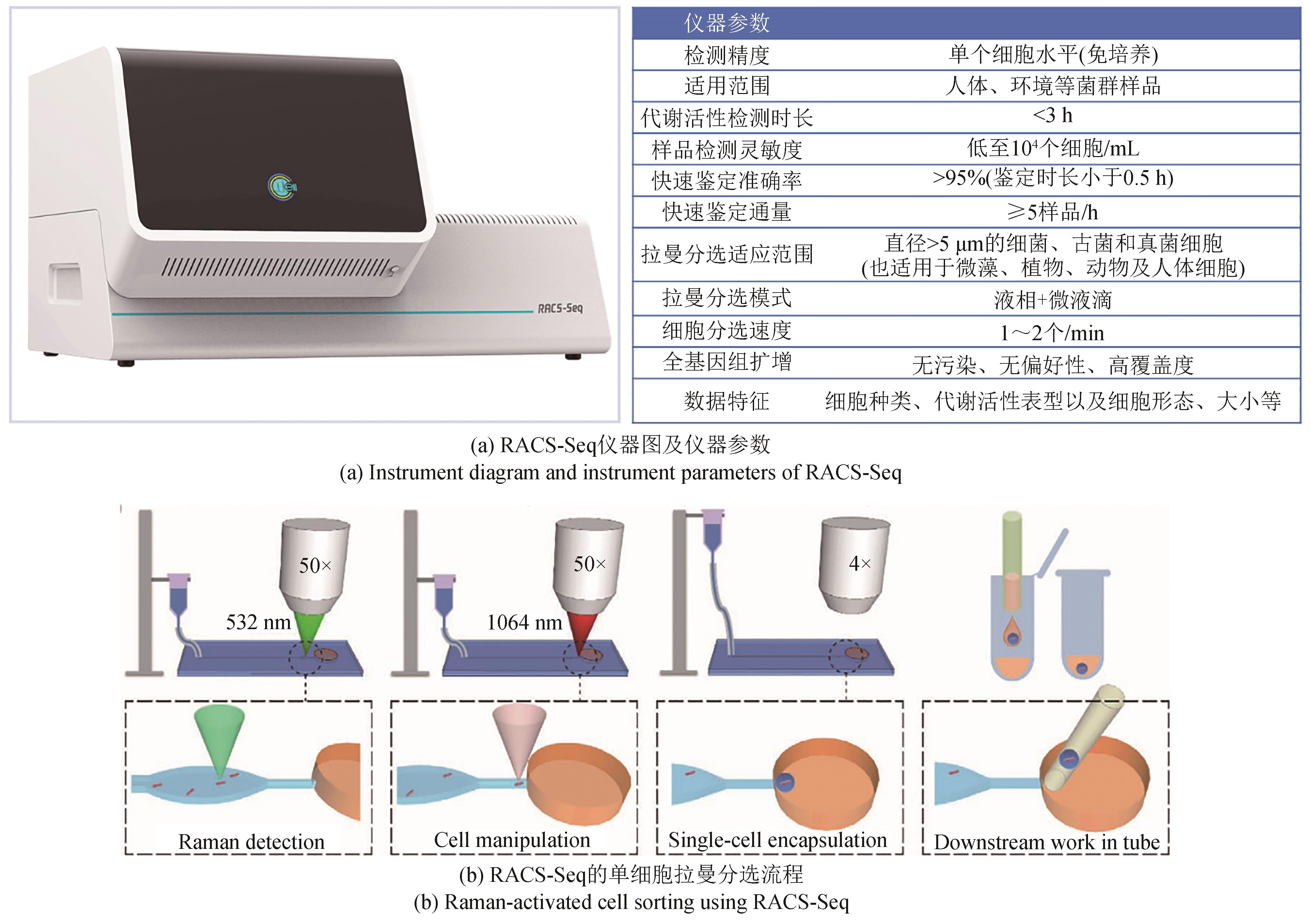
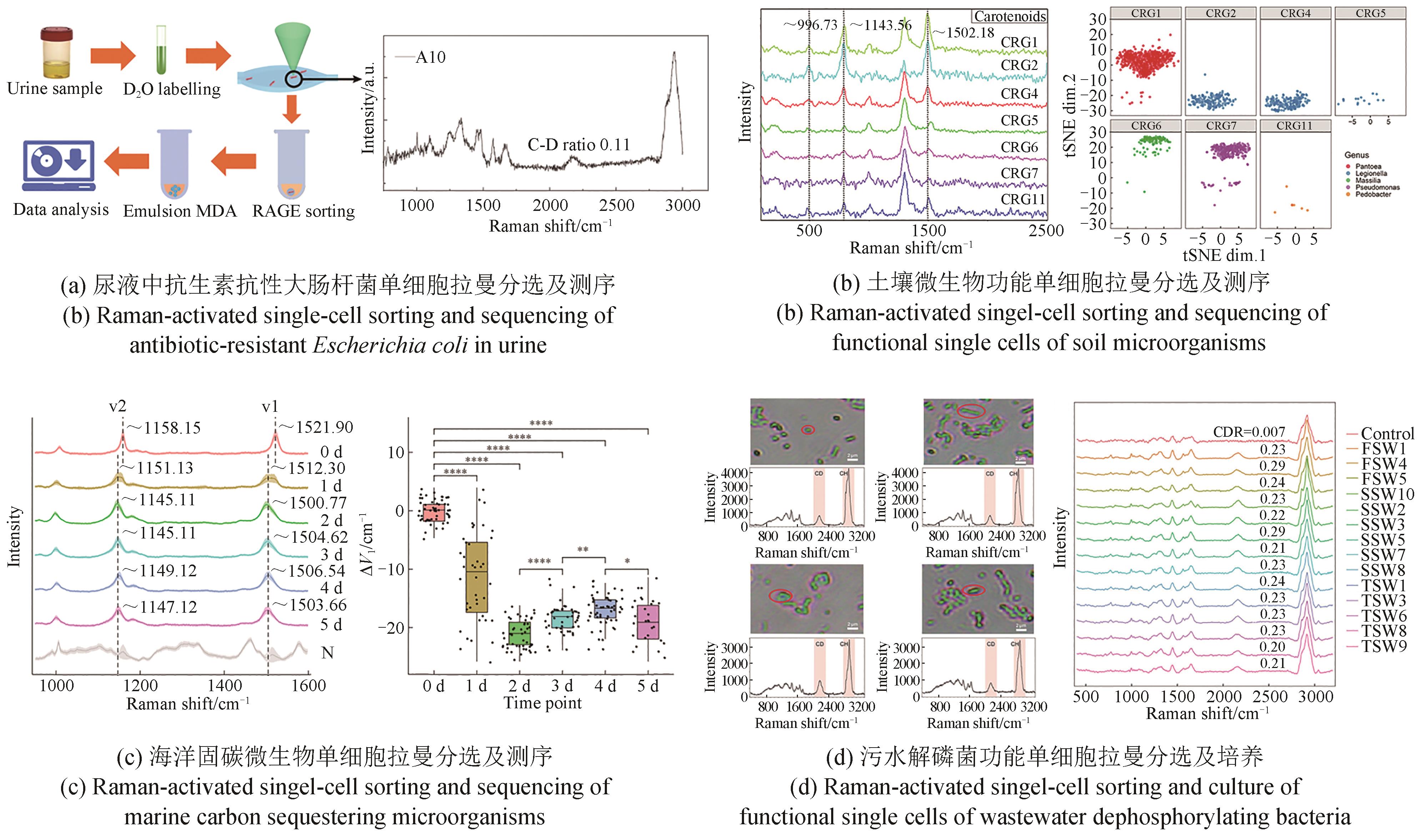
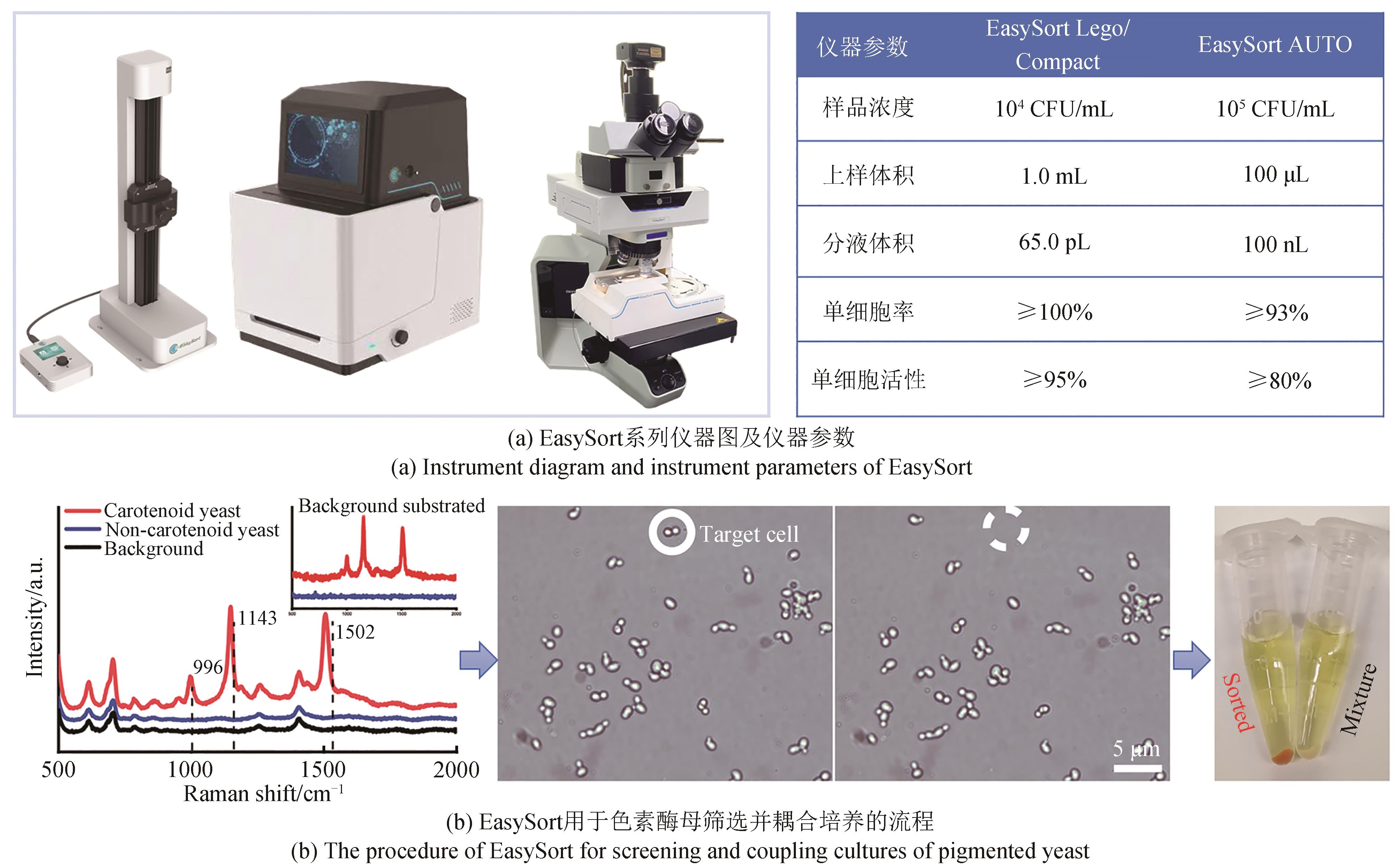
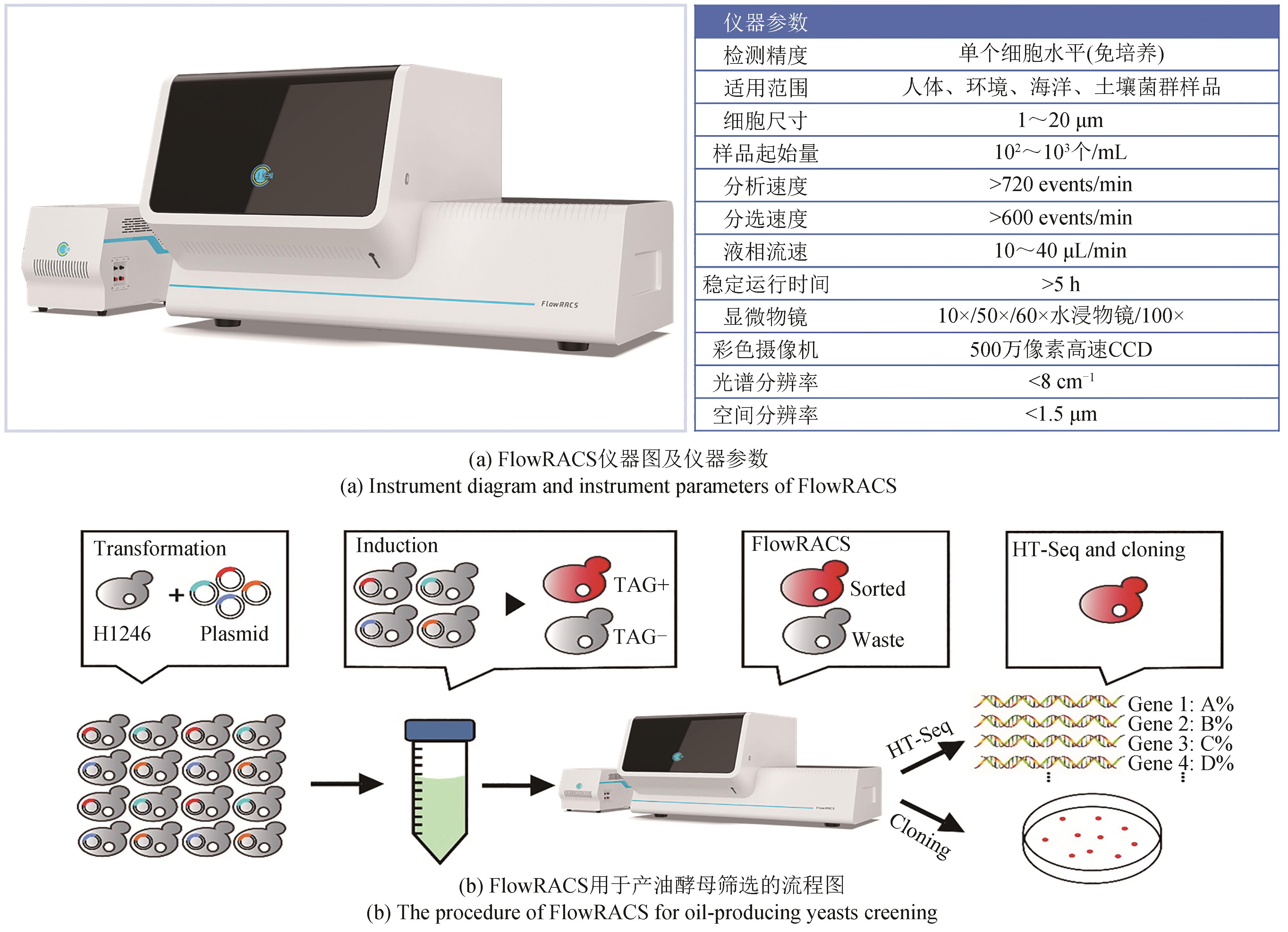
Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (5): 1020-1035.DOI: 10.12211/2096-8280.2023-025
• Invited Review • Previous Articles Next Articles
Advances and applications of single-cell Raman spectroscopy testing and sorting equipment
DIAO Zhidian, WANG Xixian, SUN Qing, XU Jian, MA Bo
- Single-cell Center,Qingdao Institute of Bioenergy and Bioprocess Technology,Chinese Academy of Sciences,Qingdao 266101,Shandong,China
-
Received:2023-03-21Revised:2022-05-17Online:2023-11-15Published:2023-10-31 -
Contact:MA Bo
单细胞拉曼光谱测试分选装备研制及应用进展
刁志钿, 王喜先, 孙晴, 徐健, 马波
- 中国科学院青岛生物能源与过程研究所,单细胞中心,山东 青岛 266101
-
通讯作者:马波 -
作者简介:刁志钿 (1995—),男,在读博士研究生。研究方向为液滴微流控技术、高通量流式拉曼分选技术等。 E-mail:diaozd@qibebt.ac.cn马波 (1976—),男,博士,研究员,博士生导师。研究方向为单细胞关键技术与仪器、微流控技术等,长期从事基于拉曼光谱的单细胞分析/分选及后续的单细胞测序等关键技术和仪器研究。E-mail:mabo@qibebt.ac.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2018YFA090290);天津市合成生物技术创新能力提升行动项目(TSBICIP-PTJS-003-05)
CLC Number:
Cite this article
DIAO Zhidian, WANG Xixian, SUN Qing, XU Jian, MA Bo. Advances and applications of single-cell Raman spectroscopy testing and sorting equipment[J]. Synthetic Biology Journal, 2023, 4(5): 1020-1035.
刁志钿, 王喜先, 孙晴, 徐健, 马波. 单细胞拉曼光谱测试分选装备研制及应用进展[J]. 合成生物学, 2023, 4(5): 1020-1035.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-025
| 1 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 2 | BENNER S A, SISMOUR A M. Synthetic biology[J]. Nature Reviews Genetics, 2005, 6(7): 533-543. |
| 3 | SCHOBER L, BÜTTNER E, LASKE C, et al. Cell dispensing in low-volume range with the immediate drop-on-demand technology (I-DOT)[J]. Journal of Laboratory Automation, 2015, 20(2): 154-163. |
| 4 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 5 | SANDER J D, JOUNG J K. CRISPR-Cas systems for editing, regulating and targeting genomes[J]. Nature Biotechnology, 2014, 32(4): 347-355. |
| 6 | SMITH H O, HUTCHISON C A, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| 7 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 8 | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 9 | WANG X X, REN L H, DIAO Z D, et al. Robust spontaneous Raman flow cytometry for single-cell metabolic phenome profiling via pDEP-DLD-RFC[J]. Advanced Science, 2023: 2207497. |
| 10 | ZINCHENKO A, DEVENISH S R A, KINTSES B, et al. One in a million: flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution[J]. Analytical Chemistry, 2014, 86(5): 2526-2533. |
| 11 | BREHM-STECHER B F, JOHNSON E A. Single-cell microbiology: tools, technologies, and applications[J]. Microbiology and Molecular Biology Reviews, 2004, 68(3): 538-559. |
| 12 | 杨建花, 苏晓岚, 朱蕾蕾. 高通量筛选系统在定向改造中的新进展[J]. 生物工程学报, 2021, 37(7): 2197-2210. |
| YANG J H, SU X L, ZHU L L. Advances of high-throughput screening system in reengineering of biological entities[J]. Chinese Journal of Biotechnology, 2021, 37(7): 2197-2210. | |
| 13 | ALI A, ABOULEILA Y, SHIMIZU Y,et al. Single-cell metabolomics by mass spectrometry: advances, challenges, and future applications[J]. TrAC Trends in Analytical Chemistry, 2019, 120: 115436. |
| 14 | SPITZER M H, NOLAN G P. Mass cytometry: single cells, many features[J]. Cell, 2016, 165(4): 780-791. |
| 15 | AMANTONICO A, URBAN P L, ZENOBI R. Analytical techniques for single-cell metabolomics: state of the art and trends[J]. Analytical and Bioanalytical Chemistry, 2010, 398(6): 2493-2504. |
| 16 | RAMAN C V, KRISHNAN K S. A new type of secondary radiation[J]. Nature, 1928, 121(3048): 501-502. |
| 17 | XU J, MA B, SU X Q, et al. Emerging trends for microbiome analysis: From single-cell functional imaging to microbiome big data[J]. Engineering, 2017, 3(1): 66-70. |
| 18 | HE Y H, WANG X X, MA B, et al. Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution[J]. Biotechnology Advances, 2019, 37(6): 107388. |
| 19 | LEE K S, LANDRY Z, PEREIRA F C, et al. Raman microspectroscopy for microbiology[J]. Nature Reviews Methods Primers, 2021, 1(80): 1-25. |
| 20 | YAN S S, QIU J X, GUO L, et al. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level[J].Applied Microbiology and Biotechnology, 2021, 105(4): 1315-1331. |
| 21 | WANG Y, JI Y T, WHARFE E S, et al. Raman activated cell ejection for isolation of single cells[J]. Analytical Chemistry, 2013, 85(22): 10697-10701. |
| 22 | HUANG W E, WARD A D, WHITELEY A S. Raman tweezers sorting of single microbial cells[J]. Environmental Microbiology Reports, 2009, 1(1): 44-49. |
| 23 | XIE C G, CHEN D, LI Y Q. Raman sorting and identification of single living micro-organisms with optical tweezers[J]. Optics Letters, 2005, 30(14): 1800-1802. |
| 24 | XU T, GONG Y H, SU X L, et al. Phenome-genome profiling of single bacterial cell by Raman-activated gravity-driven encapsulation and sequencing [J]. Small, 2020, 16(30): 2001172. |
| 25 | LEE K S, PALATINSZKY M, PEREIRA F C, et al. An automated Raman-based platform for the sorting of live cells by functional properties[J]. Nature Microbiology, 2019, 4(6): 1035-1048. |
| 26 | ZHANG P R, REN L H, ZHANG X, et al. Raman-activated cell sorting based on dielectrophoretic single-cell trap and release[J]. Analytical Chemistry, 2015, 87(4): 2282-2289. |
| 27 | WANG X X, REN L H, SU Y T, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 28 | WANG X X, XIN Y, REN L H, et al. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo [J]. Science Advances, 2020, 6(32): eabb3521. |
| 29 | NITTA N, IINO T, ISOZAKI A, et al. Raman image-activated cell sorting[J]. Nature Communications, 2020, 11(1): 3452. |
| 30 | LINDLEY M, DE PABLO J G, PETERSON W, et al. High-throughput Raman-activated cell sorting in the fingerprint region[J]. Advanced Materials Technologies, 2022, 7(10): 2101567. |
| 31 | JING X Y, GONG Y H, XU T, et al. One-cell metabolic phenotyping and sequencing of soil microbiome by Raman-activated gravity-driven encapsulation (RAGE)[J]. mSystems, 2021, 6(3): e00181-21. |
| 32 | JING X Y, GONG Y H, XU T, et al. Revealing CO2-fixing SAR11 bacteria in the ocean by Raman-based single-cell metabolic profiling and genomics[J]. BioDesign Research, 2022, 2022: 9782712. |
| 33 | JING X Y, GONG Y H, PAN H H, et al. Single-cell Raman-activated sorting and cultivation (scRACS-Culture) for assessing and mining in situ phosphate-solubilizing microbes from nature[J]. ISME Communications, 2022, 2: 106. |
| 34 | XU T, LI Y D, HAN X, et al. Versatile, facile and low-cost single-cell isolation, culture and sequencing by optical tweezer-assisted pool-screening[J]. Lab on a Chip, 2023, 23(1): 125-135. |
| 35 | DIAO Z D, KAN L Y, ZHAO Y L, et al. Artificial intelligence-assisted automatic and index-based microbial single-cell sorting system for One-Cell-One-Tube[J]. mLife, 2022, 1(4): 448-459. |
| 36 | XIN Y, SHEN C, SHE Y T, et al. Biosynthesis of triacylglycerol molecules with a tailored PUFA profile in industrial microalgae[J]. Molecular Plant, 2019, 12(4): 474-488. |
| 37 | XIN Y, LU Y D, LEE Y Y, et al. Producing designer oils in industrial microalgae by rational modulation of Co-evolving type-2 diacylglycerol acyltransferases[J]. Molecular Plant, 2017, 10(12): 1523-1539. |
| 38 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 39 | BERRY D, MADER E, LEE T K, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(2): E194-203. |
| 40 | JING X Y, GOU H L, GONG Y H, et al. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean[J]. Environmental Microbiology, 2018, 20(6): 2241-2255. |
| 41 | SONG Y Z, KASTER A K, VOLLMERS J, et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea[J]. Microbial Biotechnology, 2017, 10(1): 125-137. |
| 42 | WANG T T, JI Y T, WANG Y, et al. Quantitative dynamics of triacylglycerol accumulation in microalgae populations at single-cell resolution revealed by Raman microspectroscopy[J].Biotechnology for Biofuels, 2014, 7: 58. |
| 43 | JI Y T, HE Y H, CUI Y B, et al. Raman spectroscopy provides a rapid, non-invasive method for quantitation of starch in live, unicellular microalgae[J]. Biotechnology Journal, 2014, 9(12): 1512-1518. |
| 44 | HE Y H, ZHANG P, HUANG S, et al. Label-free, simultaneous quantification of starch, protein and triacylglycerol in single microalgal cells[J].Biotechnology for Biofuels, 2017, 10(1): 275. |
| 45 | TAO Y F, WANG Y, HUANG S, et al. Metabolic-activity-based assessment of antimicrobial effects by D2O-labeled single-cell Raman microspectroscopy[J]. Analytical Chemistry, 2017, 89(7): 4108-4115. |
| 46 | TENG L, WANG X, WANG X J, et al. Label-free, rapid and quantitative phenotyping of stress response in E. coli via ramanome[J]. Scientific Reports, 2016, 6: 34359. |
| 47 | HEKMATARA M, HEIDARI BALADEHI M, JI Y T, et al. D2O-probed Raman microspectroscopy distinguishes the metabolic dynamics of macromolecules in organellar anticancer drug response[J]. Analytical Chemistry, 2021, 93(4): 2125-2134. |
| 48 | WANG Y, SONG Y Z, TAO Y F, et al. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level[J]. Analytical Chemistry, 2016, 88(19): 9443-9450. |
| 49 | HE Y H, HUANG S, ZHANG P, et al. Intra-ramanome correlation analysis unveils metabolite conversion network from an isogenic population of cells[J]. mBio, 2021, 12(4): e0147021. |
| 50 | HEIDARI BALADEHI M, HEKMATARA M, HE Y H, et al. Culture-free identification and metabolic profiling of microalgal single cells via ensemble learning of ramanomes[J]. Analytical Chemistry, 2021, 93(25): 8872-8880. |
| [1] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [2] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||