X, et al. Combinatorial metabolic engineering of Saccharomyces cerevisiae for improved production of 7-dehydrocholesterol[J]. Engineering Microbiology, 2023, 3: 100100. |
| 95 | CHENG J, CHEN J, LIU X N, et al. The origin and evolution of the diosgenin biosynthetic pathway in yam[J]. Plant Communications, 2021, 2(1): 100079. |
| 96 | XU L P, WANG D, CHEN J, et al. Metabolic engineering of Saccharomyces cerevisiae for gram-scale diosgenin production[J]. Metabolic Engineering, 2022, 70: 115-128. |
| 97 | DU H X, XIAO W H, WANG Y, et al. Engineering Yarrowia lipolytica for campesterol overproduction[J]. PLoS One, 2016, 11(1): e0146773. |
| 98 | ZHANG Y, WANG Y, YAO M D, et al. Improved campesterol production in engineered Yarrowia lipolytica strains[J]. Biotechnology Letters, 2017, 39(7): 1033-1039. |
| 99 | XU S H, CHEN C, LI Y R. Engineering of phytosterol-producing yeast platforms for functional reconstitution of downstream biosynthetic pathways[J]. ACS Synthetic Biology, 2020, 9(11): 3157-3170. |
| 100 | XU S H, TENG X X, LI Y R. Optimization of campesterol-producing yeast strains as a feasible platform for the functional reconstitution of plant membrane-bound enzymes[J]. ACS Synthetic Biology, 2023, 12(4): 1109-1118. |
| 101 | QIAN Y D, TAN S Y, DONG G R, et al. Increased campesterol synthesis by improving lipid content in engineered Yarrowia lipolytica [J]. Applied Microbiology and Biotechnology, 2020, 104(16): 7165-7175. |
| 102 | 周武林, 高惠芳, 吴玉玲, 等. 重组酿酒酵母生物合成菜油甾醇[J]. 化工学报, 2021, 72(8): 4314-4324. |
| ZHOU W L, GAO H F, WU Y L, et al. Engineering of Saccharomyces cerevisiae for biosynthesis of campesterol[J]. CIESC Journal, 2021, 72(8): 4314-4324. | |
| 103 | GU L S, ZHANG R X, FAN X Q, et al. Development of CRISPR/Cas9-based genome editing tools for polyploid yeast Cyberlindnera jadinii and its application in engineering heterologous steroid-producing strains[J]. ACS Synthetic Biology, 2023, 12(10): 2947-2960. |
| 104 | DUPORT C, SPAGNOLI R, DEGRYSE E, et al. Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast[J]. Nature Biotechnology, 1998, 16(2): 186-189. |
| 105 | ZHANG R S, ZHANG Y, WANG Y, et al. Pregnenolone overproduction in Yarrowia lipolytica by integrative components pairing of the cytochrome P450scc system[J]. ACS Synthetic Biology, 2019, 8(12): 2666-2678. |
| 106 | XIONG L B, LIU H H, XU L Q, et al. Role identification and application of SigD in the transformation of soybean phytosterol to 9α-hydroxy-4-androstene-3,17-dione in Mycobacterium neoaurum [J]. Journal of Agricultural and Food Chemistry, 2017, 65(3): 626-631. |
| 107 | LIU M, ZHU Z T, TAO X Y, et al. Single nucleotide polymorphism analysis for the production of valuable steroid intermediates in Mycobacterium neoaurum [J]. Biotechnology Letters, 2016, 38(11): 1881-1892. |
| 108 | LIU M, ZHU Z T, TAO X Y, et al. RNA-Seq analysis uncovers non-coding small RNA system of Mycobacterium neoaurum in the metabolism of sterols to accumulate steroid intermediates[J]. Microbial Cell Factories, 2016, 15: 64. |
| 109 | XIONG L B, SUN W J, LIU Y J, et al. Enhancement of 9α-hydroxy-4-androstene-3,17-dione production from soybean phytosterols by deficiency of a regulated intramembrane proteolysis metalloprotease in Mycobacterium neoaurum [J]. Journal of Agricultural and Food Chemistry, 2017, 65(48): 10520-10525. |
| 110 | LIU M, XIONG L B, TAO X Y, et al. Integrated transcriptome and proteome studies reveal the underlying mechanisms for sterol catabolism and steroid production in Mycobacterium neoaurum [J]. Journal of Agricultural and Food Chemistry, 2018, 66(34): 9147-9157. |
| 111 | LIU M, XIONG L B, TAO X Y, et al. Metabolic adaptation of Mycobacterium neoaurum ATCC 25795 in the catabolism of sterols for producing important steroid intermediates[J]. Journal of Agricultural and Food Chemistry, 2018, 66(45): 12141-12150. |
| [1] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [2] | CHENG Zhongyu, LI Fuzhuo. Recent advances in chemoenzymatic synthesis of natural products via site- selective P450 oxidation [J]. Synthetic Biology Journal, 2024, 5(5): 960-980. |
| [3] | YANG Haoran, YE Farong, HUANG Ping, WANG Ping. Recent advances in glycoprotein synthesis [J]. Synthetic Biology Journal, 2024, 5(5): 1072-1101. |
| [4] | Liangbin XIONG, Lu SONG, Yunqiu ZHAO, Kun LIU, Yongjun LIU, Fengqing WANG, Dongzhi WEI. Green biomanufacturing of steroids: from biotransformation to de novo synthesis by microorganisms [J]. Synthetic Biology Journal, 2021, 2(6): 942-963. |
| [5] | Faguang ZHANG, Ge QU, Zhoutong SUN, Jun′an MA. From chemical synthesis to biosynthesis: trends toward total synthesis of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 674-696. |
| [6] | Shuke WU, Yi ZHOU, Wen WANG, Wei ZHANG, Pengfei GAO, Zhi LI. From single-enzyme catalysis to multienzyme cascade: inspired from Professor Daniel I.C. Wang’s pioneer work in enzyme technology [J]. Synthetic Biology Journal, 2021, 2(4): 543-558. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||