合成生物学 ›› 2023, Vol. 4 ›› Issue (3): 551-570.DOI: 10.12211/2096-8280.2023-006
基于靶标结构的环肽分子计算设计
王凡灏2, 来鲁华1,2,3, 张长胜1,2
- 1.北京大学前沿交叉学科研究院定量生物学中心,北京 100871
2.北京大学化学与分子工程学院,北京分子科学国家研究中心,北京 100871
3.北京大学-清华大学生命科学联合中心,北京 100871
-
收稿日期:2023-01-12修回日期:2023-02-23出版日期:2023-06-30发布日期:2023-07-05 -
通讯作者:张长胜 -
作者简介:王凡灏 (1998—),男,博士研究生。研究方向为基于靶标结构的多肽药物分子设计。 E-mail:wangfh2020@stu.pku.edu.cn张长胜 (1981—),男,副研究员。研究方向为蛋白质设计、计算结构生物学。 E-mail:changshengzhang@pku.edu.cn -
基金资助:国家自然科学基金(21977007)
Target structure based computational design of cyclic peptides
WANG Fanhao2, LAI Luhua1,2,3, ZHANG Changsheng1,2
- 1.Center for Quantitative Biology,Academy for Advanced Interdisciplinary Studies,Peking University,Beijing 100871,China
2.BNLMS,College of Chemistry and Molecular Engineering,Peking University,Beijing 100871,China
3.Peking -Tsinghua Center for Life Science,Peking University,Beijing 100871,China
-
Received:2023-01-12Revised:2023-02-23Online:2023-06-30Published:2023-07-05 -
Contact:ZHANG Changsheng
摘要:
环肽在调控蛋白质-蛋白质相互作用方面具有独特的优势,在新药研发领域受到了越来越多的关注。蛋白质相互作用界面一般较大而平坦,相较于小分子化合物,环肽分子更容易获得与这些靶标位点结合的高亲和力和高特异性。相较于线性多肽或蛋白质,环肽结构一般具有更大的骨架刚性,更难被酶降解,从而在代谢上更稳定,而且环肽更易于通过修饰改造增加跨膜活性,从而结合细胞内的靶标蛋白。结构数据和结构建模方法是开发基于靶标结构计算设计环肽药物的基础。本文分析了蛋白质结构数据库中环肽与靶标蛋白结合情况,介绍了目前环肽构象生成或结构预测的四类主要算法;总结了基于靶标结构计算设计环肽分子的主要方法,包括基于分子对接的虚拟筛选方法、借助于动力学模拟的设计方法、从头生成的设计方法以及具有跨膜活性的环肽设计方法;并展望了数据驱动的机器学习方法在环肽设计领域中的可能应用以及未来环肽药物分子开发的可能方向。
中图分类号:
引用本文
王凡灏, 来鲁华, 张长胜. 基于靶标结构的环肽分子计算设计[J]. 合成生物学, 2023, 4(3): 551-570.
WANG Fanhao, LAI Luhua, ZHANG Changsheng. Target structure based computational design of cyclic peptides[J]. Synthetic Biology Journal, 2023, 4(3): 551-570.
| 名称 | 结构 | 功能简介 |
|---|---|---|
| 罗米地辛(Romidepsin) | 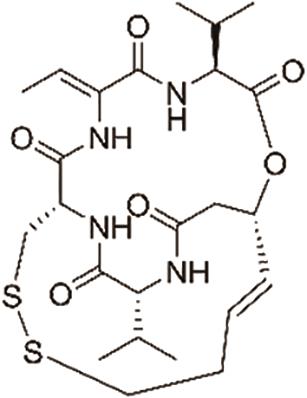 | 靶向组蛋白去乙酰化酶,具有抗癌活性,用于治疗T细胞淋巴瘤 |
伏环孢素 (Voclosporin) | 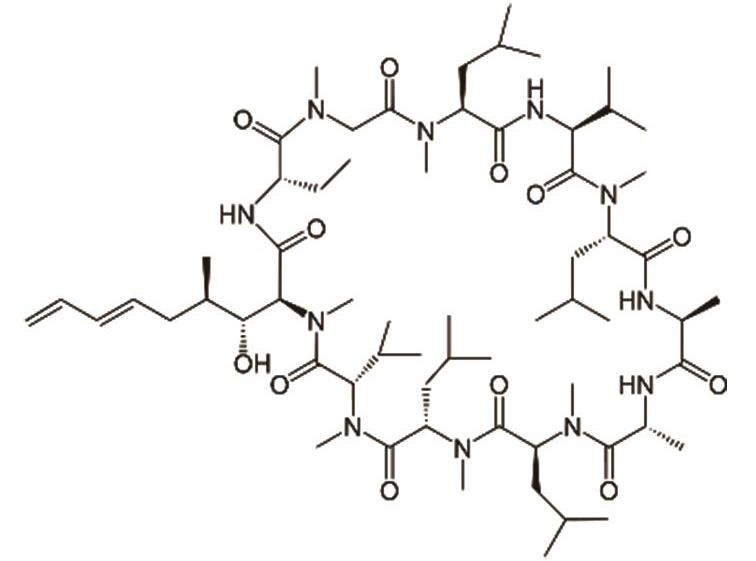 | 靶向钙调神经磷酸酶,用于治疗狼疮性肾炎 |
齐考诺肽 (Ziconotide) | 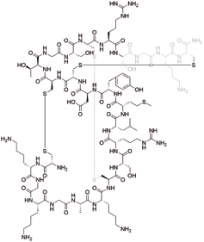 | 靶向G蛋白偶联受体,N型钙通道的有效和选择性阻滞剂 |
利那洛肽 (Linaclotide) | 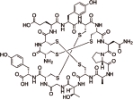 | 鸟苷酸环化酶 C(GC-C) 激动剂 |
普卡那肽 (Plecanatide) | 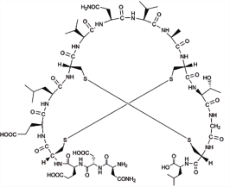 | 鸟苷酸环化酶-C(GC-C)激动剂 |
帕西瑞肽 (Pasireotide) | 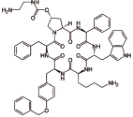 | 生长抑素类似物,靶向生长抑素受体,可以提高生长抑素受体的激动剂活性 |
兰瑞肽 (Lanreotide) | 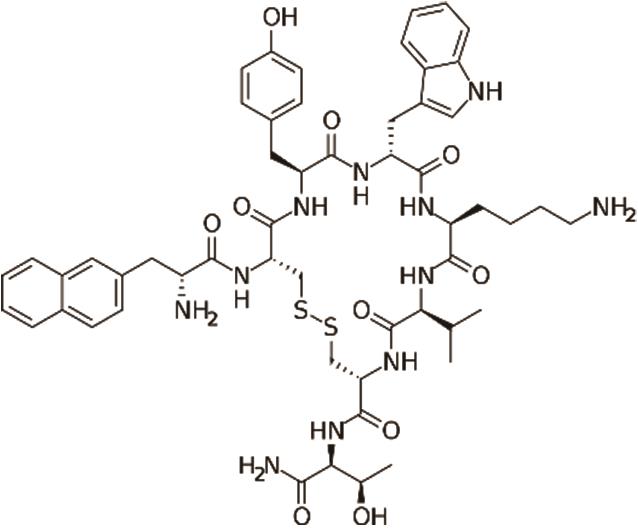 | 生长素抑制剂的第二代类似物,靶向生长抑素受体,活性更高 |
加压素 (Vasopressin) | 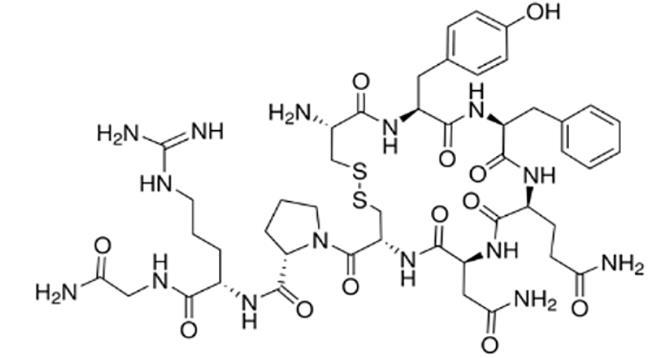 | 一种抗利尿激素,靶向血管加压素受体 |
特利加压素 (Terlipressin) | 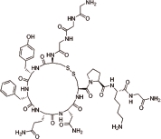 | 一种人工合成的长效抗利尿激素,靶向血管加压素受体 |
布美兰肽 (Bremelanotide) | 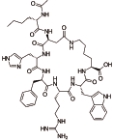 | 黑素皮质素受体的激动剂,主要作用于MC3和MC4受体 |
赛美拉肽 (Setmelanotide) | 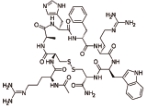 | 黑素皮质素受体的激动剂,主要作用于MC4受体 |
达托霉素 (Daptomycin) | 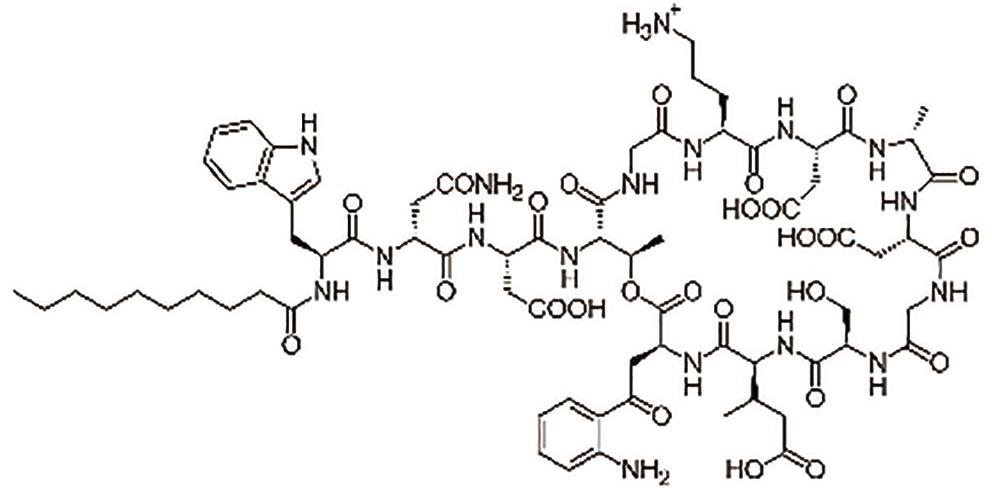 | 一种环状脂肽抗生素,破坏革兰氏阳性菌的细胞壁 |
特拉万星 (Telavancin) | 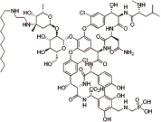 | 万古霉素的半合成衍生物,抑制革兰氏阳性菌的细胞壁合成 |
达巴万星 (Dalbavancin) | 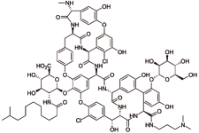 | 第二代脂糖肽抗生素药物,比万古霉素更有效,也针对细胞壁的生物合成 |
奥利万星 (Oritavancin) | 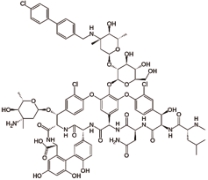 | 由氯霉素衍生的万古霉素的合成类似物,抑制细菌细胞壁的合成 |
卡泊芬净 (Caspofungin) | 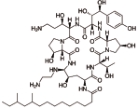 | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
米卡芬净 (Micafungin) |  | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
阿尼芬净 (Anidulafungin) |  | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
表1 已获FDA批准的18种环肽类药物汇总表
Table 1 Summary of 18 cyclic peptide drugs approved by FDA
| 名称 | 结构 | 功能简介 |
|---|---|---|
| 罗米地辛(Romidepsin) |  | 靶向组蛋白去乙酰化酶,具有抗癌活性,用于治疗T细胞淋巴瘤 |
伏环孢素 (Voclosporin) |  | 靶向钙调神经磷酸酶,用于治疗狼疮性肾炎 |
齐考诺肽 (Ziconotide) |  | 靶向G蛋白偶联受体,N型钙通道的有效和选择性阻滞剂 |
利那洛肽 (Linaclotide) |  | 鸟苷酸环化酶 C(GC-C) 激动剂 |
普卡那肽 (Plecanatide) |  | 鸟苷酸环化酶-C(GC-C)激动剂 |
帕西瑞肽 (Pasireotide) |  | 生长抑素类似物,靶向生长抑素受体,可以提高生长抑素受体的激动剂活性 |
兰瑞肽 (Lanreotide) |  | 生长素抑制剂的第二代类似物,靶向生长抑素受体,活性更高 |
加压素 (Vasopressin) |  | 一种抗利尿激素,靶向血管加压素受体 |
特利加压素 (Terlipressin) |  | 一种人工合成的长效抗利尿激素,靶向血管加压素受体 |
布美兰肽 (Bremelanotide) |  | 黑素皮质素受体的激动剂,主要作用于MC3和MC4受体 |
赛美拉肽 (Setmelanotide) |  | 黑素皮质素受体的激动剂,主要作用于MC4受体 |
达托霉素 (Daptomycin) |  | 一种环状脂肽抗生素,破坏革兰氏阳性菌的细胞壁 |
特拉万星 (Telavancin) |  | 万古霉素的半合成衍生物,抑制革兰氏阳性菌的细胞壁合成 |
达巴万星 (Dalbavancin) |  | 第二代脂糖肽抗生素药物,比万古霉素更有效,也针对细胞壁的生物合成 |
奥利万星 (Oritavancin) |  | 由氯霉素衍生的万古霉素的合成类似物,抑制细菌细胞壁的合成 |
卡泊芬净 (Caspofungin) |  | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
米卡芬净 (Micafungin) |  | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
阿尼芬净 (Anidulafungin) |  | 靶向1,3-β-葡聚糖合成酶,对侵袭性念珠菌病和其他形式的系统性真菌病有临床疗效 |
| PDB编号 | 环肽配体序列 | 肽链长度 | 环的大小 | 环肽功能 | 靶蛋白界面 埋藏面积/Å2 | 环化类型 |
|---|---|---|---|---|---|---|
| 1e4w | 8 | 8 | TGF-α抗原表位类似物,与 | 381 | 酰胺键 | |
| 1sfi | 14 | 14 | 胰蛋白酶抑制剂 | 612 | 酰胺键 | |
| 2axi | 10 | 10 | P53-HDM2配体 | 510 | 酰胺键 | |
| 3av9 | 8 | 8 | HIV整合酶重组抑制剂 | 397 | 酰胺键 | |
| 3ava | 8 | 8 | HIV整合酶重组抑制剂 | 395 | 酰胺键 | |
| 3avb | 8 | 8 | HIV整合酶重组抑制剂 | 401 | 酰胺键 | |
| 3avf | 8 | 8 | HIV整合酶重组抑制剂 | 418 | 酰胺键 | |
| 3avg | 8 | 8 | HIV整合酶重组抑制剂 | 385 | 酰胺键 | |
| 3avh | 8 | 8 | HIV整合酶重组抑制剂 | 388 | 酰胺键 | |
| 3avi | 8 | 8 | HIV整合酶重组抑制剂 | 432 | 酰胺键 | |
| 3avj | 8 | 8 | HIV整合酶重组抑制剂 | 437 | 酰胺键 | |
| 3avk | 8 | 8 | HIV整合酶重组抑制剂 | 419 | 酰胺键 | |
| 3avm | 8 | 8 | HIV整合酶重组抑制剂 | 402 | 酰胺键 | |
| 3avn | 8 | 8 | HIV整合酶重组抑制剂 | 394 | 酰胺键 | |
| 3wne | 6 | 6 | HIV整合酶天然配体 | 378 | 酰胺键 | |
| 3wng | 6 | 6 | HIV-1整合酶抑制剂 | 360 | 酰胺键 | |
| 3wnh | 6 | 6 | HIV-1整合酶抑制剂 | 367 | 酰胺键 | |
| 3zgc | 7 | 7 | 红细胞核因子配体 | 343 | 酰胺键 | |
| 4k1e | 14 | 14 | 胰蛋白酶抑制剂 | 605 | 酰胺键 | |
| 4kel | 14 | 14 | 胰蛋白酶抑制剂 | 609 | 酰胺键 | |
| 4y1d | 6 | 6 | HIV-1整合酶抑制剂 | 412 | 酰胺键 | |
| 5n99 | 5 | 5 | 链霉亲和素配体 | 371 | 酰胺键 | |
| 5xn3 | 8 | 8 | SPSB2-iNOS相互作用抑制剂 | 270 | 酰胺键 | |
| 6jwm | 7 | 7 | SPSB2-iNOS相互作用抑制剂 | 281 | 酰胺键 | |
| 6jwn | 9 | 9 | SPSB2-iNOS相互作用抑制剂 | 294 | 酰胺键 | |
| 7k2e | 7 | 7 | 人源KEAP1蛋白抑制剂 | 360 | 酰胺键 | |
| 7k2g | 7 | 7 | 人源KEAP1蛋白抑制剂 | 403 | 酰胺键 | |
| 7k2h | 7 | 7 | 人源KEAP1蛋白抑制剂 | 343 | 酰胺键 | |
| 7k2m | 7 | 7 | 人源KEAP1蛋白抑制剂 | 320 | 酰胺键 | |
| 6xbe | 8 | 8 | NDM-1金属-β-内酰胺酶抑制剂 | 379 | 酰胺键 | |
| 6xbf | 8 | 8 | NDM-2金属-β-内酰胺酶抑制剂 | 452 | 酰胺键 | |
| 1hqq | RC | 13 | 11 | 链霉亲和素配体 | 413 | 二硫键 |
| 1jbu | EEWEVL | 15 | 7 | 凝血因子Ⅶ抑制剂 | 908 | 二硫键 |
| 1smf | 9 | 9 | 胰蛋白酶抑制剂 | 414 | 二硫键 | |
| 1sle | 8 | 8 | 链霉亲和素配体 | 324 | 二硫键 | |
| 1vpp | RGWVEI | 18 | 9 | 生长素抑制剂 | 556 | 二硫键 |
| 1vwb | 6 | 6 | 链霉亲和素配体 | 306 | 二硫键 | |
| 2ck0 | 11 | 10 | 血管紧张素Ⅱ抗体结合肽 | 504 | 二硫键 | |
| 2nwn | 12 | 12 | 丝氨酸蛋白酶抑制剂 | 597 | 二硫键 | |
| 3g5v | 16 | 16 | EGFR肽段 | 603 | 二硫键 | |
| 3p72 | 11 | 11 | 血小板糖蛋白1b抑制剂 | 521 | 二硫键 | |
| 3wnf | 6 | 6 | HIV-1整合酶抑制剂 | 432 | 二硫键 | |
| 4ib5 | G | 13 | 11 | CK2beta拮抗剂 | 456 | 二硫键 |
| 4m1d | 14 | 14 | HIV-1 gp120蛋白V3域类似物 | 588 | 二硫键 | |
| 4ou3 | 6 | 5 | 猪氨肽酶N抑制剂 | 366 | 二硫键 | |
| 5djc | D | 13 | 11 | 抗体结合肽 | 610 | 二硫键 |
| 5co5 | G | 16 | 15 | 芋螺毒素 | 692 | 二硫键 |
| 5eoc | 13 | 13 | 丙型肝炎病毒E2表位Ⅰ类似物 | 468 | 二硫键 | |
| 5h5q | 13 | 11 | 人源GPX4抑制剂 | 505 | 二硫键 | |
| 5th2 | 12 | 12 | L5Q中间位变体类似物 | 709 | 二硫键 | |
| 5vb9 | 15 | 14 | IL-17抗体抑制剂 | 571 | 二硫键 | |
| 5wxr | GA | 14 | 12 | 尿激酶型纤溶酶原激活物抑制剂 | 526 | 二硫键 |
| 5xco | RRRR | 19 | 11 | K-Ras(G12D突变体)抑制剂 | 650 | 二硫键 |
| 6e5m | 9 | 9 | β-胰蛋白酶抑制剂 | 434 | 二硫键 | |
| 1bm2 | 6 | 6 | GBR2-sh2结构域高活性配体 | 336 | 其他环化 | |
| 1bzh | 7 | 7 | 酪氨酸磷酸酶抑制剂 | 398 | 其他环化 | |
| 1vwl | 9 | 8 | 链霉亲和素配体 | 300 | 其他环化 | |
| 4zjx | 8 | 7 | 肉毒杆菌神经毒素(血清型A)抑制剂 | 604 | 其他环化 | |
| 5nes | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 259 | 其他环化 | |
| 5ney | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 271 | 其他环化 | |
| 5nf0 | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 283 | 其他环化 | |
| 6b67 | 7 | 7 | PPM1A活性调节剂 | 528 | 其他环化 | |
| 6dn6 | 5 | 5 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 234 | 其他环化 | |
| 6dn7 | 7 | 7 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 254 | 其他环化 | |
| 6dn8 | 8 | 8 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 258 | 其他环化 | |
| 6nnv | 14 | 13 | PD-L1抑制剂 | 510 | 其他环化 | |
| 6u4a | 11 | 11 | BRD2-BD1抑制剂 | 587 | 其他环化 | |
| 6u74 | 14 | 14 | BRD2-BD1抑制剂 | 582 | 其他环化 | |
| 6u8m | 17 | 17 | BRD2-BD1抑制剂 | 504 | 其他环化 | |
| 6wgn | 15 | 14 | K-Ras(G12D突变体)抑制剂 | 641 | 其他环化 | |
| 6xci | 8 | 8 | NDM-3金属-β-内酰胺酶抑制剂 | 457 | 其他环化 | |
| 6xib | 12 | 11 | PCSK9抑制剂 | 546 | 其他环化 | |
| 6xic | 11 | 11 | PCSK9抑制剂 | 518 | 其他环化 | |
| 6xid | 12 | 11 | PCSK9抑制剂 | 537 | 其他环化 | |
| 6xie | 11 | 11 | PCSK9抑制剂 | 496 | 其他环化 | |
| 6xif | 11 | 11 | PCSK9抑制剂 | 528 | 其他环化 | |
| 6xs5 | 17 | 17 | 人源Vps29抑制剂,结构稳定剂 | 529 | 其他环化 | |
| 6xs7 | 17 | 17 | 人源Vps30抑制剂,结构稳定剂 | 654 | 其他环化 | |
| 6xs8 | 13 | 13 | 人源Vps31抑制剂,结构稳定剂 | 380 | 其他环化 | |
| 6xsa | 15 | 15 | 人源Vps32抑制剂,结构稳定剂 | 623 | 其他环化 | |
| 6yw1 | 14 | 14 | 促进HIF脯氨酰羟化酶2结晶的环肽配体 | 671 | 其他环化 | |
| 7bph | 14 | 13 | GNAS抑制剂 | 520 | 其他环化 | |
| 7k2k | 7 | 7 | 人源KEAP1蛋白抑制剂 | 349 | 其他环化 | |
| 7k2l | 7 | 7 | 人源KEAP1蛋白抑制剂 | 330 | 其他环化 | |
| 7k2o | 7 | 7 | 人源KEAP1蛋白抑制剂 | 346 | 其他环化 | |
| 7k2p | 7 | 7 | 人源KEAP1蛋白抑制剂 | 344 | 其他环化 | |
| 7k2r | 7 | 7 | 人源KEAP1蛋白抑制剂 | 350 | 其他环化 | |
| 7rov | 14 | 12 | K-Ras(G12D突变体)抑制剂 | 564 | 其他环化 |
表2 PDB数据库中的环肽-靶标蛋白质复合物结构数据表
Table 2 Non-redundant cyclic peptide-target protein complex structures in the PDB database
| PDB编号 | 环肽配体序列 | 肽链长度 | 环的大小 | 环肽功能 | 靶蛋白界面 埋藏面积/Å2 | 环化类型 |
|---|---|---|---|---|---|---|
| 1e4w | 8 | 8 | TGF-α抗原表位类似物,与 | 381 | 酰胺键 | |
| 1sfi | 14 | 14 | 胰蛋白酶抑制剂 | 612 | 酰胺键 | |
| 2axi | 10 | 10 | P53-HDM2配体 | 510 | 酰胺键 | |
| 3av9 | 8 | 8 | HIV整合酶重组抑制剂 | 397 | 酰胺键 | |
| 3ava | 8 | 8 | HIV整合酶重组抑制剂 | 395 | 酰胺键 | |
| 3avb | 8 | 8 | HIV整合酶重组抑制剂 | 401 | 酰胺键 | |
| 3avf | 8 | 8 | HIV整合酶重组抑制剂 | 418 | 酰胺键 | |
| 3avg | 8 | 8 | HIV整合酶重组抑制剂 | 385 | 酰胺键 | |
| 3avh | 8 | 8 | HIV整合酶重组抑制剂 | 388 | 酰胺键 | |
| 3avi | 8 | 8 | HIV整合酶重组抑制剂 | 432 | 酰胺键 | |
| 3avj | 8 | 8 | HIV整合酶重组抑制剂 | 437 | 酰胺键 | |
| 3avk | 8 | 8 | HIV整合酶重组抑制剂 | 419 | 酰胺键 | |
| 3avm | 8 | 8 | HIV整合酶重组抑制剂 | 402 | 酰胺键 | |
| 3avn | 8 | 8 | HIV整合酶重组抑制剂 | 394 | 酰胺键 | |
| 3wne | 6 | 6 | HIV整合酶天然配体 | 378 | 酰胺键 | |
| 3wng | 6 | 6 | HIV-1整合酶抑制剂 | 360 | 酰胺键 | |
| 3wnh | 6 | 6 | HIV-1整合酶抑制剂 | 367 | 酰胺键 | |
| 3zgc | 7 | 7 | 红细胞核因子配体 | 343 | 酰胺键 | |
| 4k1e | 14 | 14 | 胰蛋白酶抑制剂 | 605 | 酰胺键 | |
| 4kel | 14 | 14 | 胰蛋白酶抑制剂 | 609 | 酰胺键 | |
| 4y1d | 6 | 6 | HIV-1整合酶抑制剂 | 412 | 酰胺键 | |
| 5n99 | 5 | 5 | 链霉亲和素配体 | 371 | 酰胺键 | |
| 5xn3 | 8 | 8 | SPSB2-iNOS相互作用抑制剂 | 270 | 酰胺键 | |
| 6jwm | 7 | 7 | SPSB2-iNOS相互作用抑制剂 | 281 | 酰胺键 | |
| 6jwn | 9 | 9 | SPSB2-iNOS相互作用抑制剂 | 294 | 酰胺键 | |
| 7k2e | 7 | 7 | 人源KEAP1蛋白抑制剂 | 360 | 酰胺键 | |
| 7k2g | 7 | 7 | 人源KEAP1蛋白抑制剂 | 403 | 酰胺键 | |
| 7k2h | 7 | 7 | 人源KEAP1蛋白抑制剂 | 343 | 酰胺键 | |
| 7k2m | 7 | 7 | 人源KEAP1蛋白抑制剂 | 320 | 酰胺键 | |
| 6xbe | 8 | 8 | NDM-1金属-β-内酰胺酶抑制剂 | 379 | 酰胺键 | |
| 6xbf | 8 | 8 | NDM-2金属-β-内酰胺酶抑制剂 | 452 | 酰胺键 | |
| 1hqq | RC | 13 | 11 | 链霉亲和素配体 | 413 | 二硫键 |
| 1jbu | EEWEVL | 15 | 7 | 凝血因子Ⅶ抑制剂 | 908 | 二硫键 |
| 1smf | 9 | 9 | 胰蛋白酶抑制剂 | 414 | 二硫键 | |
| 1sle | 8 | 8 | 链霉亲和素配体 | 324 | 二硫键 | |
| 1vpp | RGWVEI | 18 | 9 | 生长素抑制剂 | 556 | 二硫键 |
| 1vwb | 6 | 6 | 链霉亲和素配体 | 306 | 二硫键 | |
| 2ck0 | 11 | 10 | 血管紧张素Ⅱ抗体结合肽 | 504 | 二硫键 | |
| 2nwn | 12 | 12 | 丝氨酸蛋白酶抑制剂 | 597 | 二硫键 | |
| 3g5v | 16 | 16 | EGFR肽段 | 603 | 二硫键 | |
| 3p72 | 11 | 11 | 血小板糖蛋白1b抑制剂 | 521 | 二硫键 | |
| 3wnf | 6 | 6 | HIV-1整合酶抑制剂 | 432 | 二硫键 | |
| 4ib5 | G | 13 | 11 | CK2beta拮抗剂 | 456 | 二硫键 |
| 4m1d | 14 | 14 | HIV-1 gp120蛋白V3域类似物 | 588 | 二硫键 | |
| 4ou3 | 6 | 5 | 猪氨肽酶N抑制剂 | 366 | 二硫键 | |
| 5djc | D | 13 | 11 | 抗体结合肽 | 610 | 二硫键 |
| 5co5 | G | 16 | 15 | 芋螺毒素 | 692 | 二硫键 |
| 5eoc | 13 | 13 | 丙型肝炎病毒E2表位Ⅰ类似物 | 468 | 二硫键 | |
| 5h5q | 13 | 11 | 人源GPX4抑制剂 | 505 | 二硫键 | |
| 5th2 | 12 | 12 | L5Q中间位变体类似物 | 709 | 二硫键 | |
| 5vb9 | 15 | 14 | IL-17抗体抑制剂 | 571 | 二硫键 | |
| 5wxr | GA | 14 | 12 | 尿激酶型纤溶酶原激活物抑制剂 | 526 | 二硫键 |
| 5xco | RRRR | 19 | 11 | K-Ras(G12D突变体)抑制剂 | 650 | 二硫键 |
| 6e5m | 9 | 9 | β-胰蛋白酶抑制剂 | 434 | 二硫键 | |
| 1bm2 | 6 | 6 | GBR2-sh2结构域高活性配体 | 336 | 其他环化 | |
| 1bzh | 7 | 7 | 酪氨酸磷酸酶抑制剂 | 398 | 其他环化 | |
| 1vwl | 9 | 8 | 链霉亲和素配体 | 300 | 其他环化 | |
| 4zjx | 8 | 7 | 肉毒杆菌神经毒素(血清型A)抑制剂 | 604 | 其他环化 | |
| 5nes | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 259 | 其他环化 | |
| 5ney | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 271 | 其他环化 | |
| 5nf0 | 12 | 12 | 靶向铜绿假单胞菌糖蛋白的抗菌肽 | 283 | 其他环化 | |
| 6b67 | 7 | 7 | PPM1A活性调节剂 | 528 | 其他环化 | |
| 6dn6 | 5 | 5 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 234 | 其他环化 | |
| 6dn7 | 7 | 7 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 254 | 其他环化 | |
| 6dn8 | 8 | 8 | iNOS-SPSB蛋白-蛋白相互作用抑制剂 | 258 | 其他环化 | |
| 6nnv | 14 | 13 | PD-L1抑制剂 | 510 | 其他环化 | |
| 6u4a | 11 | 11 | BRD2-BD1抑制剂 | 587 | 其他环化 | |
| 6u74 | 14 | 14 | BRD2-BD1抑制剂 | 582 | 其他环化 | |
| 6u8m | 17 | 17 | BRD2-BD1抑制剂 | 504 | 其他环化 | |
| 6wgn | 15 | 14 | K-Ras(G12D突变体)抑制剂 | 641 | 其他环化 | |
| 6xci | 8 | 8 | NDM-3金属-β-内酰胺酶抑制剂 | 457 | 其他环化 | |
| 6xib | 12 | 11 | PCSK9抑制剂 | 546 | 其他环化 | |
| 6xic | 11 | 11 | PCSK9抑制剂 | 518 | 其他环化 | |
| 6xid | 12 | 11 | PCSK9抑制剂 | 537 | 其他环化 | |
| 6xie | 11 | 11 | PCSK9抑制剂 | 496 | 其他环化 | |
| 6xif | 11 | 11 | PCSK9抑制剂 | 528 | 其他环化 | |
| 6xs5 | 17 | 17 | 人源Vps29抑制剂,结构稳定剂 | 529 | 其他环化 | |
| 6xs7 | 17 | 17 | 人源Vps30抑制剂,结构稳定剂 | 654 | 其他环化 | |
| 6xs8 | 13 | 13 | 人源Vps31抑制剂,结构稳定剂 | 380 | 其他环化 | |
| 6xsa | 15 | 15 | 人源Vps32抑制剂,结构稳定剂 | 623 | 其他环化 | |
| 6yw1 | 14 | 14 | 促进HIF脯氨酰羟化酶2结晶的环肽配体 | 671 | 其他环化 | |
| 7bph | 14 | 13 | GNAS抑制剂 | 520 | 其他环化 | |
| 7k2k | 7 | 7 | 人源KEAP1蛋白抑制剂 | 349 | 其他环化 | |
| 7k2l | 7 | 7 | 人源KEAP1蛋白抑制剂 | 330 | 其他环化 | |
| 7k2o | 7 | 7 | 人源KEAP1蛋白抑制剂 | 346 | 其他环化 | |
| 7k2p | 7 | 7 | 人源KEAP1蛋白抑制剂 | 344 | 其他环化 | |
| 7k2r | 7 | 7 | 人源KEAP1蛋白抑制剂 | 350 | 其他环化 | |
| 7rov | 14 | 12 | K-Ras(G12D突变体)抑制剂 | 564 | 其他环化 |
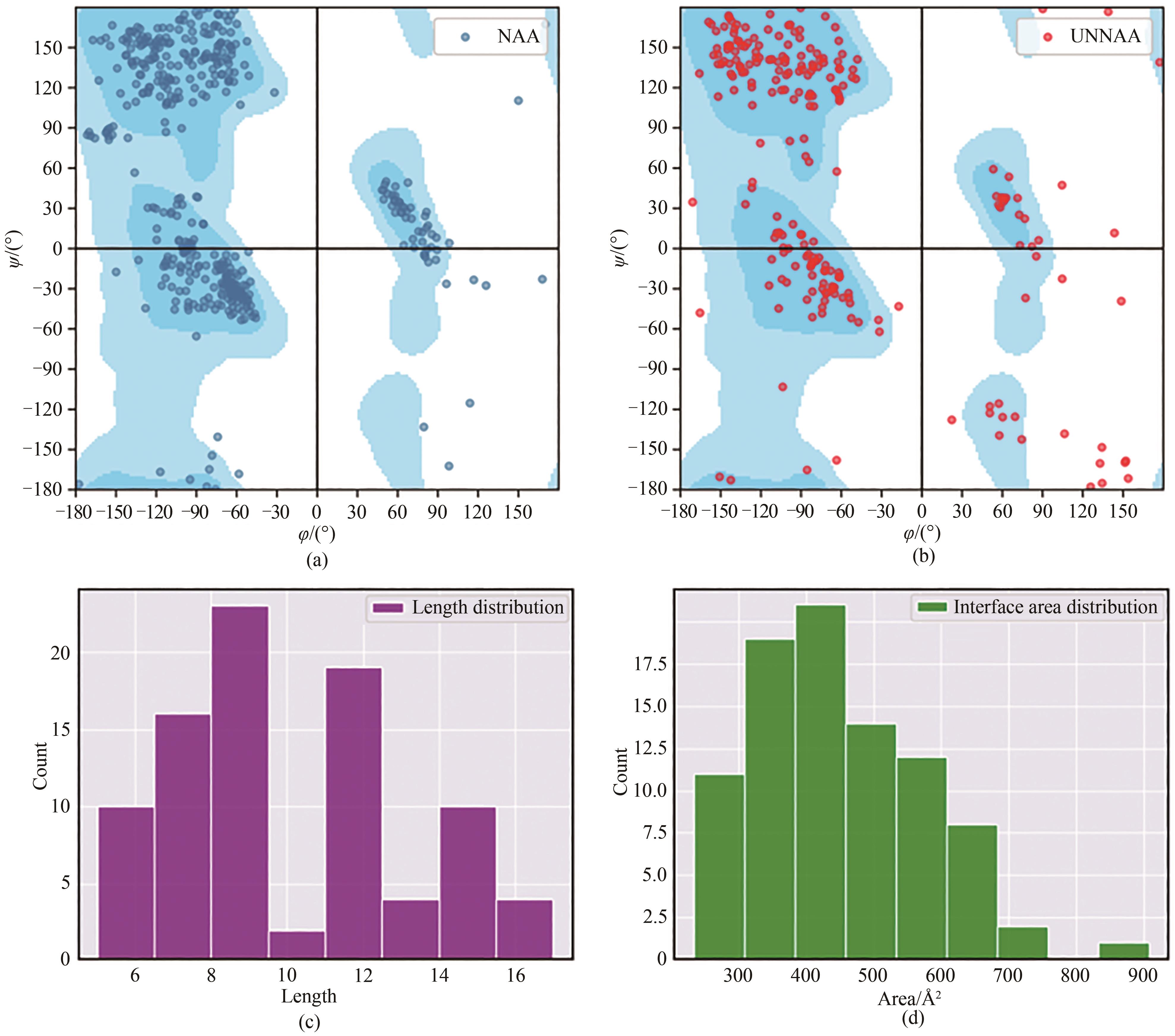
图1 PDB中环肽-靶标复合物数据集(见表2)中环肽配体的参数统计图(a)、(b)中蓝色背景分布为天然蛋白质体系的氨基酸残基扭转角ψ/φ的分布。(a)仅含天然氨基酸残基的环肽配体主链扭转角分布图(ψ/φ);(b)存在非天然氨基酸残基的环肽配体主链扭转角分布图(ψ/φ);(c)数据集中所有环肽配体环序列长度分布图;(d)数据集中所有环肽配体与靶标之间界面面积分布图
Fig. 1 Parameters of cyclic peptide ligands with the cyclic peptide-target complex data set (see Table 1) in PDBDistribution of torsion angles of the main chain of cyclic peptide ligands containing natural (a) and non-natural (b) amino acid residues (ψ/φ), in which blue cloud highlights the distribution of the torsion angle ψ/φ of amino acid residues in natural proteins; Length distribution of the loop sequences of all cyclic peptide ligands in the data set (c); Distribution of the interface area between all cyclic peptide ligands and targets (d)
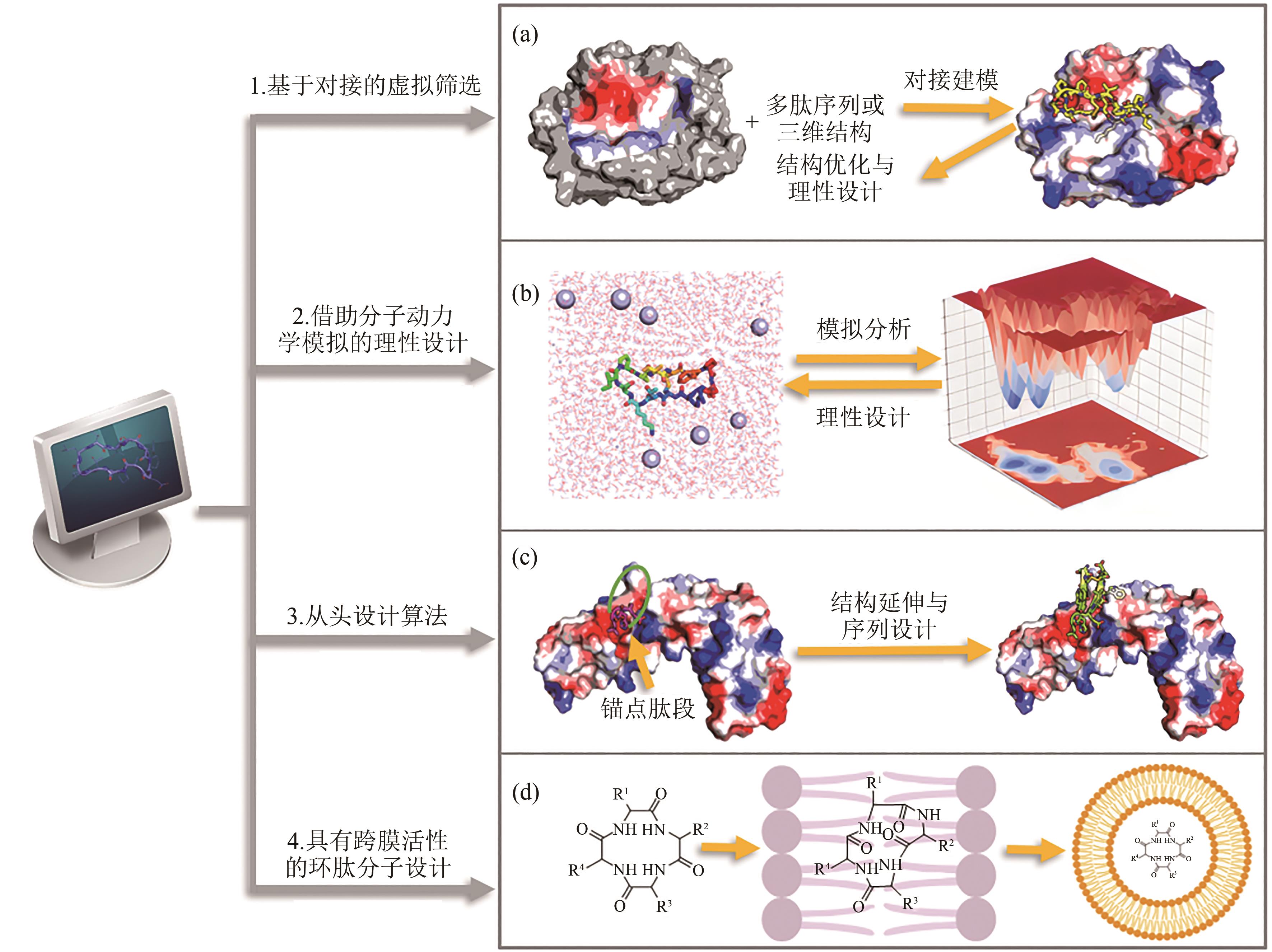
图3 基于靶标结构的环肽设计算法(a)基于分子对接的虚拟筛选算法;(b)基于分子动力学模拟的理性设计算法;(c)从头设计算法;(d)跨膜环肽分子的设计算法
Fig. 3 Overview of computational methods for target structure based cyclic peptide design(a) Virtual screening algorithms based on molecular docking; (b) Rational design algorithms based on molecular dynamics simulation; (c) De novo design algorithms;(d) Design algorithms for transmembrane cyclic peptides
| 类型 | 代表算法 | 算法简介 | 算法目的 |
|---|---|---|---|
| 基于对接的虚拟 筛选算法 | AutoDock CrankPep[ | 将环肽结构库中的分子与靶标蛋白对接,基于对接打分富集可能结合靶标的环肽。对接打分是对环肽和靶标蛋白结合的粗略但高效的评估 | 从现有环肽、已知结构环肽或某一类环肽等环肽库中富集可能结合靶标蛋白的分子,为实验发现未知的结合环肽提供候选分子 |
| 借助分子动力学 模拟的设计算法 | REMD[ | 借助于分子动力学模拟增强采样算法对环肽以及环肽与靶标蛋白的复合物结构进行采样。通过动力学采样对目标环肽和靶标蛋白的结合能进行精细的计算和比较 | 对已知结合的环肽分子进行分析和改造设计,估算结合自由能,研究各残基对结合的贡献,找到对环肽进行改进的可能方案 |
| 从头设计算法 | Rosetta Anchor Extension[ | 以某个对结合能贡献较大的基团为起点,在靶标蛋白表面通过主链优化采样与序列设计生长出全新的环肽 | 不受限于已知结合环肽或是环肽结构库,针对靶标蛋白从头设计环肽配体,并能引入化学修饰与非天然氨基酸 |
| 具有跨膜活性的 环肽分子设计 | Rosetta[ | 引入主链酰胺键的N-甲基化修饰或降低侧链极性的非天然氨基酸 | 在设计中加入环肽跨膜活性的考量 |
表3 基于靶标结构的环肽分子计算设计算法
Table 3 Structure based computational design algorithms of cyclic peptides
| 类型 | 代表算法 | 算法简介 | 算法目的 |
|---|---|---|---|
| 基于对接的虚拟 筛选算法 | AutoDock CrankPep[ | 将环肽结构库中的分子与靶标蛋白对接,基于对接打分富集可能结合靶标的环肽。对接打分是对环肽和靶标蛋白结合的粗略但高效的评估 | 从现有环肽、已知结构环肽或某一类环肽等环肽库中富集可能结合靶标蛋白的分子,为实验发现未知的结合环肽提供候选分子 |
| 借助分子动力学 模拟的设计算法 | REMD[ | 借助于分子动力学模拟增强采样算法对环肽以及环肽与靶标蛋白的复合物结构进行采样。通过动力学采样对目标环肽和靶标蛋白的结合能进行精细的计算和比较 | 对已知结合的环肽分子进行分析和改造设计,估算结合自由能,研究各残基对结合的贡献,找到对环肽进行改进的可能方案 |
| 从头设计算法 | Rosetta Anchor Extension[ | 以某个对结合能贡献较大的基团为起点,在靶标蛋白表面通过主链优化采样与序列设计生长出全新的环肽 | 不受限于已知结合环肽或是环肽结构库,针对靶标蛋白从头设计环肽配体,并能引入化学修饰与非天然氨基酸 |
| 具有跨膜活性的 环肽分子设计 | Rosetta[ | 引入主链酰胺键的N-甲基化修饰或降低侧链极性的非天然氨基酸 | 在设计中加入环肽跨膜活性的考量 |
| 算法名称 | 目标任务 | 测试体系或数据集 | 测试集规模 | 环肽序列长度 | 算法性能 |
|---|---|---|---|---|---|
| MODPEP 2.0[ | 仅SS-环肽结构建模 | 非冗余SSCP库 | 193 | 约30 | 前100构象与前10构象的平均Cα RMSD为2.20 Å和1.66 Å,优于mETKDG与PEP-FOLD |
| Peplook[ | 环肽结构建模 | 环肽建模困难数据集 | 38 | 5~30 | 平均BB-RMSD为3.8 Å,优于PEP-FOLD |
| PEPstrMOD[ | 环肽结构建模 | CyclicPep | 34 | 10~30 | 平均Cα-RMSD为4.06 Å,优于PEP-FOLD |
| PEP-FOLD[ | 环肽结构建模 | 环肽建模困难数据集 | 34 | 10~30 | 平均RMS为2.75 Å |
| I-TASSER[ | 环肽结构建模 | 环肽建模困难数据集 | 35 | 10~30 | 平均BB-RMSD为2.5 Å |
| mETKDG[ | 环肽结构构象生成 | mc-PEP-set | 2 | 10,11 | 最优BB-RMSD分布均值分别为0.60 Å,1.27 Å |
| AutoDock CrankPep[ | 环肽配体对接 | PDB数据库中非冗余环肽-蛋白复合物 | CNCP: 18; SSCP: 20 | CNCP: 6~14; SSCP: 6~20 | 对于CNCP,SSCP的最优平均fnc分别为0.86和0.70 |
| HADDOCK 2.4[ | 环肽配体对接 | PDB数据库中非冗余环肽-蛋白复合物 | CNCP: 18; SSCP: 12 | CNCP: 6~14; SSCP: 6~14 | CNCP的平均RMSD为1.5~2.0 Å(略优于AutoDock CrankPep),SSCP的最优平均RMSD为3.0 Å |
| DES3PI[ | 环肽配体从头设计 | 靶标蛋白:Ras, Mcl-1,Aβ protofibril | — | — | 生成环肽配体的AutoDock CrankPep对接打分可以达到-9~-12 kcal/mol |
| Rosetta | 环肽单体理性 从头设计[ | — | — | 7~14 | 生成了200种环肽结构,其中部分进行了结构解析,BB-RMSD<1.6 Å |
| 环肽配体 从头设计[ | 靶标蛋白:HDAC2,HDAC6 | — | 7~10 | 设计环肽配体实验测得最优IC50为:HDAC2 9.1 nmol/L;HDAC6 5.4 nmol/L | |
| 跨膜活性环肽 从头设计[ | — | — | 6~12 | 生成了具有跨膜活性的环肽结构,与实验解析结构的BB-RMSD<1.5 Å,具有跨膜活性与较好的口服利用率 |
表4 本文所涉及的算法总体简介
Table 4 A brief introduction of algorithms included
| 算法名称 | 目标任务 | 测试体系或数据集 | 测试集规模 | 环肽序列长度 | 算法性能 |
|---|---|---|---|---|---|
| MODPEP 2.0[ | 仅SS-环肽结构建模 | 非冗余SSCP库 | 193 | 约30 | 前100构象与前10构象的平均Cα RMSD为2.20 Å和1.66 Å,优于mETKDG与PEP-FOLD |
| Peplook[ | 环肽结构建模 | 环肽建模困难数据集 | 38 | 5~30 | 平均BB-RMSD为3.8 Å,优于PEP-FOLD |
| PEPstrMOD[ | 环肽结构建模 | CyclicPep | 34 | 10~30 | 平均Cα-RMSD为4.06 Å,优于PEP-FOLD |
| PEP-FOLD[ | 环肽结构建模 | 环肽建模困难数据集 | 34 | 10~30 | 平均RMS为2.75 Å |
| I-TASSER[ | 环肽结构建模 | 环肽建模困难数据集 | 35 | 10~30 | 平均BB-RMSD为2.5 Å |
| mETKDG[ | 环肽结构构象生成 | mc-PEP-set | 2 | 10,11 | 最优BB-RMSD分布均值分别为0.60 Å,1.27 Å |
| AutoDock CrankPep[ | 环肽配体对接 | PDB数据库中非冗余环肽-蛋白复合物 | CNCP: 18; SSCP: 20 | CNCP: 6~14; SSCP: 6~20 | 对于CNCP,SSCP的最优平均fnc分别为0.86和0.70 |
| HADDOCK 2.4[ | 环肽配体对接 | PDB数据库中非冗余环肽-蛋白复合物 | CNCP: 18; SSCP: 12 | CNCP: 6~14; SSCP: 6~14 | CNCP的平均RMSD为1.5~2.0 Å(略优于AutoDock CrankPep),SSCP的最优平均RMSD为3.0 Å |
| DES3PI[ | 环肽配体从头设计 | 靶标蛋白:Ras, Mcl-1,Aβ protofibril | — | — | 生成环肽配体的AutoDock CrankPep对接打分可以达到-9~-12 kcal/mol |
| Rosetta | 环肽单体理性 从头设计[ | — | — | 7~14 | 生成了200种环肽结构,其中部分进行了结构解析,BB-RMSD<1.6 Å |
| 环肽配体 从头设计[ | 靶标蛋白:HDAC2,HDAC6 | — | 7~10 | 设计环肽配体实验测得最优IC50为:HDAC2 9.1 nmol/L;HDAC6 5.4 nmol/L | |
| 跨膜活性环肽 从头设计[ | — | — | 6~12 | 生成了具有跨膜活性的环肽结构,与实验解析结构的BB-RMSD<1.5 Å,具有跨膜活性与较好的口服利用率 |
| 1 | IVANOV A A, KHURI F R, FU H. Targeting protein-protein interactions as an anticancer strategy[J]. Trends in Pharmacological Sciences, 2013, 34(7): 393-400. |
| 2 | SCOTT D E, BAYLY A R, ABELL C, et al. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge[J]. Nature Reviews Drug Discovery, 2016, 15(8): 533-550. |
| 3 | JONES S, THORNTON J M. Principles of protein-protein interactions[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(1): 13-20. |
| 4 | NOOREN I M A, THORNTON J M. Diversity of protein-protein interactions[J]. The EMBO Journal, 2003, 22(14): 3486-3492. |
| 5 | REYNÈS C, HOST H, CAMPROUX A C, et al. Designing focused chemical libraries enriched in protein-protein interaction inhibitors using machine-learning methods[J]. PLoS Computational Biology, 2010, 6(3): e1000695. |
| 6 | ROGNAN D. Rational design of protein-protein interaction inhibitors[J]. MedChemComm, 2015, 6(1): 51-60. |
| 7 | MORELLI X, BOURGEAS R, ROCHE P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I)[J]. Current Opinion in Chemical Biology, 2011, 15(4): 475-481. |
| 8 | SPERANDIO O, REYNÈS C H, CAMPROUX A C, et al. Rationalizing the chemical space of protein-protein interaction inhibitors[J]. Drug Discovery Today, 2010, 15(5/6): 220-229. |
| 9 | LAGASSÉ H A, ALEXAKI A, SIMHADRI V L, et al. Recent advances in (therapeutic protein) drug development[J]. F1000Research, 2017, 6: 113. |
| 10 | CHENG S S, QU Y Q, WU J, et al. Inhibition of the CDK9-cyclin T1 protein-protein interaction as a new approach against triple-negative breast cancer[J]. Acta Pharmaceutica Sinica B, 2022, 12(3): 1390-1405. |
| 11 | NICOLAOU K C, BODDY C N, BRÄSE S, et al. Chemistry, biology, and medicine of the glycopeptide antibiotics[J]. Angewandte Chemie International Edition, 1999, 38(15): 2096-2152. |
| 12 | BODANSZKY M. Principles of Peptide Synthesis[M]. Heidelberg: Springer, 2012. |
| 13 | MERRIFIELD R B. Solid phase peptide synthesis.Ⅰ. The synthesis of a tetrapeptide[J]. Journal of the American Chemical Society, 1963, 85(14): 2149-2154. |
| 14 | BEHRENDT R, WHITE P, OFFER J. Advances in Fmoc solid-phase peptide synthesis[J]. Journal of Peptide Science, 2016, 22(1): 4-27. |
| 15 | AMBLARD M, FEHRENTZ J A, MARTINEZ J, et al. Methods and protocols of modern solid phase peptide synthesis[J]. Molecular Biotechnology, 2006, 33(3): 239-254. |
| 16 | CHAN W C, WHITE P D. Fmoc solid phase peptide synthesis: a practical approach[M]. New York: Oxford University Press, 2000 |
| 17 | LEADER B, BACA Q J, GOLAN D E. Protein therapeutics: a summary and pharmacological classification[J]. Nature Reviews Drug Discovery, 2008, 7(1): 21-39. |
| 18 | SANTOS G B, GANESAN A, EMERY F S. Oral administration of peptide-based drugs: beyond Lipinski's rule[J]. ChemMedChem, 2016, 11(20): 2245-2251. |
| 19 | DANHO W, SWISTOK J, KHAN W, et al. Opportunities and challenges of developing peptide drugs in the pharmaceutical industry[C]// The Proceedings of the 20th American Peptide Symposium-Peptides for youth: Advances in Experimental Medicine and Biology, 2009, 611: 467-469. |
| 20 | COOPER B M, IEGRE J, , O'DONOVAN D H, et al. Peptides as a platform for targeted therapeutics for cancer: peptide-drug conjugates (PDCs)[J]. Chemical Society Reviews, 2021, 50(3): 1480-1494. |
| 21 | WANG J, YADAV V, SMART A L, et al. Stability of peptide drugs in the colon[J]. European Journal of Pharmaceutical Sciences, 2015, 78: 31-36. |
| 22 | VALEUR E, GUÉRET S M, ADIHOU H, et al. New modalities for challenging targets in drug discovery[J]. Angewandte Chemie International Edition, 2017, 56(35): 10294-10323. |
| 23 | BRUZZONI-GIOVANELLI H, ALEZRA V, WOLFF N, et al. Interfering peptides targeting protein-protein interactions: the next generation of drugs?[J]. Drug Discovery Today, 2018, 23(2): 272-285. |
| 24 | JENSEN K K, ANDREATTA M, MARCATILI P, et al. Improved methods for predicting peptide binding affinity to MHC classⅡ molecules[J]. Immunology, 2018, 154(3): 394-406. |
| 25 | BEUMING T, FARID R, SHERMAN W. High-energy water sites determine peptide binding affinity and specificity of PDZ domains[J]. Protein Science, 2009, 18(8): 1609-1619. |
| 26 | ZHOU H X, GILSON M K. Theory of free energy and entropy in noncovalent binding[J]. Chemical Reviews, 2009, 109(9): 4092-4107. |
| 27 | MURPHY K P, XIE D, THOMPSON K S, et al. Entropy in biological binding processes: estimation of translational entropy loss[J]. Proteins: Structure, Function, and Bioinformatics, 1994, 18(1): 63-67. |
| 28 | VERTERAMO M L, STENSTRÖM O, IGNJATOVIĆ M M, et al. Interplay between conformational entropy and solvation entropy in protein-ligand binding[J]. Journal of the American Chemical Society, 2019, 141(5): 2012-2026. |
| 29 | EKBERG V, RYDE U. On the use of interaction entropy and related methods to estimate binding entropies[J]. Journal of Chemical Theory and Computation, 2021, 17(8): 5379-5391. |
| 30 | PAGE M I. Entropy, binding energy, and enzymic catalysis[J]. Angewandte Chemie International Edition, 1977, 16(7): 449-459. |
| 31 | HUIGENS III R W, MORRISON K C, HICKLIN R W, et al. A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products[J]. Nature Chemistry, 2013, 5(3): 195-202. |
| 32 | YOON S, EOM G H. HDAC and HDAC inhibitor: from cancer to cardiovascular diseases[J]. Chonnam Medical Journal, 2016, 52(1): 1-11. |
| 33 | WEST A C, JOHNSTONE R W. New and emerging HDAC inhibitors for cancer treatment[J]. The Journal of Clinical Investigation, 2014, 124(1): 30-39. |
| 34 | HOSSEINZADEH P, WATSON P R, CRAVEN T W, et al. Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites[J]. Nature Communications, 2021, 12(1): 3384. |
| 35 | KLING A, LUKAT P, ALMEIDA D V, et al. Targeting DnaN for tuberculosis therapy using novel griselimycins[J]. Science, 2015, 348(6239): 1106-1112. |
| 36 | DEMMER O, FRANK A O, HAGN F, et al. A conformationally frozen peptoid boosts CXCR4 affinity and anti-HIV activity[J]. Angewandte Chemie International Edition, 2012, 51(32): 8110-8113. |
| 37 | XIONG J P, STEHLE T, ZHANG R G, et al. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand[J]. Science, 2002, 296(5565): 151-155. |
| 38 | MIRANDA E, NORDGREN I K, MALE A L, et al. A cyclic peptide inhibitor of HIF-1 heterodimerization that inhibits hypoxia signaling in cancer cells[J]. Journal of the American Chemical Society, 2013, 135(28): 10418-10425. |
| 39 | BUCKTON L K, RAHIMI M N, MCALPINE S R. Cyclic peptides as drugs for intracellular targets: the next frontier in peptide therapeutic development[J]. Chemistry, 2021, 27(5): 1487-1513. |
| 40 | WANG Y, CHEETHAM A G, ANGACIAN G, et al. Peptide-drug conjugates as effective prodrug strategies for targeted delivery[J]. Advanced Drug Delivery Reviews, 2017, 110/111: 112-126. |
| 41 | CROXTALL J D, SCOTT L J. Lanreotide autogel® [J]. Drugs, 2008, 68(5): 711-723. |
| 42 | CAPLIN M E, PAVEL M, ĆWIKŁA J B, et al. Lanreotide in metastatic enteropancreatic neuroendocrine tumors[J]. The New England Journal of Medicine, 2014, 371(3): 224-233. |
| 43 | ANGLADE E, ASPESLET L J, WEISS S L. A new agent for the treatment of noninfectious uveitis: rationale and design of three LUMINATE (Lux Uveitis Multicenter Investigation of a New Approach to Treatment) trials of steroid-sparing voclosporin[J]. Clinical Ophthalmology, 2008, 2(4): 693-702. |
| 44 | KUGLSTATTER A, MUELLER F, KUSZNIR E, et al. Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin)[J]. Acta Crystallographica Section D, Biological Crystallography, 2011, 67(Pt 2): 119-123. |
| 45 | HEO Y A. Voclosporin: first approval[J]. Drugs, 2021, 81(5): 605-610. |
| 46 | BECK J G, CHATTERJEE J, LAUFER B, et al. Intestinal permeability of cyclic peptides: common key backbone motifs identified[J]. Journal of the American Chemical Society, 2012, 134(29): 12125-12133. |
| 47 | MEIRELES L M C, MUSTATA G. Discovery of modulators of protein-protein interactions: current approaches and limitations[J]. Current Topics in Medicinal Chemistry, 2011, 11(3): 248-257. |
| 48 | WANG L, LI L, FU W T, et al. Optimization and bioevaluation of Cdc37-derived peptides: an insight into Hsp90-Cdc37 protein-protein interaction modulators[J]. Bioorganic & Medicinal Chemistry, 2017, 25(1): 233-240. |
| 49 | BEAUFAYS J, LINS L, THOMAS A, et al. In silico predictions of 3D structures of linear and cyclic peptides with natural and non-proteinogenic residues[J]. Journal of Peptide Science, 2012, 18(1): 17-24. |
| 50 | TAO H Y, WU Q L, ZHAO X J, et al. Efficient 3D conformer generation of cyclic peptides formed by a disulfide bond[J]. Journal of Cheminformatics, 2022, 14(1): 26. |
| 51 | RILEY B T, ILYICHOVA O, COSTA M G S, et al. Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics[J]. Scientific Reports, 2016, 6: 35385. |
| 52 | WANG S Z, WITEK J, LANDRUM G A, et al. Improving conformer generation for small rings and macrocycles based on distance geometry and experimental torsional-angle preferences[J]. Journal of Chemical Information and Modeling, 2020, 60(4): 2044-2058. |
| 53 | ANDREWS M D, DACK K N, DE GROOT M J, et al. Discovery of an oral, rule of 5 compliant, interleukin 17A protein-protein interaction modulator for the potential treatment of psoriasis and other inflammatory diseases[J]. Journal of Medicinal Chemistry, 2022, 65(13): 8828-8842. |
| 54 | SOUROUJON M C, MOCHLY-ROSEN D. Peptide modulators of protein-protein interactions in intracellular signaling[J]. Nature Biotechnology, 1998, 16(10): 919-924. |
| 55 | HOSSEINZADEH P, BHARDWAJ G, MULLIGAN V K, et al. Comprehensive computational design of ordered peptide macrocycles[J]. Science, 2017, 358(6369): 1461-1466. |
| 56 | BHARDWAJ G, O'CONNOR J, RETTIE S, et al. Accurate de novo design of membrane-traversing macrocycles[J]. Cell, 2022, 185(19): 3520-3532.e26. |
| 57 | MCHUGH S M, ROGERS J R, YU H T, et al. Insights into how cyclic peptides switch conformations[J]. Journal of Chemical Theory and Computation, 2016, 12(5): 2480-2488. |
| 58 | GAVENONIS J, SHENEMAN B A, SIEGERT T R, et al. Comprehensive analysis of loops at protein-protein interfaces for macrocycle design[J]. Nature Chemical Biology, 2014, 10(9): 716-722. |
| 59 | DUFFY F J, O'DONOVAN D, DEVOCELLE M, et al. Virtual screening using combinatorial cyclic peptide libraries reveals protein interfaces readily targetable by cyclic peptides[J]. Journal of Chemical Information and Modeling, 2015, 55(3): 600-613. |
| 60 | DUFFY F, MAHESHWARI N, BUCHETE N V, et al. Computational opportunities and challenges in finding cyclic peptide modulators of protein-protein interactions[M]. Methods in Molecular Biology: Cyclic Peptide Design. New York: Springer New York, 2019: 73-95. |
| 61 | CHE Y. Design of cyclic peptides as protein recognition motifs[M]//Methods in Molecular Biology: Cyclic Peptide Design. New York: Springer New York, 2019: 97-106. |
| 62 | SANTINI B L, ZACHARIAS M. Rapid in silico design of potential cyclic peptide binders targeting protein-protein interfaces[J]. Frontiers in Chemistry, 2020, 8: 573259. |
| 63 | SCOTT C P, ABEL-SANTOS E, WALL M, et al. Production of cyclic peptides and proteins in vivo [J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(24): 13638-13643. |
| 64 | GENG H, JIANG F, WU Y D. Accurate structure prediction and conformational analysis of cyclic peptides with residue-specific force fields[J]. The Journal of Physical Chemistry Letters, 2016, 7(10): 1805-1810. |
| 65 | LINDORFF-LARSEN K, PIANA S, PALMO K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field[J]. Proteins: Structure, Function, and Bioinformatics, 2010, 78(8): 1950-1958. |
| 66 | JIANG F, ZHOU C Y, WU Y D. Residue-specific force field based on the protein coil library. RSFF1: modification of OPLS-AA/L[J]. The Journal of Physical Chemistry B, 2014, 118(25): 6983-6998. |
| 67 | ZHOU C Y, JIANG F, WU Y D. Residue-specific force field based on protein coil library. RSFF2: modification of AMBER ff99SB[J]. The Journal of Physical Chemistry B, 2015, 119(3): 1035-1047. |
| 68 | SLOUGH D P, YU H T, MCHUGH S M, et al. Toward accurately modeling N-methylated cyclic peptides[J]. Physical Chemistry Chemical Physics, 2017, 19(7): 5377-5388. |
| 69 | SUGITA Y, OKAMOTO Y. Replica-exchange molecular dynamics method for protein folding[J]. Chemical Physics Letters, 1999, 314(1/2): 141-151. |
| 70 | RHEE Y M, PANDE V S. Multiplexed-replica exchange molecular dynamics method for protein folding simulation[J]. Biophysical Journal, 2003, 84(2): 775-786. |
| 71 | MELI M, COLOMBO G. A Hamiltonian replica exchange molecular dynamics (MD) method for the study of folding, based on the analysis of the stabilization determinants of proteins[J]. International Journal of Molecular Sciences, 2013, 14(6): 12157-12169. |
| 72 | LAIO A, PARRINELLO M. Escaping free-energy minima[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(20): 12562-12566. |
| 73 | PIANA S, LAIO A. A bias-exchange approach to protein folding[J]. The Journal of Physical Chemistry B, 2007, 111(17): 4553-4559. |
| 74 | TAYLOR R D, REY-CARRIZO M, PASSIOURA T, et al. Identification of nonstandard macrocyclic peptide ligands through display screening[J]. Drug Discovery Today Technologies, 2017, 26: 17-23. |
| 75 | YAN Y M, ZHANG D, HUANG S Y. Efficient conformational ensemble generation of protein-bound peptides[J]. Journal of Cheminformatics, 2017, 9(1): 59. |
| 76 | ZHANG Y. I-TASSER server for protein 3D structure prediction[J]. BMC Bioinformatics, 2008, 9: 40. |
| 77 | SINGH S, SINGH H, TUKNAIT A, et al. PEPstrMOD: structure prediction of peptides containing natural, non-natural and modified residues[J]. Biology Direct, 2015, 10(1): 73. |
| 78 | DAMAS J M, FILIPE L C S, CAMPOS S R R, et al. Predicting the thermodynamics and kinetics of helix formation in a cyclic peptide model[J]. Journal of Chemical Theory and Computation, 2013, 9(11): 5148-5157. |
| 79 | RAZAVI A M, WUEST W M, VOELZ V A. Computational screening and selection of cyclic peptide hairpin mimetics by molecular simulation and kinetic network models[J]. Journal of Chemical Information and Modeling, 2014, 54(5): 1425-1432. |
| 80 | THÉVENET P, SHEN Y M, MAUPETIT J, et al. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides[J]. Nucleic Acids Research, 2012, 40(W1): W288-W293. |
| 81 | BINETTE V, MOUSSEAU N, TUFFERY P. A generalized attraction-repulsion potential and revisited fragment library improves PEP-FOLD peptide structure prediction[J]. Journal of Chemical Theory and Computation, 2022, 18(4): 2720-2736. |
| 82 | ZHAO D X, LU K, LIU G B, et al. PEP-FOLD design, synthesis, and characteristics of finger-like polypeptides[J]. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2020, 224: 117401. |
| 83 | SHEN Y M, MAUPETIT J, DERREUMAUX P, et al. Improved PEP-FOLD approach for peptide and miniprotein structure prediction[J]. Journal of Chemical Theory and Computation, 2014, 10(10): 4745-4758. |
| 84 | MANDELL D J, COUTSIAS E A, KORTEMME T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling[J]. Nature Methods, 2009, 6(8): 551-552. |
| 85 | COUTSIAS E A, SEOK C, WESTER M J, et al. Resultants and loop closure[J]. International Journal of Quantum Chemistry, 2006, 106(1): 176-189. |
| 86 | GO N, SCHERAGA H A. Ring closure and local conformational deformations of chain molecules[J]. Macromolecules, 1970, 3(2): 178-187. |
| 87 | THOMAS A, DESHAYES S, DECAFFMEYER M, et al. PepLook: an innovative in silico tool for determination of structure, polymorphism and stability of peptides[J]. Advances in Experimental Medicine and Biology, 2009, 611: 459-460. |
| 88 | CRAVEUR P, JOSEPH A P, ESQUE J, et al. Protein flexibility in the light of structural alphabets[J]. Frontiers in Molecular Biosciences, 2015, 2: 20. |
| 89 | JOSEPH A P, AGARWAL G, MAHAJAN S, et al. A short survey on protein blocks[J]. Biophysical Reviews, 2010, 2(3): 137-145. |
| 90 | ETCHEBEST C, BENROS C, HAZOUT S, et al. A structural alphabet for local protein structures: improved prediction methods[J]. Proteins: Structure, Function, and Bioinformatics, 2005, 59(4): 810-827. |
| 91 | THOMAS A, DESHAYES S, DECAFFMEYER M, et al. Prediction of peptide structure: how far are we?[J]. Proteins: Structure, Function, and Bioinformatics, 2006, 65(4): 889-897. |
| 92 | RINIKER S, LANDRUM G A. Better informed distance geometry: using what we know to improve conformation generation[J]. Journal of Chemical Information and Modeling, 2015, 55(12): 2562-2574. |
| 93 | ZHANG Y Q, SANNER M F. Docking flexible cyclic peptides with AutoDock CrankPep [J]. Journal of Chemical Theory and Computation, 2019, 15(10): 5161-5168. |
| 94 | CHARITOU V, VAN KEULEN S C, BONVIN A M J J. Cyclization and docking protocol for cyclic peptide-protein modeling using HADDOCK2.4 [J]. Journal of Chemical Theory and Computation, 2022, 18(6): 4027-4040. |
| 95 | DUFFY F J, VERNIERE M, DEVOCELLE M, et al. CycloPs: generating virtual libraries of cyclized and constrained peptides including nonnatural amino acids[J]. Journal of Chemical Information and Modeling, 2011, 51(4): 829-836. |
| 96 | ZHANG Y Q, SANNER M F. AutoDock CrankPep: combining folding and docking to predict protein-peptide complexes[J]. Bioinformatics, 2019, 35(24): 5121-5127. |
| 97 | FORLI S, HUEY R, PIQUE M E, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite[J]. Nature Protocols, 2016, 11(5): 905-919. |
| 98 | LENSINK M F, BRYSBAERT G, MAURI T, et al. Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment[J]. Proteins, 2021, 89(12): 1800-1823. |
| 99 | LASHKOV A A, TOLMACHEV I V, EISTRIKH-HELLER P A, et al. PyFepRestr: Plugin to PyMOL molecular graphics system for calculating the free energy of ligand-receptor binding[J]. Crystallography Reports, 2021, 66(5): 861-865. |
| 100 | MOZAFFARI S, SALEHI D, MAHDIPOOR P, et al. Design and application of hybrid cyclic-linear peptide-doxorubicin conjugates as a strategy to overcome doxorubicin resistance and toxicity[J]. European Journal of Medicinal Chemistry, 2021, 226: 113836. |
| 101 | YEDVABNY E, NERENBERG P S, SO C, et al. Disordered structural ensembles of vasopressin and oxytocin and their mutants[J]. The Journal of Physical Chemistry B, 2015, 119(3): 896-905. |
| 102 | SPITALERI A, GHITTI M, MARI S, et al. Use of metadynamics in the design of isoDGR-based αvβ3 antagonists to fine-tune the conformational ensemble[J]. Angewandte Chemie International Edition, 2011, 50(8): 1832-1836. |
| 103 | KIM H J, BAE S C. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs[J]. American Journal of Translational Research, 2011, 3(2): 166-179. |
| 104 | RITO M, SILVA F, FELIX A. Histone Deacetylase (HDAC) inhibitors as potential anticancer drugs in Neuroendocrine Carcinomas (NEC) of cervix and endometrium[J]. Virchows Archiv, 2015, 467: S 15-S16. |
| 105 | DELAUNAY M, HA-DUONG T. Des3PI: a fragment-based approach to design cyclic peptides targeting protein-protein interactions[J]. Journal of Computer-Aided Molecular Design, 2022, 36(8): 605-621. |
| 106 | SINDHIKARA D, BORRELLI K. High throughput evaluation of macrocyclization strategies for conformer stabilization[J]. Scientific Reports, 2018, 8: 6585. |
| 107 | WHITTY A, ZHONG M Q, VIARENGO L, et al. Quantifying the chameleonic properties of macrocycles and other high-molecular-weight drugs[J]. Drug Discovery Today, 2016, 21(5): 712-717. |
| 108 | AHLBACH C L, LEXA K W, BOCKUS A T, et al. Beyond cyclosporine A: conformation-dependent passive membrane permeabilities of cyclic peptide natural products[J]. Future Medicinal Chemistry, 2015, 7(16): 2121-2130. |
| 109 | WANG C K, SWEDBERG J E, HARVEY P J, et al. Conformational flexibility is a determinant of permeability for cyclosporin[J]. The Journal of Physical Chemistry B, 2018, 122(8): 2261-2276. |
| 110 | ROSSI SEBASTIANO M, DOAK B C, BACKLUND M, et al. Impact of dynamically exposed polarity on permeability and solubility of chameleonic drugs beyond the rule of 5[J]. Journal of Medicinal Chemistry, 2018, 61(9): 4189-4202. |
| 111 | ALFORD R F, LEAVER-FAY A, JELIAZKOV J R, et al. The Rosetta all-atom energy function for macromolecular modeling and design[J]. Journal of Chemical Theory and Computation, 2017, 13(6): 3031-3048. |
| 112 | MUTTENTHALER M, KING G F, ADAMS D J, et al. Trends in peptide drug discovery[J]. Nature Reviews Drug Discovery, 2021, 20(4): 309-325. |
| 113 | WATSON J L, JUERGENS D, BENNETT N R, et al. Broadly applicable and accurate protein design by integrating structure prediction networks and diffusion generative models[EB/OL]. bioRxiv, 2022[2022-12-20]. . |
| 114 | INGRAHAM J, BARANOV M, COSTELLO Z, et al. Illuminating protein space with a programmable generative model[EB/OL]. bioRxiv, 2022[2022-12-20]. . |
| 115 | SKALIC M, SABBADIN D, SATTAROV B, et al. From target to drug: generative modeling for the multimodal structure-based ligand design[J]. Molecular Pharmaceutics, 2019, 16(10): 4282-4291. |
| 116 | LI Y B, PEI J F, LAI L H. Structure-based de novo drug design using 3D deep generative models[J]. Chemical Science, 2021, 12(41): 13664-13675. |
| 117 | SUBTELNY A O, HARTMAN M C T, SZOSTAK J W. Ribosomal synthesis of N-methyl peptides[J]. Journal of the American Chemical Society, 2008, 130(19): 6131-6136. |
| 118 | KAWAKAMI T, MURAKAMI H, SUGA H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides[J]. Chemistry & Biology, 2008, 15(1): 32-42. |
| 119 | GOTO Y, MURAKAMI H, SUGA H. Initiating translation with D-amino acids[J]. RNA, 2008, 14(7): 1390-1398. |
| 120 | FUJINO T, GOTO Y, SUGA H, et al. Reevaluation of the D-amino acid compatibility with the elongation event in translation[J]. Journal of the American Chemical Society, 2013, 135(5): 1830-1837. |
| 121 | ACHENBACH J, JAHNZ M, BETHGE L, et al. Outwitting EF-Tu and the ribosome: translation with d-amino acids[J]. Nucleic Acids Research, 2015, 43(12): 5687-5698. |
| 122 | HUANG Y J, LIU T. Therapeutic applications of genetic code expansion[J]. Synthetic and Systems Biotechnology, 2018, 3(3): 150-158. |
| 123 | YOUNG D D, SCHULTZ P G. Playing with the molecules of life[J]. ACS Chemical Biology, 2018, 13(4): 854-870. |
| 124 | ARRANZ-GIBERT P, VANDERSCHUREN K, ISAACS F J. Next-generation genetic code expansion[J]. Current Opinion in Chemical Biology, 2018, 46: 203-211. |
| 125 | LOMBARDI A, MARASCO D, MAGLIO O, et al. Miniaturized metalloproteins: application to iron-sulfur proteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(22): 11922-11927. |
| 126 | QUARTARARO J S, ESHELMAN M R, PERARO L, et al. A bicyclic peptide scaffold promotes phosphotyrosine mimicry and cellular uptake[J]. Bioorganic & Medicinal Chemistry, 2014, 22(22): 6387-6391. |
| [1] | 张成辛. 基于文本数据挖掘的蛋白功能预测的机遇与挑战[J]. 合成生物学, 2025, (): 1-14. |
| [2] | 李倩, James E. Ferrell, 陈于平. 细胞质浓度:细胞生物学的老问题、新参数[J]. 合成生物学, 2025, (): 1-18. |
| [3] | 姜百翼, 钱珑. 活细胞记录器在细胞谱系追踪中的应用和前景[J]. 合成生物学, 2025, (): 1-17. |
| [4] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [5] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [6] | 张日新, 田晓军. 合成基因回路面临的细胞‘经济学窘境’[J]. 合成生物学, 2025, (): 1-14. |
| [7] | 谭骁天, 李睿涵, 杨慧. 生物分子传感中的抗体探针:由碳基计算走向硅基计算[J]. 合成生物学, 2025, (): 1-9. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 刘建明, 张炽坚, 张冰, 曾安平. 巴氏梭菌作为工业底盘细胞高效生产1,3-丙二醇——从代谢工程和菌种进化到过程工程和产品分离[J]. 合成生物学, 2024, 5(6): 1386-1403. |
| [10] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [11] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [12] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2024, (): 1-21. |
| [13] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2024, (): 1-36. |
| [14] | 郑梦梦, 刘犇犇, 林芝, 瞿旭东. 重要甾体化合物的化学酶法合成研究进展[J]. 合成生物学, 2024, 5(5): 941-959. |
| [15] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

