Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (4): 651-661.DOI: 10.12211/2096-8280.2021-016
Creation of non-natural cofactor-dependent methanol dehydrogenase
WANG Junting1,2, GUO Xiaojia1, LI Qing1,2, WAN Li1,2, ZHAO Zongbao1
- 1.Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian 116023,Liaoning,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2021-02-04Revised:2021-03-16Online:2021-09-10Published:2021-08-31 -
Contact:ZHAO Zongbao
创制非天然辅酶偏好型甲醇脱氢酶
王俊婷1,2, 郭潇佳1, 李青1,2, 万里1,2, 赵宗保1
- 1.中国科学院大连化学物理研究所,辽宁 大连 116023
2.中国科学院大学,北京 100049
-
通讯作者:赵宗保 -
作者简介:王俊婷 (1993—),女,博士研究生。研究方向为非天然辅酶偏好的酶催化反应。E-mail:wangjt@dicp.ac.cn赵宗保 (1968—),男,研究员,博士生导师,研究组组长。研究方向为能源生物技术、合成微生物学和化学生物学等。E-mail:zhaozb@dicp.ac.cn -
基金资助:国家自然科学基金面上项目(21877112)
CLC Number:
Cite this article
WANG Junting, GUO Xiaojia, LI Qing, WAN Li, ZHAO Zongbao. Creation of non-natural cofactor-dependent methanol dehydrogenase[J]. Synthetic Biology Journal, 2021, 2(4): 651-661.
王俊婷, 郭潇佳, 李青, 万里, 赵宗保. 创制非天然辅酶偏好型甲醇脱氢酶[J]. 合成生物学, 2021, 2(4): 651-661.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-016
| Enzyme | NAD | NCD | NCD preference [(kcat/Km)NCD/ (kcat/Km)NAD] | |||||
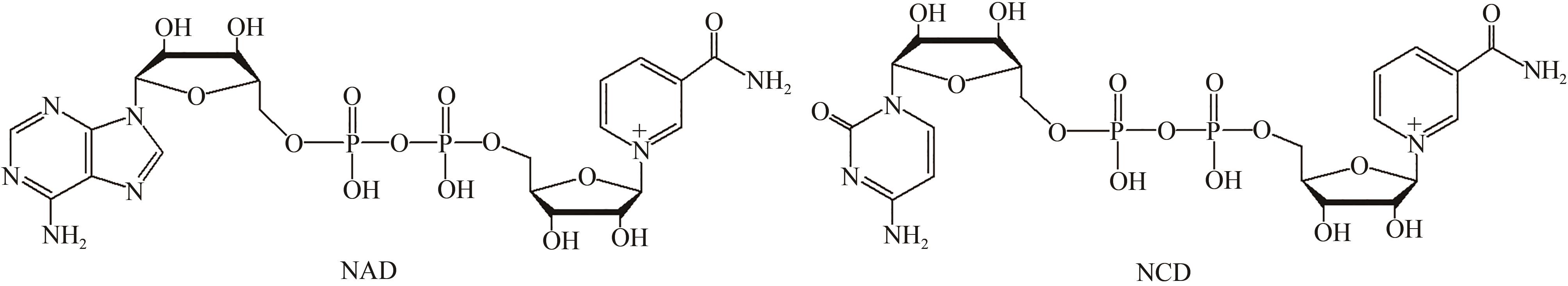
|---|---|---|---|---|---|---|---|---|
| Km/(μmol/L) | kcat/s-1 | kcat/Km /[L/(mol·s)] | Km/(μmol/L) | kcat/s-1 | kcat/Km /[L/(mol·s)] | |||
| WT | 25.5±4.4 | 6.1×10-2±0.1×10-2 | 2.4×103 | 328.6±35.8 | 2.5×10-2±0.1×10-2 | 76.1 | 3.2×10-2 | |
| 3D2 | 6.6±0.5 | 1.3×10-2±0.0 | 2.0×103 | 115.6±11.4 | 2.0×10-2±0.0 | 173.0 | 8.7×10-2 | |
| 3F1 | 121.5±24.4 | 1.0×10-3±0.0 | 8.2 | 77.2±11.3 | 6.0×10-3±0.0 | 77.7 | 9.5 | |
| 3-5 | 5.3±2.5 | 1.0×10-3±0.0 | 188.7 | 47.1±7.7 | 6.0×10-3±0.0 | 127.4 | 0.7 | |
| 3-1 | 728.1±95.4 | 9.0×10-3±0.0 | 12.4 | 44.3±6.6 | 1.3×10-2±0.0 | 293.5 | 23.7 | |
| 9D1 | 1 018.2±257.1 | 2.0×10-3±0.0 | 2.0 | 33.8±10.2 | 2.9×10-2±0.0 | 858.0 | 4.3×102 | |
Tab. 1 Kinetic data of methanol oxidation catalyzed by MDH variants
| Enzyme | NAD | NCD | NCD preference [(kcat/Km)NCD/ (kcat/Km)NAD] | |||||
|---|---|---|---|---|---|---|---|---|
| Km/(μmol/L) | kcat/s-1 | kcat/Km /[L/(mol·s)] | Km/(μmol/L) | kcat/s-1 | kcat/Km /[L/(mol·s)] | |||
| WT | 25.5±4.4 | 6.1×10-2±0.1×10-2 | 2.4×103 | 328.6±35.8 | 2.5×10-2±0.1×10-2 | 76.1 | 3.2×10-2 | |
| 3D2 | 6.6±0.5 | 1.3×10-2±0.0 | 2.0×103 | 115.6±11.4 | 2.0×10-2±0.0 | 173.0 | 8.7×10-2 | |
| 3F1 | 121.5±24.4 | 1.0×10-3±0.0 | 8.2 | 77.2±11.3 | 6.0×10-3±0.0 | 77.7 | 9.5 | |
| 3-5 | 5.3±2.5 | 1.0×10-3±0.0 | 188.7 | 47.1±7.7 | 6.0×10-3±0.0 | 127.4 | 0.7 | |
| 3-1 | 728.1±95.4 | 9.0×10-3±0.0 | 12.4 | 44.3±6.6 | 1.3×10-2±0.0 | 293.5 | 23.7 | |
| 9D1 | 1 018.2±257.1 | 2.0×10-3±0.0 | 2.0 | 33.8±10.2 | 2.9×10-2±0.0 | 858.0 | 4.3×102 | |
| 1 | WHITAKER W B, SANDOVAL N R, BENNETT R K, et al. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization [J]. Current Opinion in Biotechnology, 2015, 33: 165-175. |
| 2 | CHANG K, WANG T F, CHEN J G. Hydrogenation of CO2 to methanol over CuCeTiO x catalysts [J]. Applied Catalysis B-Environmental, 2017, 206: 704-711. |
| 3 | DU X L, JIANG Z, SU D S, et al. Research progress on the indirect hydrogenation of carbon dioxide to methanol [J]. ChemSusChem, 2016, 9(4): 322-332. |
| 4 | COTTON C A R, CLAASSENS N J, BENITO-VAQUERIZO S, et al. Renewable methanol and formate as microbial feedstocks [J]. Current Opinion in Biotechnology, 2020, 62: 168-180. |
| 5 | ANTONIEWICZ M R. Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production[J]. Current Opinion in Biotechnology, 2019, 59: 165-174. |
| 6 | ZHANG W M, ZHANG T, WU S H, et al. Guidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy[J]. RSC Advances, 2017, 7(7): 4083-4091. |
| 7 | SHEEHAN M C, BAILEY C J, DOWDS B C A, et al. A new alcohol dehydrogense, reactive towards methanol, from Bacillus stearothermophilus [J]. Biochemical Journal, 1988, 252(3): 661-666. |
| 8 | WHITAKER W B, JONES J A, BENNETT R K, et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli [J]. Metabolic Engineering, 2017, 39: 49-59. |
| 9 | WU T-Y, CHEN C-T, LIU J T-J, et al. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1[J]. Applied Microbiology and Biotechnology, 2016, 100(11): 4969-4983. |
| 10 | 凡立稳, 王钰, 郑平, 等. 一碳代谢关键酶——甲醇脱氢酶的研究进展与展望[J]. 生物工程学报, 2021, 37(2): 530-540. |
| FAN L W, WANG Y, ZHENG P, et al. Methanol dehydrogenase, a key enzyme of one-carbon metabolism: a review [J]. Chinese Journal of Biotechnology, 2021, 37(2): 530-540. | |
| 11 | TUYISHIME P, WANG Y, FAN L W, et al. Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production [J]. Metabolic Engineering, 2018, 49: 220-231. |
| 12 | KIM S, LINDNER S N, ASLAN S, et al. Growth of E. coli on formate and methanol via the reductive glycine pathway [J]. Nature Chemical Biology, 2020, 16(5): 538-545. |
| 13 | KIM Y H, CAMPBELL E, YU J, et al. Complete oxidation of methanol in biobattery devices using a hydrogel created from three modified dehydrogenases[J]. Angewandte Chemie International Edition, 2013, 52(5): 1437-1440. |
| 14 | WANG Y P, SAN K Y, BENNETT G N. Cofactor engineering for advancing chemical biotechnology[J]. Current Opinion in Biotechnology, 2013, 24(6): 994-999. |
| 15 | KARA S, SCHRITTWIESER J H, HOLLMANN F, et al. Recent trends and novel concepts in cofactor-dependent biotransformations[J]. Applied Microbiology and Biotechnology, 2014, 98(4): 1517-1529. |
| 16 | XIAO W S, WANG R-S, HANDY D E, et al. NAD(H) and NADP(H) redox couples and cellular energy metabolism [J]. Antioxidants & Redox Signaling, 2018, 28(3): 251-272. |
| 17 | GRAY K A, RICHARDSON T H, KRETZ K, et al. Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase [J]. Advanced Synthesis & Catalysis, 2001, 343(6/7): 607-617. |
| 18 | MAMPEL J, BUESCHER J M, MEURER G, et al. Coping with complexity in metabolic engineering [J]. Trends in Biotechnology, 2013, 31(1): 52-60. |
| 19 | PAUL C E, ARENDS I W C E, HOLLMANN F. Is simpler better? synthetic nicotinamide cofactor analogues for redox chemistry [J]. ACS Catalysis, 2014, 4(3): 788-797. |
| 20 | KNAUS T, PAUL C E, LEVY C W, et al. Better than nature: nicotinamide biomimetics that outperform natural coenzymes [J]. Journal of the American Chemical Society, 2016, 138(3): 1033-1039. |
| 21 | WEUSTHUIS R A, FOLCH P L, POZO-RODRÍGUEZ A, et al. Applying non-canonical redox cofactors in fermentation processes [J]. iScience, 2020, 23(9): 101471. |
| 22 | BLACK W B, ZHANG L Y, MAK W S, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis [J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| 23 | AGARWAL P K, WEBB S P, HAMMES-SCHIFFER S. Computational studies of the mechanism for proton and hydride transfer in liver alcohol dehydrogenase [J]. Journal of the American Chemical Society, 2000, 122(19): 4803-4812. |
| 24 | JI D B, WANG L, LIU W J, et al. Synthesis of NAD analogs to develop bioorthogonal redox system[J]. Science China - Chemistry, 2013, 56(3): 296-300. |
| 25 | HOU S H, LIU W J, JI D B, et al. Synthesis of 1,2,3-triazole moiety-containing NAD analogs and their potential as redox cofactors [J]. Tetrahedron Letters, 2011, 52(44): 5855-5857. |
| 26 | HOU S H, LIU W J, ZHAO Z B. Synthesis of novel nicotinamide adenine dinucleotide (NAD) analogs and their coenzyme activities [J]. Chinese Journal of Organic Chemistry, 2012, 32(2): 349-353. |
| 27 | JI D B, WANG L, HOU S H, et al. Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide [J]. Journal of the American Chemical Society, 2011, 133(51): 20857-20862. |
| 28 | WANG L, JI D B, LIU Y X, et al. Synthetic cofactor-linked metabolic circuits for selective energy transfer [J]. ACS Catalysis, 2017, 7(3): 1977-1983. |
| 29 | GUO X J, LIU Y X, WANG Q, et al. Non-natural cofactor and formate-driven reductive carboxylation of pyruvate [J]. Angewandte Chemie International Edition, 2020, 59(8): 3143-3146. |
| 30 | LIAO Z P, YANG X T, FU H X, et al. The significance of aspartate on NAD(H) biosynthesis and ABE fermentation in Clostridium acetobutylicum ATCC 824 [J]. AMB Express, 2019, 9(1): 142. |
| 31 | 刘美霞, 李强子, 孟冬冬, 等. 烟酰胺类辅酶依赖型氧化还原酶的辅酶偏好性改造及其在合成生物学中的应用[J]. 合成生物学, 2020, 1(5): 570-582. |
| LIU M X, LI Q Z, MENG D D, et al. Protein engineering of nicotinamide coenzyme-dependent oxidoreductases for coenzyme preference and its application in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(5): 570-582. | |
| 32 | GUO X J, FENG Y B, WANG X Y, et al. Characterization of the substrate scope of an alcohol dehydrogenase commonly used as methanol dehydrogenase [J]. Bioorganic & Medicinal Chemistry Letters, 2019, 29(12): 1446-1449. |
| 33 | WATERHOUSE A, BERTONI M, BIENERT S, et al. SWISS-MODEL: homology modelling of protein structures and complexes [J]. Nucleic Acids Research, 2018, 46(W1): W296-W303. |
| 34 | LILL M A, DANIELSON M L. Computer-aided drug design platform using PyMOL [J]. Journal of Computer-Aided Molecular Design, 2011, 25(1): 13-19. |
| 35 | RUI L Y, CAO L, CHEN W, et al. Active site engineering of the epoxide hydrolase from Agrobacterium radiobacter AD1 to enhance aerobic mineralization of cis-1,2-dichloroethylene in cells expressing an evolved toluene ortho-monooxygenase [J]. Journal of Biological Chemistry, 2004, 279(45): 46810-46817. |
| 36 | VAN DEN ENT F, LÖWE J. RF cloning: A restriction-free method for inserting target genes into plasmids [J]. Journal of Biochemical and Biophysical Methods, 2006, 67(1): 67-74. |
| 37 | WU T K, LIU Y T, CHANG C H, et al. Site-saturated mutagenesis of histidine 234 of Saccharomyces cerevisiae oxidosqualene-lanosterol cyclase demonstrates dual functions in cyclization and rearrangement reactions [J]. Journal of the American Chemical Society, 2006, 128(19): 6414-6419. |
| 38 | GRAGEROV A, HORIE K, PAVLOVA M, et al. Large-scale, saturating insertional mutagenesis of the mouse genome [J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(36): 14406-14411. |
| 39 | ELHAWRANI A S, SESSIONS R B, MORETON K M, et al. Guided evolution of enzymes with new substrate specificities [J]. Journal of Molecular Biology, 1996, 264(1): 97-110. |
| 40 | WANG L, ZHOU Y J, JI D B, et al. Identification of UshA as a major enzyme for NAD degradation in Escherichia coli [J]. Enzyme and Microbial Technology, 2014, 58/59: 75-79. |
| 41 | REETZ M T, CARBALLEIRA J D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes [J]. Nature Protocols, 2007, 2(4): 891-903. |
| 42 | LAMZIN V S, DAUTER Z, POPOV V O, et al. High resolution structures of holo and apo formate dehydrogense [J]. Journal of Molecular Biology, 1994, 236(3): 759-785. |
| 43 | REETZ M T, WANG L W, BOCOLA M. Directed evolution of enantioselective enzymes: Iterative cycles of CASTing for probing protein-sequence space [J]. Angewandte Chemie International Edition, 2006, 45(8): 1236-1241. |
| 44 | SUN Z T, LONSDALE R, ILIE A, et al. Catalytic asymmetric reduction of difficult-to-reduce ketones: triple-code saturation mutagenesis of an alcohol dehydrogenase [J]. ACS Catalysis, 2016, 6(3): 1598-1605. |
| 45 | LIU Y X, FENG Y B, WANG L, et al. Structural insights into phosphite dehydrogenase variants favoring a non-natural redox cofactor [J]. ACS Catalysis, 2019, 9(3): 1883-1887. |
| 46 | GUO X J, WANG X Y, LIU Y X, et al. Structure-guided design of formate dehydrogenase for regeneration of a non-natural redox cofactor [J]. Chemistry a European Journal, 2020, 26(70): 16611-16615. |
| [1] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [2] | CHENG Feng, ZOU Shuping, XU Jianmiao, TANG Heng, XUE Yaping, ZHENG Yuguo. BioHPP®: a benchmark of biomanufacturing for high optically pure L-phosphinothricin [J]. Synthetic Biology Journal, 2024, 5(6): 1404-1418. |
| [3] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [4] | FU Yu, ZHONG Fangrui. Recent advances in chemically driven enantioselective photobiocatalysis [J]. Synthetic Biology Journal, 2024, 5(5): 1021-1049. |
| [5] | QI Yanping, ZHU Jin, ZHANG Kai, LIU Tong, WANG Yajie. Recent development of directed evolution in protein engineering [J]. Synthetic Biology Journal, 2022, 3(6): 1081-1108. |
| [6] | WANG Huibin, CHE Changli, YOU Song. Recent advances of enzymatic synthesis of organohalogens catalyzed by Fe/αKG-dependent halogenases [J]. Synthetic Biology Journal, 2022, 3(3): 545-566. |
| [7] | LOU Yujiao, XU Jian, WU Qi. Progress of biocatalytic deuteration of inert carbon-hydrogen bonds [J]. Synthetic Biology Journal, 2022, 3(3): 530-544. |
| [8] | YANG Lu, QU Xudong. Application of imine reductase in the synthesis of chiral amines [J]. Synthetic Biology Journal, 2022, 3(3): 516-529. |
| [9] | XIONG Liangbin, SONG Lu, ZHAO Yunqiu, LIU Kun, LIU Yongjun, WANG Fengqing, WEI Dongzhi. Green biomanufacturing of steroids: from biotransformation to de novo synthesis by microorganisms [J]. Synthetic Biology Journal, 2021, 2(6): 942-963. |
| [10] | ZHANG Faguang, QU Ge, SUN Zhoutong, MA Jun′an. From chemical synthesis to biosynthesis: trends toward total synthesis of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 674-696. |
| [11] | TANG Heng, HAN Xin, ZOU Shuping, ZHENG Yuguo. Application of multi-enzyme catalytic system in the synthesis of pharmaceutical chemicals [J]. Synthetic Biology Journal, 2021, 2(4): 559-576. |
| [12] | WU Shuke, ZHOU Yi, WANG Wen, ZHANG Wei, GAO Pengfei, LI Zhi. From single-enzyme catalysis to multienzyme cascade: inspired from Professor Daniel I.C. Wang’s pioneer work in enzyme technology [J]. Synthetic Biology Journal, 2021, 2(4): 543-558. |
| [13] | YUAN Feiyan, YU Yang, LI Chun. Artificial enzyme designs and its application based on non-natural structural elements [J]. Synthetic Biology Journal, 2020, 1(6): 685-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||