合成生物学 ›› 2022, Vol. 3 ›› Issue (3): 602-615.DOI: 10.12211/2096-8280.2021-047
• 研究论文 • 上一篇
Caulobactercrescentus蔗糖水解酶受体亚位点分子改造及其在松二糖制备中的应用
王蕾, 邢晨晨, 郭志勇, 宿玲恰, 吴敬
- 江南大学食品科学与技术国家重点实验室,江南大学生物工程学院,江苏 无锡 214122
-
收稿日期:2021-04-21修回日期:2021-06-03出版日期:2022-06-30发布日期:2022-07-13 -
通讯作者:吴敬 -
作者简介:王蕾 (1987—),男,助理研究员。研究方向为酶工程与技术。E-mail: leiwang_enzyme@jiangnan.edu.cn吴敬 (1969—),女,博士生导师,教授。研究方向为酶工程技术与发酵工程。 E-mail: jingwu@jiangnan.edu.cn -
基金资助:国家自然科学基金(31730067);国家自然科学基金青年科学基金(32001637)
Molecular modification of acceptor subsite in sucrose hydrolase from Calobacter crescentus and its application in producing turanose
WANG Lei, XING Chenchen, GUO Zhiyong, SU Lingqia, WU Jing
- State Key Laboratory of Food Science and Technology,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2021-04-21Revised:2021-06-03Online:2022-06-30Published:2022-07-13 -
Contact:WU Jing
摘要:
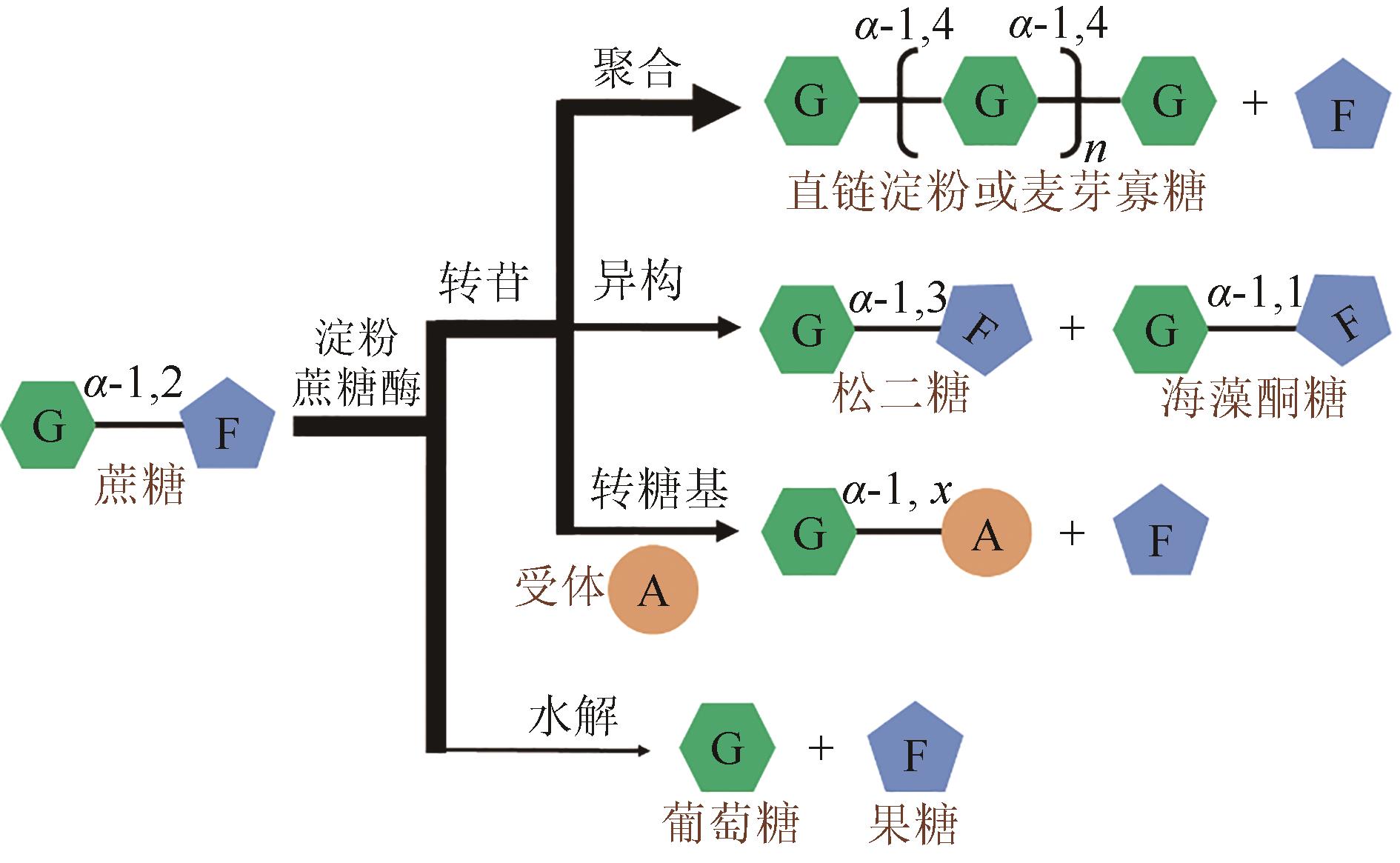
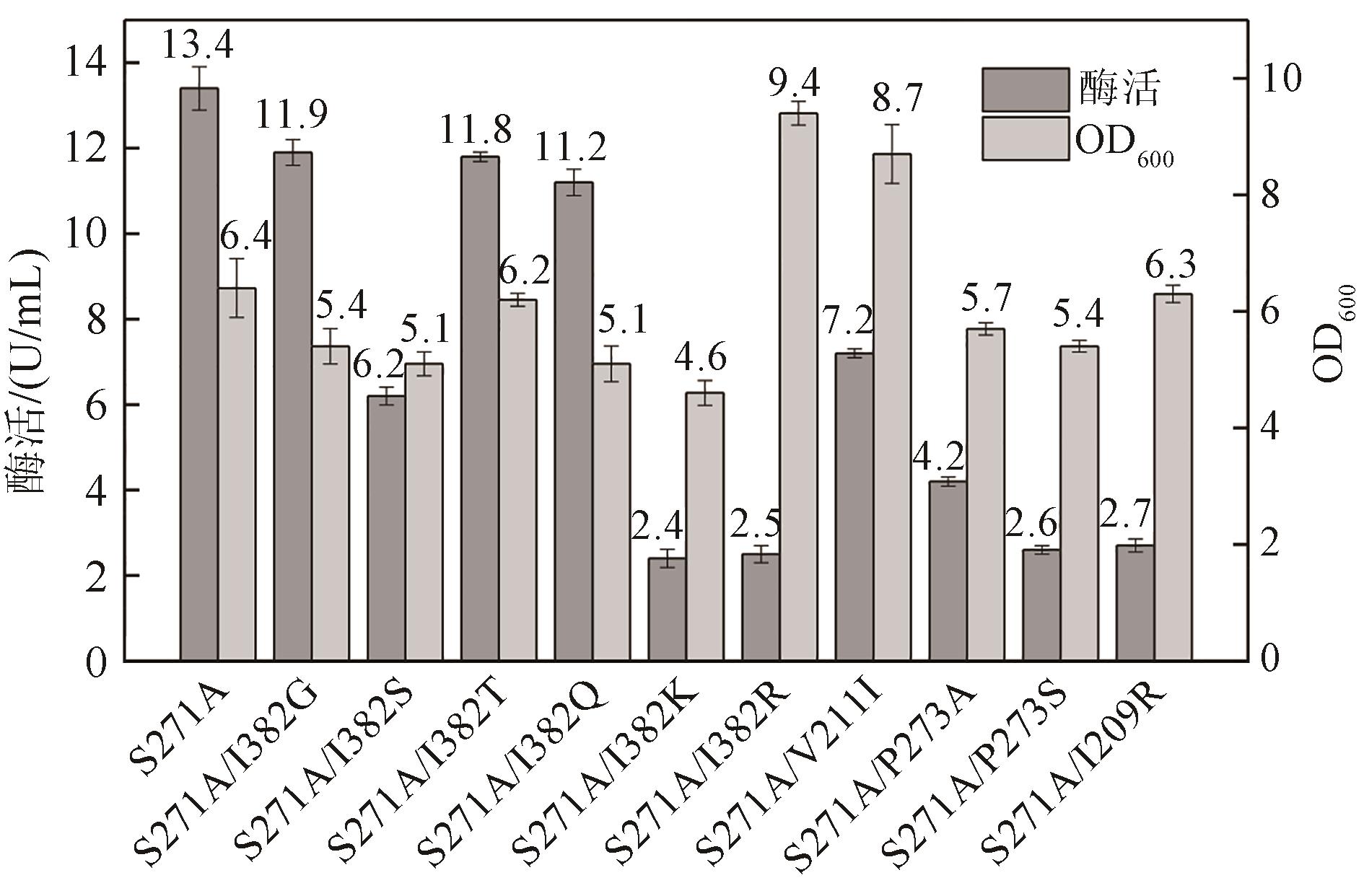
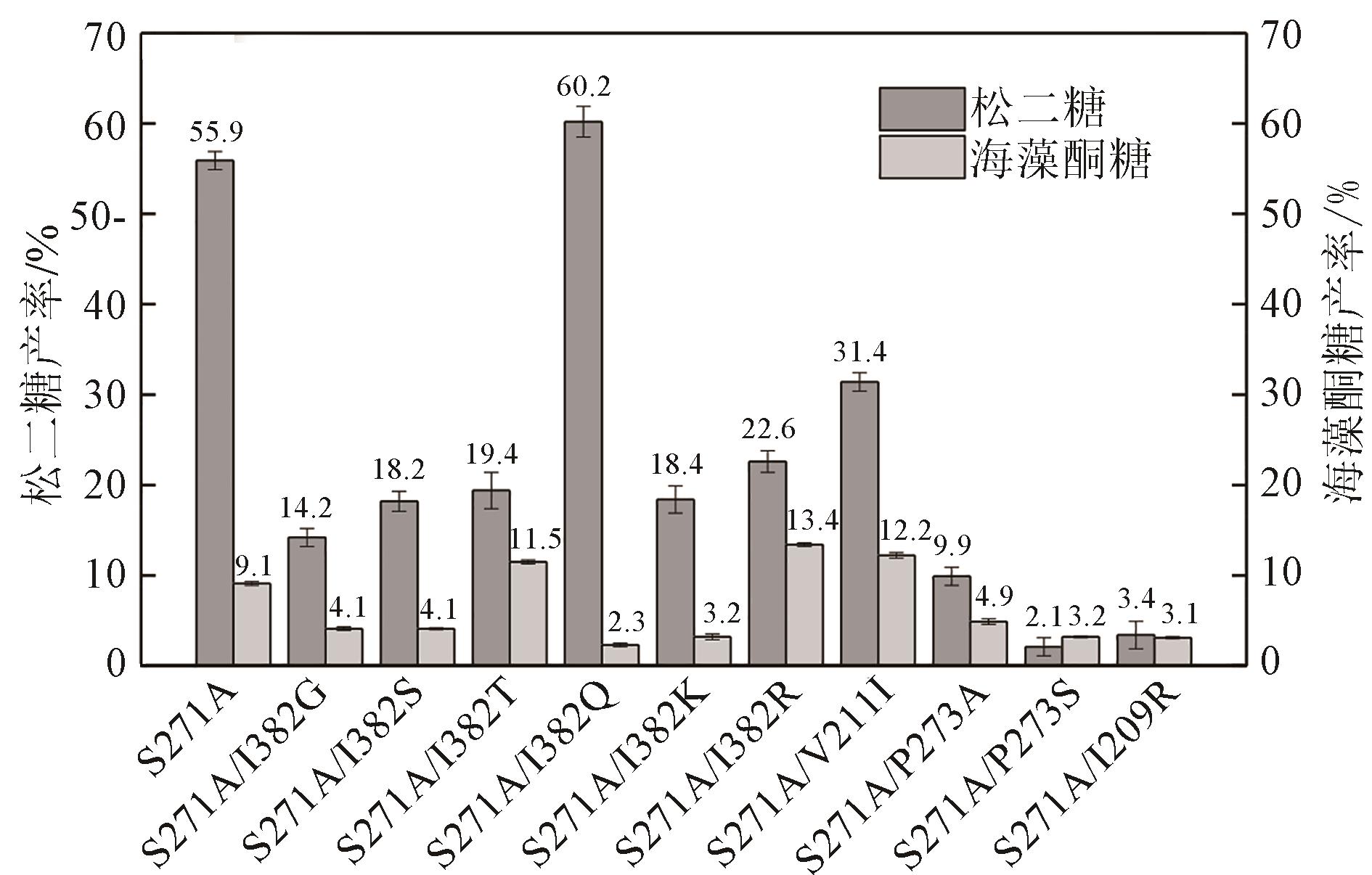
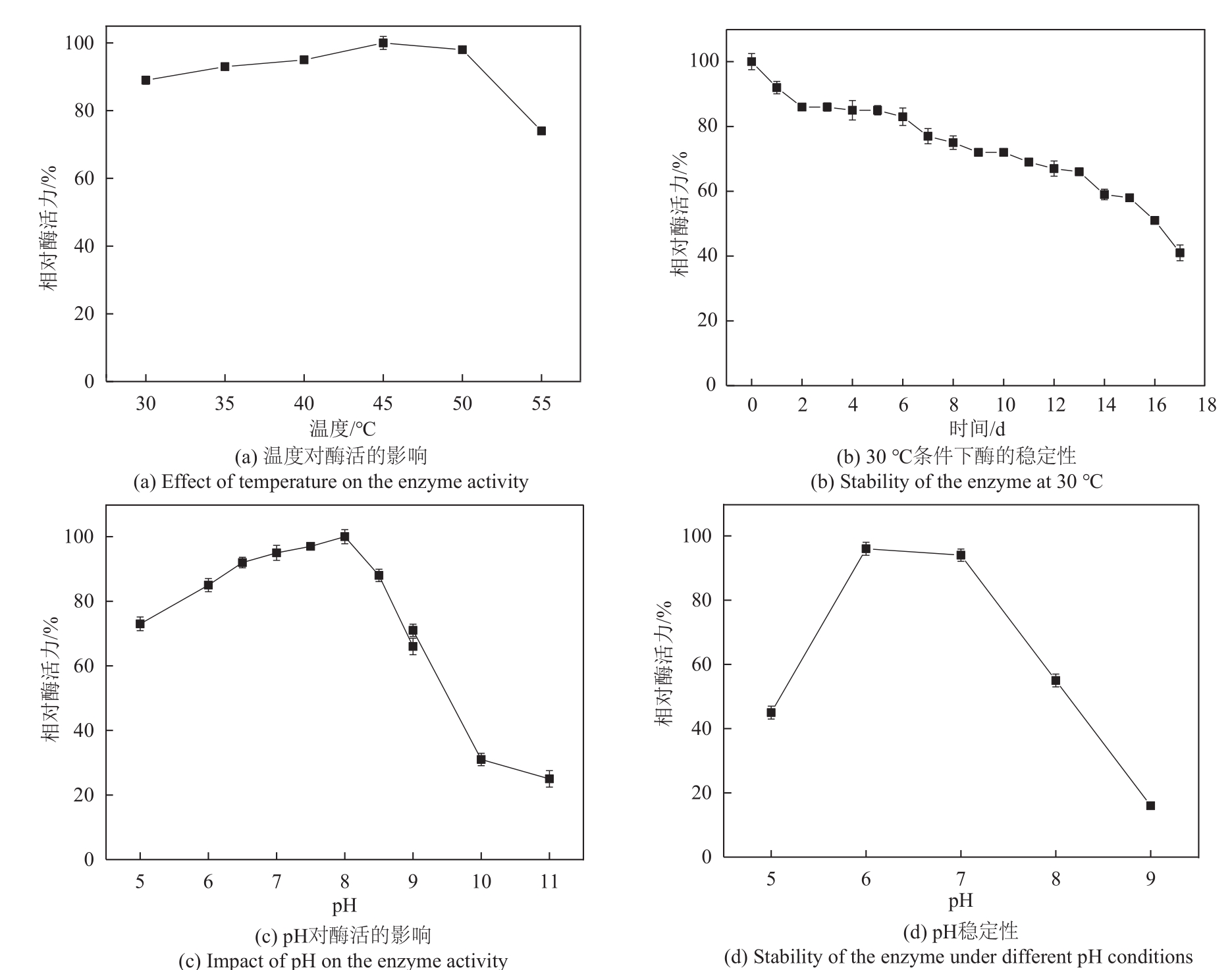
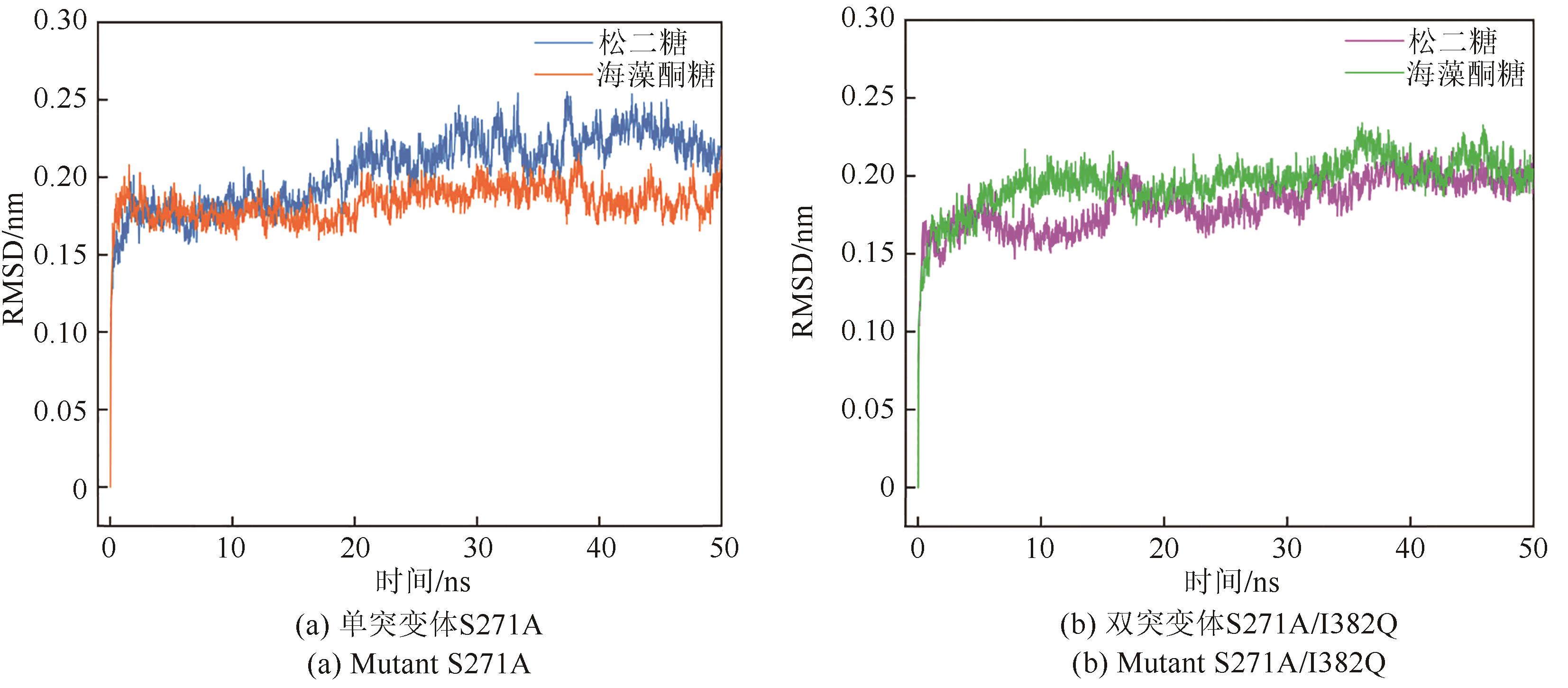
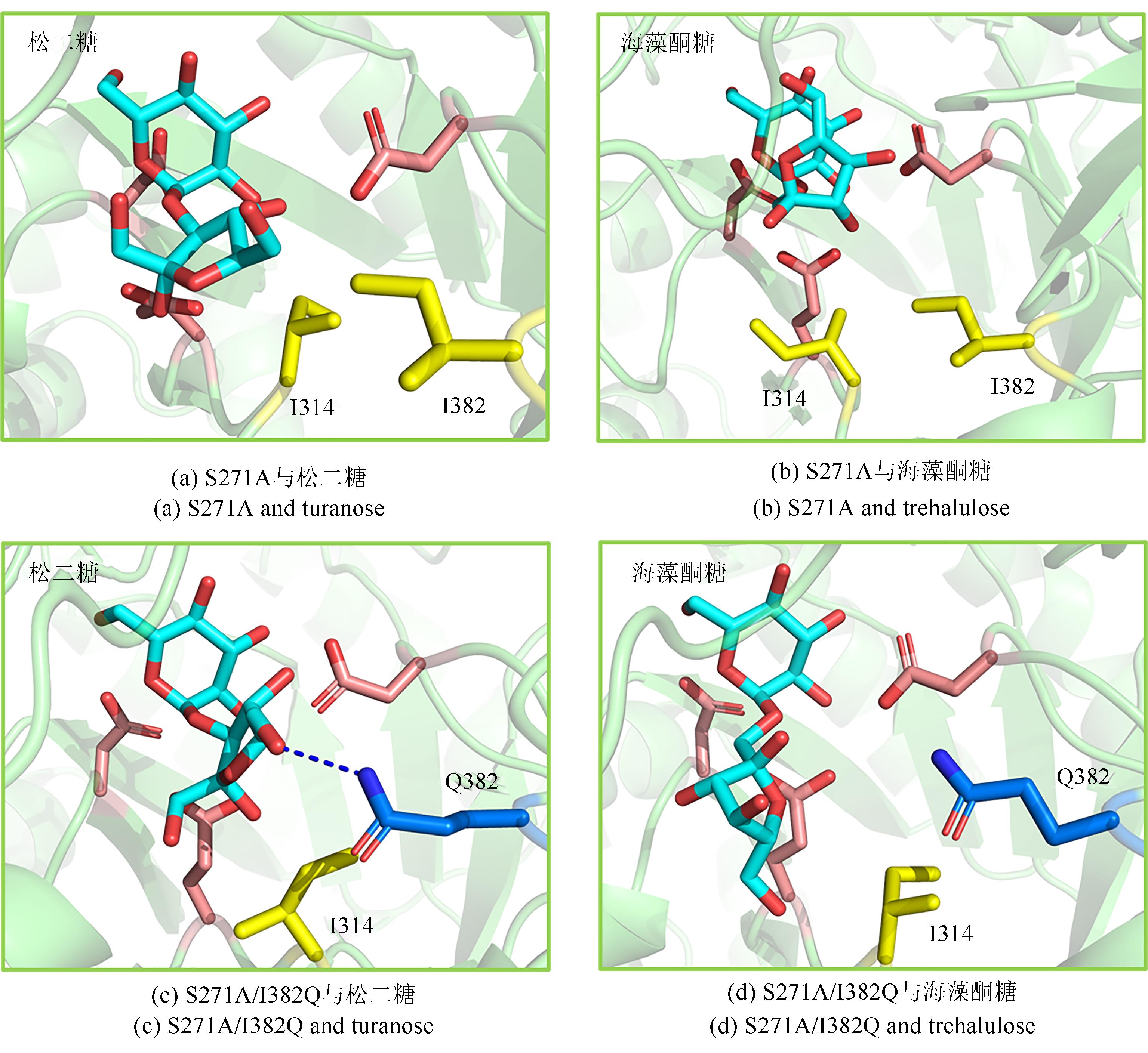
松二糖是由葡萄糖与果糖以α-1,3糖苷键连接而成的还原性二糖,具有代替蔗糖成为新型功能性甜味剂的潜力,在食品工业中应用前景广阔。淀粉蔗糖酶能够以蔗糖为底物催化异构(分子内转苷)反应制备松二糖,产率高但易产生副产物麦芽寡糖和海藻酮糖。为解决这一问题,选用前期获得的松二糖产率高并且不产副产物麦芽寡糖的Caulobactercrescentus蔗糖水解酶突变体S271A为研究对象,进一步通过受体亚位点分子改造,获得了反应特异性和松二糖产率提升的突变体S271A/I382Q。在此基础上进行了酶转化条件优化,当以2 mol/L蔗糖溶液为底物,加酶量为40 U/mL,在pH 5.0、30 ℃的条件下,松二糖的产率达到最高为70.3%,松二糖的浓度为480 g/L,并且反应产物中不含副产物海藻酮糖。分子动力学模拟表明,突变体S271A/I382Q可通过氢键相互作用稳定受体果糖参与形成α-1,3糖苷键时的构象,从而更有利于生成松二糖。本研究创新性地将蔗糖水解酶改造为键型特异性强的转苷酶,获得的松二糖产率为目前报道的最高水平,为松二糖的规模化制备与应用奠定了理论和技术基础。
中图分类号:
引用本文
王蕾, 邢晨晨, 郭志勇, 宿玲恰, 吴敬. Caulobactercrescentus蔗糖水解酶受体亚位点分子改造及其在松二糖制备中的应用[J]. 合成生物学, 2022, 3(3): 602-615.
WANG Lei, XING Chenchen, GUO Zhiyong, SU Lingqia, WU Jing. Molecular modification of acceptor subsite in sucrose hydrolase from Calobacter crescentus and its application in producing turanose[J]. Synthetic Biology Journal, 2022, 3(3): 602-615.
| 引物名称 | 引物序列(5′—3′) |
|---|---|
| S271A/I382G-F | TGTCATGATGATTTA |
| S271A/I382G-R | TGCTAATGCATTCCA |
| S271A/I382S-F | TGTCATGATGATTTA |
| S271A/I382S-R | TGCTAATGCATTCCA |
| S271A/I382T-F | TGTCATGATGATTTA |
| S271A/I382T-R | TGCTAATGCATTCCA |
| S271A/I382Q-F | TGTCATGATGATTTA |
| S271A/I382Q-R | TGCTAATGCATTCCA |
| S271A/I382K-F | TGTCATGATGATTTA |
| S271A/I382K-R | TGCTAATGCATTCCA |
| S271A/I382R-F | TGTCATGATGATTTA |
| S271A/I382R-R | TGCTAATGCATTCCA |
| S271A/V211I-F | CGCGAATTAATCGAT |
| S271A/V211I-R | GGTATCCGGAAA |
| S271A/P273A-F | CGCTTAGATGCCGCA |
| S271A/P273A-R | CTGTTTCCACAGAAA |
| S271A/P273S-F | CGCTTAGATGCCGCA |
| S271A/P273S-R | CTGTTTCCACAGAAA |
| S271A/I209R-F | GATCGCGAATTA |
| S271A/I209R-R | CGGAAACACATC |
表1 定点突变引物
Tab. 1 Primers used for the site-directed mutation
| 引物名称 | 引物序列(5′—3′) |
|---|---|
| S271A/I382G-F | TGTCATGATGATTTA |
| S271A/I382G-R | TGCTAATGCATTCCA |
| S271A/I382S-F | TGTCATGATGATTTA |
| S271A/I382S-R | TGCTAATGCATTCCA |
| S271A/I382T-F | TGTCATGATGATTTA |
| S271A/I382T-R | TGCTAATGCATTCCA |
| S271A/I382Q-F | TGTCATGATGATTTA |
| S271A/I382Q-R | TGCTAATGCATTCCA |
| S271A/I382K-F | TGTCATGATGATTTA |
| S271A/I382K-R | TGCTAATGCATTCCA |
| S271A/I382R-F | TGTCATGATGATTTA |
| S271A/I382R-R | TGCTAATGCATTCCA |
| S271A/V211I-F | CGCGAATTAATCGAT |
| S271A/V211I-R | GGTATCCGGAAA |
| S271A/P273A-F | CGCTTAGATGCCGCA |
| S271A/P273A-R | CTGTTTCCACAGAAA |
| S271A/P273S-F | CGCTTAGATGCCGCA |
| S271A/P273S-R | CTGTTTCCACAGAAA |
| S271A/I209R-F | GATCGCGAATTA |
| S271A/I209R-R | CGGAAACACATC |
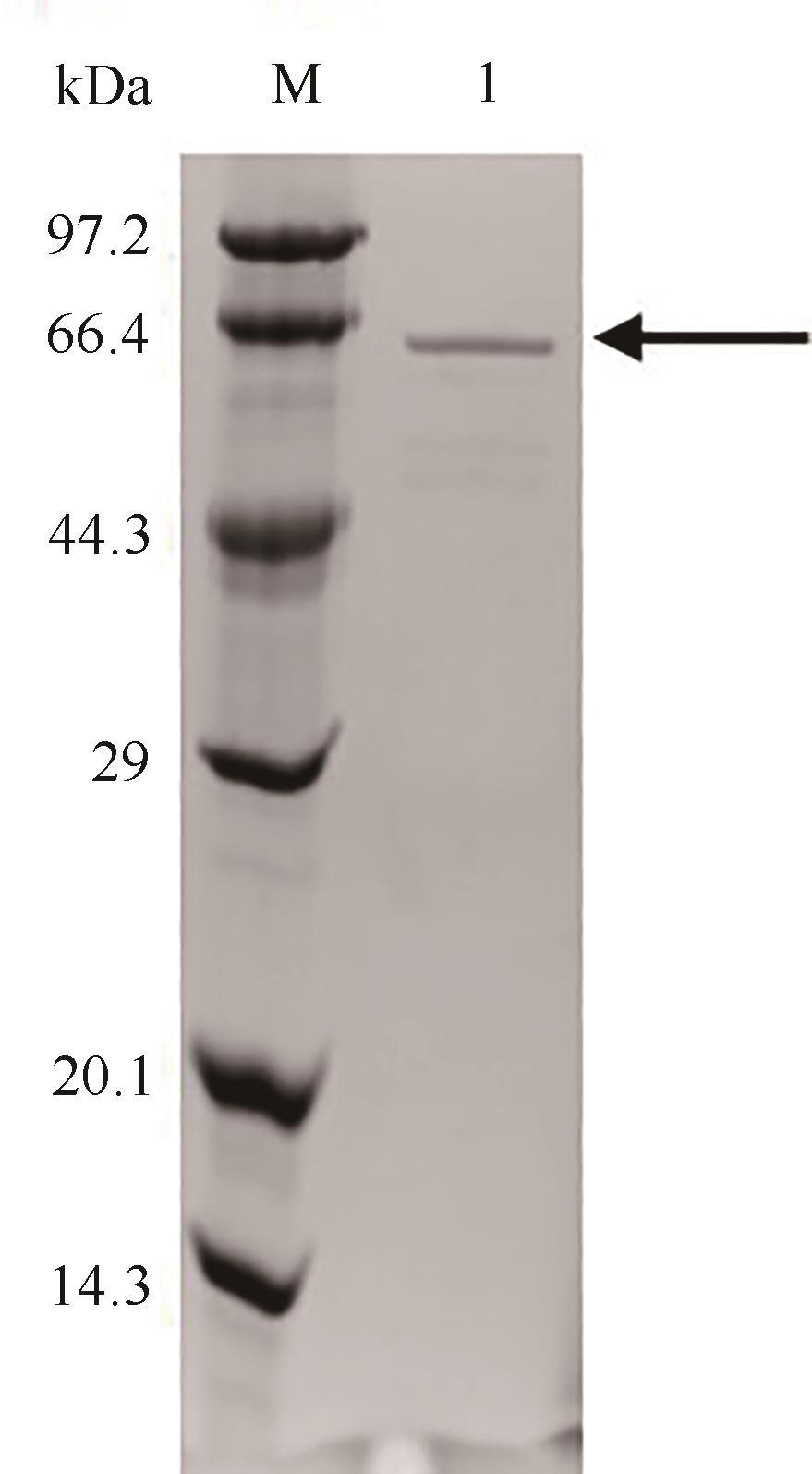
图5 纯化后突变体S271A/I382Q的SDS-PAGE分析M—低分子量蛋白Marker;1—S271A/I382Q
Fig. 5 SDS-PAGE analysis of the purified S271A/I382QM—Low molecular weight protein marker; 1—Purified S271A/I382Q
| 步骤 | 总酶活 /U | 总蛋白 含量/mg | 比活 /(U/mg) | 纯化 倍数 | 回收率 /% |
|---|---|---|---|---|---|
| 粗酶液 | 2918.5 | 653.0 | 4.5 | 1.0 | 100.0 |
| 镍柱纯化 | 573.3 | 49.8 | 11.5 | 2.6 | 20.0 |
表2 S271A/I382Q纯化过程参数
Tab. 2 Parameter for S271A/I382Q purification
| 步骤 | 总酶活 /U | 总蛋白 含量/mg | 比活 /(U/mg) | 纯化 倍数 | 回收率 /% |
|---|---|---|---|---|---|
| 粗酶液 | 2918.5 | 653.0 | 4.5 | 1.0 | 100.0 |
| 镍柱纯化 | 573.3 | 49.8 | 11.5 | 2.6 | 20.0 |
| 1 | DONER L W. The sugars of honey—a review[J]. Journal of the Science of Food and Agriculture, 1977, 28(5): 443-456. |
| 2 | COSTA LEITE J M DA, TRUGO L C, COSTA L S M, et al. Determination of oligosaccharides in Brazilian honeys of different botanical origin[J]. Food Chemistry, 2000, 70(1): 93-98. |
| 3 | PARK M O, LEE B H, LIM E, et al. Enzymatic process for high-yield turanose production and its potential property as an adipogenesis regulator[J]. Journal of Agricultural and Food Chemistry, 2016, 64(23): 4758-4764. |
| 4 | HAN D J, LEE B H, YOO S H. Physicochemical properties of turanose and its potential applications as a sucrose substitute[J]. Food Science and Biotechnology, 2021, 30(3): 433-441. |
| 5 | TIAN Y Q, DENG Y, ZHANG W L, et al. Sucrose isomers as alternative sweeteners: properties, production, and applications[J]. Applied Microbiology and Biotechnology, 2019, 103(21/22): 8677-8687. |
| 6 | SHIBUYA T, MANDAI T, KUBOTA M, et al. Production of turanose by cyclomaltodextrin glucanotransferase from Bacillus stearothermophilus [J]. Journal of Applied Glycoscience, 2004, 51(3): 223-227. |
| 7 | TIAN Y Q, XU W, ZHANG W L, et al. Amylosucrase as a transglucosylation tool: from molecular features to bioengineering applications[J]. Biotechnology Advances, 2018, 36(5): 1540-1552. |
| 8 | GUÉRIN F, BARBE S, PIZZUT-SERIN S, et al. Structural investigation of the thermostability and product specificity of amylosucrase from the bacterium Deinococcus geothermalis [J]. Journal of Biological Chemistry, 2012, 287(9): 6642-6654. |
| 9 | WANG R, BAE J S, KIM J H, et al. Development of an efficient bioprocess for turanose production by sucrose isomerisation reaction of amylosucrase[J]. Food Chemistry, 2012, 132(2): 773-779. |
| 10 | SU L Q, ZHAO Y Q, WU D, et al. Heterogeneous expression, molecular modification of amylosucrase from Neisseria polysaccharea, and its application in the preparation of turanose[J]. Food Chemistry, 2020, 314: 126212. |
| 11 | 赵雅琪. 松二糖的酶法制备研究[D]. 无锡: 江南大学, 2019. |
| ZHAO Y Q. Enzymatic synthesis of turanose[D]. Wuxi: Jiangnan University, 2019. | |
| 12 | SEO D H, YOO S H, CHOI S J, et al. Versatile biotechnological applications of amylosucrase, a novel glucosyltransferase[J]. Food Science and Biotechnology, 2019, 29(1): 1-16. |
| 13 | POTOCKI DE MONTALK G, REMAUD-SIMEON M, WILLEMOT R M, et al. Amylosucrase from Neisseria polysaccharea: novel catalytic properties[J]. FEBS Letters, 2000, 471(2/3): 219-223. |
| 14 | ALBENNE C, SKOV L K, MIRZA O, et al. Molecular basis of the amylose-like polymer formation catalyzed by Neisseria polysaccharea amylosucrase[J]. Journal of Biological Chemistry, 2004, 279(1): 726-734. |
| 15 | STAM M R, DANCHIN E G J, RANCUREL C, et al. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins[J]. Protein Engineering, Design & Selection, 2006, 19(12): 555-562. |
| 16 | LOMBARD V, GOLACONDA RAMULU H, DRULA E, et al. The carbohydrate-active enzymes database (CAZy) in 2013[J]. Nucleic Acids Research, 2013, 42(D1): D490-D495. |
| 17 | KIM M I, KIM H S, JUNG J, et al. Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold[J]. Journal of Molecular Biology, 2008, 380(4): 636-647. |
| 18 | 邢晨晨, 王蕾, 郭志勇, 等. Caulobacter crescentus蔗糖水解酶突变体S271A的重组表达及其转化蔗糖制备松二糖的研究[J]. 食品与发酵工业, 2022, 48(2): 20-25. |
| XING C C, WANG L, GUO Z Y, et al. Recombinant expression of Caulobacter crescentus sucrose hydrolase mutant S271A and its conversion of sucrose to turanose[J]. Food and Fermentation Industries, 2022, 48(2): 20-25. | |
| 19 | ARNOLD K, BORDOLI L, KOPP J, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling[J]. Bioinformatics, 2006, 22(2): 195-201. |
| 20 | WANG Z, SUN H Y, YAO X J, et al. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power[J]. Physical Chemistry Chemical Physics, 2016, 18(18): 12964-12975. |
| 21 | OLSSON M H M, SØNDERGAARD C R, ROSTKOWSKI M, et al. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions[J]. Journal of Chemical Theory and Computation, 2011, 7(2): 525-537. |
| 22 | MAIER J A, MARTINEZ C, KASAVAJHALA K, et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB[J]. Journal of Chemical Theory and Computation, 2015, 11(8): 3696-3713. |
| 23 | KIRSCHNER K N, YONGYE A B, TSCHAMPEL S M, et al. GLYCAM06: a generalizable biomolecular force field. carbohydrates[J]. Journal of Computational Chemistry, 2008, 29(4): 622-655. |
| 24 | JORGENSEN W L, CHANDRASEKHAR J, MADURA J D, et al. Comparison of simple potential functions for simulating liquid water[J]. The Journal of Chemical Physics, 1983, 79(2): 926-935. |
| 25 | RYCKAERT J P, CICCOTTI G, BERENDSEN H J C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes[J]. Journal of Computational Physics, 1977, 23(3): 327-341. |
| 26 | DARDEN T, YORK D, PEDERSEN L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems[J]. The Journal of Chemical Physics, 1993, 98(12): 10089-10092. |
| 27 | HUMPHREY W, DALKE A, SCHULTEN K. VMD: Visual molecular dynamics[J]. Journal of Molecular Graphics, 1996, 14(1): 33-38. |
| 28 | ROE D R, CHEATHAM T E 3rd. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data[J]. Journal of Chemical Theory and Computation, 2013, 9(7): 3084-3095. |
| 29 | BENKERT P, BIASINI M, SCHWEDE T. Toward the estimation of the absolute quality of individual protein structure models[J]. Bioinformatics, 2011, 27(3): 343-350. |
| 30 | BIASINI M, BIENERT S, WATERHOUSE A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information[J]. Nucleic Acids Research, 2014, 42(W1): W252-W258. |
| 31 | WATERHOUSE A, BERTONI M, BIENERT S, et al. SWISS-MODEL: homology modelling of protein structures and complexes[J]. Nucleic Acids Research, 2018, 46(W1): W296-W303. |
| 32 | SKOV L K, MIRZA O, SPROGØE D, et al. Oligosaccharide and sucrose complexes of amylosucrase: structural implications for the polymerase activity[J]. Journal of Biological Chemistry, 2002, 277(49): 47741-47747. |
| 33 | NIELSEN J E, BORCHERT T V, VRIEND G. The determinants of α-amylase pH-activity profiles[J]. Protein Engineering, Design and Selection, 2001, 14(7): 505-512. |
| 34 | NIELSEN J E, VRIEND G. Optimizing the hydrogen-bond network in Poisson-Boltzmann equation-based pKa calculations[J]. Proteins, 2001, 43(4): 403-412. |
| [1] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, (): 1-19. |
| [2] | 董玲玲, 李斐煊, 雷航彬, 宋启迪, 王世珍. 仿生分区室固定化多酶体系[J]. 合成生物学, 2024, 5(6): 1518-1529. |
| [3] | 雷航彬, 何宁, 李斐煊, 董玲玲, 王世珍. 氢化酶固定化研究进展[J]. 合成生物学, 2024, 5(6): 1485-1497. |
| [4] | 王子渊, 杨立荣, 吴坚平, 郑文隆. 酶促合成手性氨基酸的研究进展[J]. 合成生物学, 2024, 5(6): 1319-1349. |
| [5] | 张阿磊, 魏国光, 张弛, 陈磊, 周奚, 刘伟, 陈可泉. 几丁质资源生物降解和高值转化的研究进展[J]. 合成生物学, 2024, 5(6): 1279-1299. |
| [6] | 刘晚秋, 季向阳, 许慧玲, 卢屹聪, 李健. 限制性内切酶的无细胞快速制备研究[J]. 合成生物学, 2023, 4(4): 840-851. |
| [7] | 曾涛, 巫瑞波. 数据驱动的酶反应预测与设计[J]. 合成生物学, 2023, 4(3): 535-550. |
| [8] | 康里奇, 谈攀, 洪亮. 人工智能时代下的酶工程[J]. 合成生物学, 2023, 4(3): 524-534. |
| [9] | 赖铭元, 韦健, 许建和, 郁惠蕾. 非特异性过加氧酶(UPO)的研究综述[J]. 合成生物学, 2022, 3(6): 1235-1249. |
| [10] | 王盼盼, 于洪巍. 酶催化在维生素及其衍生物制备中的应用[J]. 合成生物学, 2022, 3(3): 500-515. |
| [11] | 汤恒, 韩鑫, 邹树平, 郑裕国. 多酶催化体系在医药化学品合成中的应用[J]. 合成生物学, 2021, 2(4): 559-576. |
| [12] | 吴淑可, 周颐, 王文, 张巍, 高鹏飞, 李智. 从单酶催化到多酶级联催化——从王义翘教授在酶技术领域的贡献说开去[J]. 合成生物学, 2021, 2(4): 543-558. |
| [13] | 袁飞燕, 于洋, 李春. 基于非天然结构组件的人工酶设计与应用[J]. 合成生物学, 2020, 1(6): 685-696. |
| [14] | 史然, 江正强. 2'-岩藻糖基乳糖的酶法合成研究进展和展望[J]. 合成生物学, 2020, 1(4): 481-494. |
| [15] | 马田, 邓子新, 刘天罡. 维生素E的“前世”和“今生”[J]. 合成生物学, 2020, 1(2): 174-186. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||