• 特约评述 •
活体生物药在代谢类疾病中的研究
匡家奇1, 章素秀1, 江晗2, 魏韬1
- 1.华南农业大学 食品学院,广东 广州 510642
2.华南农业大学 动物与科学学院,广东 广州 510640
-
收稿日期:2025-03-24修回日期:2025-06-16出版日期:2025-06-18 -
通讯作者:魏韬 -
作者简介:匡家奇 (2004—),男,本科生。研究方向为利用合成生物学方法开发活体生物药。E-mail:kjq200416@gmail.com章素秀 (2004—),女,本科生。研究方向为基因组规模代谢网络模型开发。E-mail:Eoxlotl@outlook.com魏韬 (1989—),男,讲师,硕士生导师。研究方向为利用计算生物学与合成生物学方法开发活体生物药等。E-mail:weitao@scau.edu.cn
第一联系人:共同第一作者
Research on live biotherapeutic products in metabolic diseases
KUANG Jiaqi1, ZHANG Suxiu1, JIANG Han2, WEI Tao1
- 1.College of Food Science,South China Agricultural University,Guangzhou 510642,Guangdong,China
2.College of Animal Science,South China Agricultural University,Guangzhou 510640 Guangdong,China
-
Received:2025-03-24Revised:2025-06-16Online:2025-06-18 -
Contact:WEI Tao
摘要:
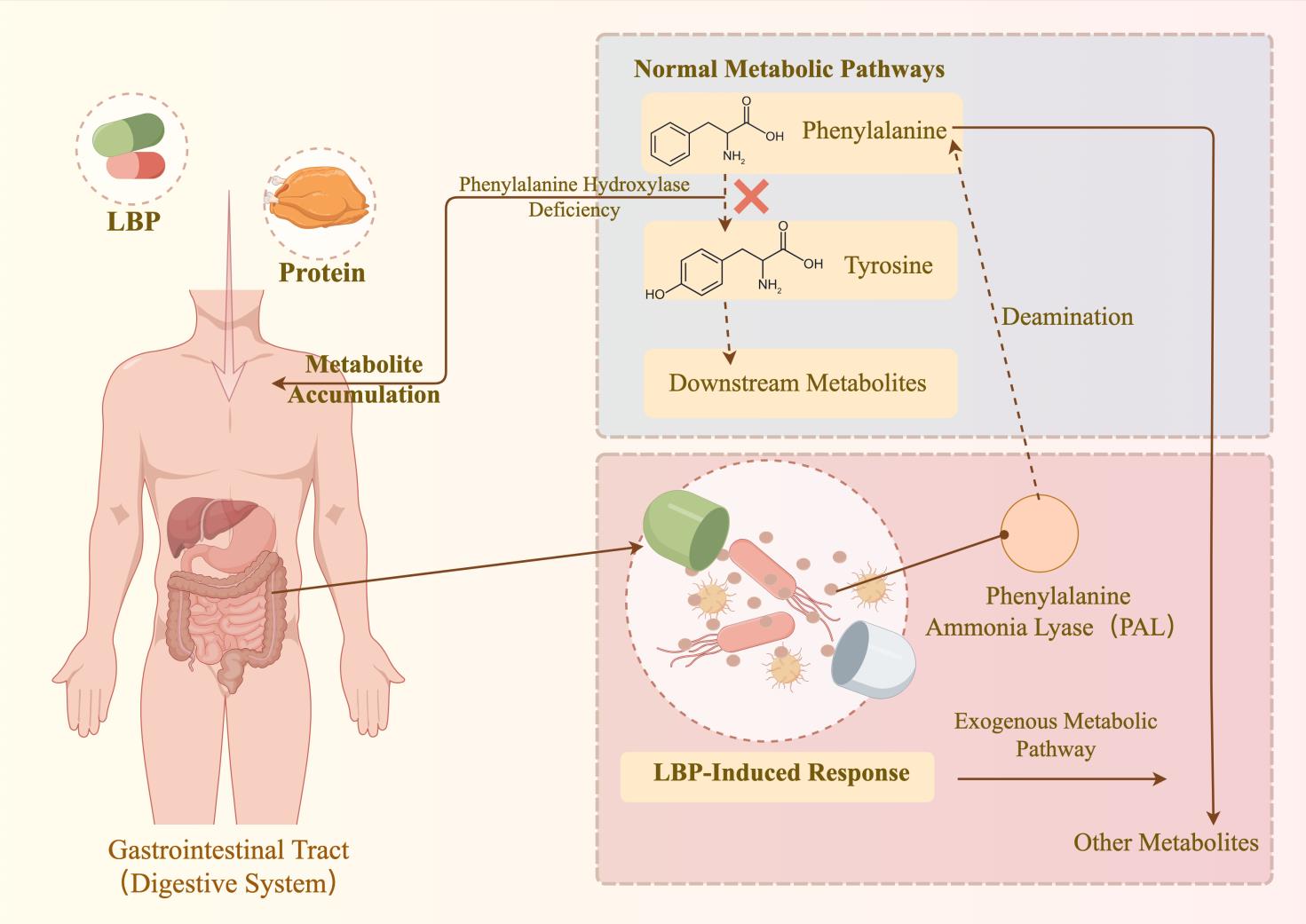
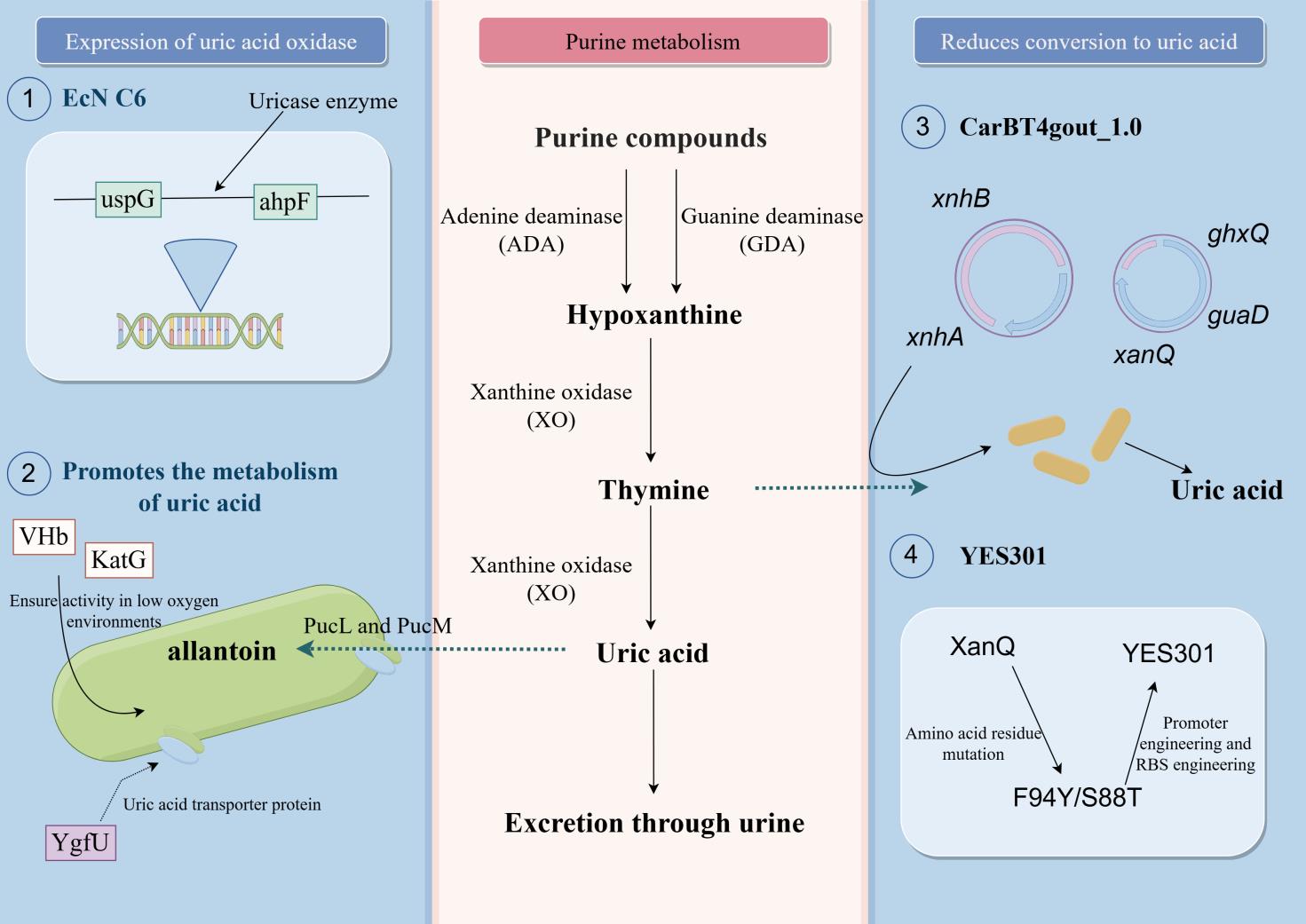
合成生物学技术的持续发展促进了益生菌的改造,进而推动了活体生物药(live biotherapeutic products, LBP)在疾病治疗领域的研究。近年来,LBP在多种疾病治疗剂的开发上有所应用。目前,选择合适的底盘细胞并进行工程化改造,以及为特定疾病设计专门的功能基因模块,已成为活体生物药开发的一般流程。尽管临床实验中针对代谢性疾病的LBP治疗结果尚未完全达到预期,但相关疗法正逐步向临床治疗迈进。本文全面综述了活体生物药在代谢性疾病治疗领域的最新进展,详细阐述了苯丙酮尿症、高尿酸血症和肠源性高草酸尿症等疾病的治疗剂开发。同时,还讨论了基于合成生物学技术的LBP开发策略。最后,总结了活体生物药当前面临的问题,包括安全性、疗效和个体化差异等,并提出了合理的展望,如拓展活体生物药概念、开发个性化活体生物药等。
中图分类号:
引用本文
匡家奇, 章素秀, 江晗, 魏韬. 活体生物药在代谢类疾病中的研究[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-024.
KUANG Jiaqi, ZHANG Suxiu, JIANG Han, WEI Tao. Research on live biotherapeutic products in metabolic diseases[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-024.
| 公司名称 | 所应用的适应症 | 产品名称 | 现状 | 参考文献 |
|---|---|---|---|---|
| Synlogic | 苯丙酮尿症 | SYNB1618和SYNB1934 | 已在美国完成单臂2期研究和Ⅰ期临床试验,目前已停产 | [ |
| 同型胱氨酸尿症 | SYNB1353 | 完成研发,已在美国完成Ⅰ期临床试验 | [ | |
| 高氨血症 | SYNB1020 | 完成研发,已在美国完成Ⅰ期临床试验,目前已停产 | [ | |
| 肠源性高草酸尿症 | SYNB8802 | 完成研发,已在美国完成1b临床研究 | [ | |
| 痛风 | SYNB2081 | 研发中 | [ | |
| 胱氨酸尿症 | 尚未公布 | 尚未公布 | [ | |
| ActoBio Therapeutics | 糖尿病 | AG019 | 完成研发,已在美国完成1b/2a期临床实验,最新进展未公布 | [ |
| 和度生物医药(上海)有限公司 | 胆固醇过高 | 完成研发,未公布最新进展 | [ | |
| 苯丙酮尿症 | 完成研发,正开展临床研究 | [ | ||
| 苏州优信合生技术有限公司 | 完成研发,正开展临床前研究 | [ | ||
| 完成研发,正开展临床前研究 | [ | |||
| 完成研发,正开展临床前研究 | [ |
表1 治疗代谢病活体生物药产业化现状
Table 1 Current status of industrialization of live biotherapeutic products
| 公司名称 | 所应用的适应症 | 产品名称 | 现状 | 参考文献 |
|---|---|---|---|---|
| Synlogic | 苯丙酮尿症 | SYNB1618和SYNB1934 | 已在美国完成单臂2期研究和Ⅰ期临床试验,目前已停产 | [ |
| 同型胱氨酸尿症 | SYNB1353 | 完成研发,已在美国完成Ⅰ期临床试验 | [ | |
| 高氨血症 | SYNB1020 | 完成研发,已在美国完成Ⅰ期临床试验,目前已停产 | [ | |
| 肠源性高草酸尿症 | SYNB8802 | 完成研发,已在美国完成1b临床研究 | [ | |
| 痛风 | SYNB2081 | 研发中 | [ | |
| 胱氨酸尿症 | 尚未公布 | 尚未公布 | [ | |
| ActoBio Therapeutics | 糖尿病 | AG019 | 完成研发,已在美国完成1b/2a期临床实验,最新进展未公布 | [ |
| 和度生物医药(上海)有限公司 | 胆固醇过高 | 完成研发,未公布最新进展 | [ | |
| 苯丙酮尿症 | 完成研发,正开展临床研究 | [ | ||
| 苏州优信合生技术有限公司 | 完成研发,正开展临床前研究 | [ | ||
| 完成研发,正开展临床前研究 | [ | |||
| 完成研发,正开展临床前研究 | [ |
| 底盘细胞 | 拉丁文学名 | 所开发LBP的适应症 | 开发程度 |
|---|---|---|---|
| 大肠杆菌Nissle 1917 | Escherichia coli Nissle 1917 | 苯丙酮尿症、高尿酸血症、肠源性高草酸尿症、慢性肾病、高氨酸血症、同型胱氨酸尿症、肥胖、果糖诱发的代谢综合征及糖尿病等 | 完成基因组注释[ |
| 乳酸杆菌 | Lactobacillus | 苯丙酮尿症[ | 开发多种基因白编辑工具,如CRISPR系统和cre-lox系统[ |
| 丁酸梭菌 | Clostridium butyricum | 糖尿病[ | 开发CRISPR-Cas系统[ |
| 布拉氏酵母 | Saccharomyces boulardii | 高尿酸血症[ | 修复乳糖代谢途径,构建多种诱导表达系统[ |
表2 不同底盘细胞开发LBP现状
Table 2 Current status of LBP development in different chassis cells
| 底盘细胞 | 拉丁文学名 | 所开发LBP的适应症 | 开发程度 |
|---|---|---|---|
| 大肠杆菌Nissle 1917 | Escherichia coli Nissle 1917 | 苯丙酮尿症、高尿酸血症、肠源性高草酸尿症、慢性肾病、高氨酸血症、同型胱氨酸尿症、肥胖、果糖诱发的代谢综合征及糖尿病等 | 完成基因组注释[ |
| 乳酸杆菌 | Lactobacillus | 苯丙酮尿症[ | 开发多种基因白编辑工具,如CRISPR系统和cre-lox系统[ |
| 丁酸梭菌 | Clostridium butyricum | 糖尿病[ | 开发CRISPR-Cas系统[ |
| 布拉氏酵母 | Saccharomyces boulardii | 高尿酸血症[ | 修复乳糖代谢途径,构建多种诱导表达系统[ |
| 1 | PANT A, DAS B. Chapter Ten - Microbiome-based therapeutics: Opportunity and challenges[M]//DAS B, SINGH V. Progress in Molecular Biology and Translational Science: 卷 191. PressAcademic, 2022: 229-262. |
| 2 | SUEZ J, ELINAV E. The path towards microbiome-based metabolite treatment[J]. Nature Microbiology, 2017, 2(6): 1-5. |
| 3 | FAN Y, PEDERSEN O. Gut microbiota in human metabolic health and disease[J]. Nature Reviews Microbiology, 2021, 19(1): 55-71. |
| 4 | KIM D Y, LEE S Y, LEE J Y, et al. Gut microbiome therapy: fecal microbiota transplantation vs live biotherapeutic products[J]. Gut Microbes, 2024. |
| 5 | O of the COMMISSIONER. Early Clinical Trials With Live Biotherapeutic Products: Chemistry, Manufacturing, and Control Information[EB/OL]. FDA, 2021. . |
| 6 | ROUANET A, BOLCA S, BRU A, et al. Live Biotherapeutic Products, A Road Map for Safety Assessment[J]. Frontiers in Medicine, 2020, 7. |
| 7 | BOBER J R, BEISEL C L, NAIR N U. Synthetic Biology Approaches to Engineer Probiotics and Members of the Human Microbiota for Biomedical Applications[J]. Annual Review of Biomedical Engineering, 2018, 20(Volume 20, 2018): 277-300. |
| 8 | VAN SPRONSEN F J, BLAU N, HARDING C, et al. Phenylketonuria[J]. Nature Reviews Disease Primers, 2021, 7(1): 36. |
| 9 | DE GROOT M J, HOEKSMA M, BLAU N, et al. Pathogenesis of cognitive dysfunction in phenylketonuria: Review of hypotheses[J]. Molecular Genetics and Metabolism, 2010, 99: S86-S89. |
| 10 | 杨君, 吴昭英, 张丽丽, 等. 成人苯丙酮尿症[J]. 罕见疾病杂志, 2023, 30(8): 1-2. |
| 11 | BILDER D A, NOEL J K, BAKER E R, et al. Systematic Review and Meta-Analysis of Neuropsychiatric Symptoms and Executive Functioning in Adults With Phenylketonuria[J]. Developmental Neuropsychology, 2016, 41(4): 245-260. |
| 12 | ISABELLA V M, HA B N, CASTILLO M J, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria[J]. Nature Biotechnology, 2018, 36(9): 857-864. |
| 13 | ADOLFSEN K J, CALLIHAN I, MONAHAN C E, et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering[J]. Nature Communications, 2021, 12(1): 6215. |
| 14 | ADOLFSEN K J, CALLIHAN I, MONAHAN C E, et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering[J]. Nature Communications, 2021, 12(1): 6215. |
| 15 | PUURUNEN M K, VOCKLEY J, SEARLE S L, et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study[J]. Nature Metabolism, 2021, 3(8): 1125-1132. |
| 16 | VOCKLEY J, SONDHEIMER N, PUURUNEN M, et al. Efficacy and safety of a synthetic biotic for treatment of phenylketonuria: a phase 2 clinical trial[J]. Nature Metabolism, 2023, 5(10): 1685-1690. |
| 17 | Synlogic Announces Decision to Discontinue Synpheny-3 Study and Provides Corporate Update - Synlogic[EB/OL]. [2025-01-07]. . |
| 18 | KGI-ADMIN. CBT102-A by CommBio Therapeutics for Phenylketonuria (PKU): Likelihood of Approval[EB/OL]. (2024-01-23)[2024-12-20]. . |
| 19 | 王立, 孔繁智, 王斗, 等. 一种重组肠杆菌及其在降解酪氨酸和苯丙氨酸中的应用: CN116790468A[P]. 2023-09-22. |
| 20 | 邹丹阳, 董雨萌, 陈晶瑜. 活体生物药:生物技术推动的创新药研发前沿[J]. 生物工程学报, 2023, 39(4): 1275-1289. |
| 21 | DU L, ZONG Y, LI H, et al. Hyperuricemia and its related diseases: mechanisms and advances in therapy[J]. Signal Transduction and Targeted Therapy, 2024, 9(1): 212. |
| 22 | SORENSEN L B. Role of the intestinal tract in the elimination of uric acid[J]. Arthritis & Rheumatism, 1965, 8(4): 694-706. |
| 23 | GLIOZZI M, MALARA N, MUSCOLI S, et al. The treatment of hyperuricemia[J]. International Journal of Cardiology, 2016, 213: 23-27. |
| 24 | ZHAO R, LI Z, SUN Y, et al. Engineered Escherichia coli Nissle 1917 with urate oxidase and an oxygen-recycling system for hyperuricemia treatment[J]. Gut Microbes, 2022, 14(1): 2070391. |
| 25 | HE L, TANG W, HUANG L, et al. Rational design of a genome-based insulated system in facilitates heterologous uricase expression for hyperuricemia treatment[J]. Bioengineering & Translational Medicine, 2023, 8(2): e10449. |
| 26 | TONG Y, WEI Y, JU Y, et al. Anaerobic purinolytic enzymes enable dietary purine clearance by engineered gut bacteria[J]. Cell Chemical Biology, 2023, 30(9): 1104-1114.e7. |
| 27 | ZOU Z P, LI J L, ZHANG Y F, et al. Empowering probiotics with high xanthine transport for effective hyperuricemia management[J]. Gut Microbes, 2024. |
| 28 | WITTING C, LANGMAN C B, ASSIMOS D, et al. Pathophysiology and Treatment of Enteric Hyperoxaluria[J]. Clinical Journal of the American Society of Nephrology, 2021, 16(3): 487. |
| 29 | HIREMATH S, VISWANATHAN P. Oxalobacter formigenes: A new hope as a live biotherapeutic agent in the management of calcium oxalate renal stones[J]. Anaerobe, 2022, 75: 102572. |
| 30 | LUBKOWICZ D, HORVATH N G, JAMES M J, et al. An engineered bacterial therapeutic lowers urinary oxalate in preclinical models and in silico simulations of enteric hyperoxaluria[J]. Molecular Systems Biology, 2022, 18(3): e10539. |
| 31 | HOPPE B, NIAUDET P, SALOMON R, et al. A randomised Phase I/II trial to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria[J]. Pediatric Nephrology, 2017, 32(5): 781-790. |
| 32 | KALANTAR-ZADEH K, JAFAR T H, NITSCH D, et al. Chronic kidney disease[J]. The Lancet, 2021, 398(10302): 786-802. |
| 33 | BRACK Y, SUN C, YI D, et al. Discovery of Novel Tyrosine Ammonia Lyases for the Enzymatic Synthesis of p-Coumaric Acid[J]. ChemBioChem, 2022, 23(10): e202200062. |
| 34 | LIU Y, XU W, XU W. Production of Trans-Cinnamic and p-Coumaric Acids in Engineered E. coli[J]. Catalysts, 2022, 12(10): 1144. |
| 35 | LUBKOWICZ D, HAVA D L, LEWIS K, et al. Rational Engineering of Escherichia coli Nissle 1917 as Live Biotherapeutic to Degrade Uremic Toxin Precursors[J]. ACS Synthetic Biology, 2024: acssynbio.3c00686. |
| 36 | KRAUS J P, JANOSK M, KO?ICH V, et al. Cystathionine β-synthase mutations in homocystinuria[J]. Human Mutation, 1999, 13(5): 362-375. |
| 37 | KUMAR T, SHARMA G S, SINGH L R. Homocystinuria: Therapeutic approach[J]. Clinica Chimica Acta, 2016, 458: 55-62. |
| 38 | PERREAULT M, MEANS J, GERSON E, et al. The live biotherapeutic SYNB1353 decreases plasma methionine via directed degradation in animal models and healthy volunteers[J]. Cell Host & Microbe, 2024, 32(3): 382-395.e10. |
| 39 | CHARBONNEAU M R, ISABELLA V M, LI N, et al. Developing a new class of engineered live bacterial therapeutics to treat human diseases[J]. Nature Communications, 2020, 11(1): 1738. |
| 40 | BUECHERL L, MYERS C J. Engineering genetic circuits: advancements in genetic design automation tools and standards for synthetic biology[J]. Current Opinion in Microbiology, 2022, 68: 102155. |
| 41 | Synlogic Announces Publication of Synpheny-1 Phase 2 Study of Synthetic Biotic for Phenylketonuria in Nature Metabolism - Synlogic[EB/OL]. [2025-01-07]. . |
| 42 | Synlogic Announces Publication of Preclinical and Clinical Data for SYNB 1353 as a Potential Treatment for Classical Homocystinuria-Synlogic[EB/OL]. [2025-01-07]. . |
| 43 | KURTZ C B, MILLET Y A, PUURUNEN M K, et al An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans[J]. Science Translational Medicine, 2019, 11(475): eaau7975. |
| 44 | Synlogic Discontinues Development of SYNB 1020 to Treat Hyperammonemia-Synlogic[EB/OL]. [2025-01-07]. . |
| 45 | Synlogic Announces Achievement of Proof of Concept for SYNB8802 in Enteric Hyperoxaluria Based on Urinary Oxalate Lowering in Phase 1b Study-Synlogic[EB/OL]. [2025-01-07]. . |
| 46 | BIOWORKS G. Synlogic Announces Synthetic Biotic for Gout Developed in Partnership with Ginkgo Bioworks[EB/OL]. [2025-01-07]. . |
| 47 | Pipeline[EB/OL]. [2025-01-07]. . |
| 48 | Precigen ActoBio Announces Additional Positive Interim Data from Phase 1b/2 Study of AG a 019 ActoBioticsTM , A Novel Therapy Designed to Address the Underlying Cause of Type 1 Diabetes-Precigen[EB/OL]. [2025-01-07]. . |
| 49 | 刘彦强, 左方圆, 向斌. 一种用于催化胆固醇硫酸酯合成的工程微生物及其应用: CN118853719A[P]. 2024-10-29. |
| 50 | KGI-ADMIN. CBT102-A by CommBio Therapeutics for Phenylketonuria (PKU): Likelihood of Approval[EB/OL]. (2024-01-23)[2025-01-07]. . |
| 51 | 优信合生-产品管线[EB/OL]. [2025-01-07]. . |
| 52 | THURSBY E, JUGE N. Introduction to the human gut microbiota[J]. Biochemical Journal, 2017, 474(11): 1823-1836. |
| 53 | SONNENBORN U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties[J]. FEMS microbiology letters, 2016, 363(19): fnw212. |
| 54 | DERIU E, LIU J Z, PEZESHKI M, et al. Probiotic Bacteria Reduce Salmonella Typhimurium Intestinal Colonization by Competing for Iron[J]. Cell Host & Microbe, 2013, 14(1): 26-37. |
| 55 | FÁBREGA M J, RODRÍGUEZ-NOGALES A, GARRIDO-MESA J, 等. Intestinal Anti-inflammatory Effects of Outer Membrane Vesicles from Escherichia coli Nissle 1917 in DSS-Experimental Colitis in Mice[J]. Frontiers in Microbiology, 2017, 8: 1274. |
| 56 | REISTER M, HOFFMEIER K, KREZDORN N, et al. Complete genome sequence of the Gram-negative probiotic Escherichia coli strain Nissle 1917[J]. Journal of Biotechnology, 2014, 187: 106-107. |
| 57 | KAN A, GELFAT I, EMANI S, et al. Plasmid Vectors for in Vivo Selection-Free Use with the Probiotic E. coli Nissle 1917[J]. ACS Synthetic Biology, 2021, 10(1): 94-106. |
| 58 | ZAINUDDIN H S, BAI Y, MANSELL T J. CRISPR‐based curing and analysis of metabolic burden of cryptic plasmids in Escherichia coli Nissle 1917[J]. Engineering in Life Sciences, 2019, 19(6): 478-485. |
| 59 | ZHOU S, ZHAO L, ZUO W, et al. Minimizing Endogenous Cryptic Plasmids to Construct Antibiotic-Free Expression Systems for Escherichia Coli Nissle 1917[R]. SSRN, 2023. |
| 60 | KAN A, GELFAT I, EMANI S, et al. Plasmid Vectors for in Vivo Selection-Free Use with the Probiotic E. coli Nissle 1917[J]. ACS Synthetic Biology, 2021, 10(1): 94-106. |
| 61 | YU X, LIN C, YU J, et al. Bioengineered Escherichia coli Nissle 1917 for tumour-targeting therapy[J]. Microbial Biotechnology, 2020, 13(3): 629-636. |
| 62 | SAEZ-LARA M J, GOMEZ-LLORENTE C, PLAZA-DIAZ J, et al. The Role of Probiotic Lactic Acid Bacteria and Bifidobacteria in the Prevention and Treatment of Inflammatory Bowel Disease and Other Related Diseases: A Systematic Review of Randomized Human Clinical Trials[J]. BioMed Research International, 2015, 2015(1): 505878. |
| 63 | LU K, DONG S, WU X, et al. Probiotics in Cancer[J]. Frontiers in Oncology, 2021, 11: 638148. |
| 64 | WANG C, CUI Y, QU X. Optimization of electrotransformation (ETF) conditions in lactic acid bacteria (LAB)[J]. Journal of Microbiological Methods, 2020, 174: 105944. |
| 65 | O'SULLIVAN D J, KLAENHAMMER T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening[J]. Gene, 1993, 137(2): 227-231. |
| 66 | BAO S, ZHU L, ZHUANG Q, et al. Distribution Dynamics of Recombinant Lactobacillus in the Gastrointestinal Tract of Neonatal Rats[J]. PLOS ONE, 2013, 8(3): e60007. |
| 67 | WALKER D C, KLAENHAMMER T R. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp[J]. Journal of Bacteriology, 1994, 176(17): 5330-5340. |
| 68 | ENYEART P J, CHIRIELEISON S M, DAO M N, et al. Generalized bacterial genome editing using mobile group II introns and Cre‐lox[J]. Molecular Systems Biology, 2013, 9(1): 685. |
| 69 | ZHU D, ZHAO K, XU H, et al. Construction of thyA deficient Lactococcus lactis using the Cre-loxP recombination system[J]. Annals of Microbiology, 2015, 65(3): 1659-1665. |
| 70 | BERLEC A, ŠKRLEC K, KOCJAN J, et al. Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis[J]. Scientific Reports, 2018, 8(1): 1009. |
| 71 | KIELISZEK M, POBIEGA K, PIWOWAREK K, et al. Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria[J]. Molecules, 2021, 26(7): 1858. |
| 72 | KONGO J M. Lactic Acid Bacteria: R[M]. BoD-Books on Demand, 2013. |
| 73 | ZIELIŃSKA D, KOLOŻYN-KRAJEWSKA D. Food-Origin Lactic Acid Bacteria May Exhibit Probiotic Properties: Review[J]. BioMed Research International, 2018, 2018(1): 1-15. |
| 74 | PINTO A, BARBOSA J, ALBANO H, et al. Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics[J]. Microorganisms, 2020, 8(3): 393. |
| 75 | SOFI M H, WU Y, TICER T, et al. A single strain of Bacteroides fragilis protects gut integrity and reduces GVHD[J]. JCI Insight, 2021, 6(3): e136841. |
| 76 | LUO X, KONG Q, WANG Y, et al. Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition[J]. Microorganisms, 2021, 9(8): 1573. |
| 77 | ZHAO X, YANG J, JU Z, et al. Clostridium butyricum Ameliorates Salmonella Enteritis Induced Inflammation by Enhancing and Improving Immunity of the Intestinal Epithelial Barrier at the Intestinal Mucosal Level[J]. Frontiers in Microbiology, 2020, 11: 299. |
| 78 | WANG C, LI W, WANG H, et al. Saccharomyces boulardii alleviates ulcerative colitis carcinogenesis in mice by reducing TNF-α and IL-6 levels and functions and by rebalancing intestinal microbiota[J]. BMC Microbiology, 2019, 19(1): 246. |
| 79 | STOEVA M K, GARCIA-SO J, JUSTICE N, et al. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease[J]. Gut Microbes, 2021, 13(1): 1907272. |
| 80 | LEE S M, DONALDSON G P, MIKULSKI Z, et al. Bacterial colonization factors control specificity and stability of the gut microbiota[J]. Nature, 2013, 501(7467): 426-429. |
| 81 | CHARBONNEAU M R, ISABELLA V M, LI N, et al. Developing a new class of engineered live bacterial therapeutics to treat human diseases[J]. Nature Communications, 2020, 11(1): 1738. |
| 82 | LIU C H, CHANG J H, CHANG Y C, et al. Treatment of murine colitis by Saccharomyces boulardii secreting atrial natriuretic peptide[J]. Journal of Molecular Medicine, 2020, 98(12): 1675-1687. |
| 83 | MA M, ZHAO Z, LIANG Q, et al. Overexpression of pEGF improved the gut protective function of Clostridium butyricum partly through STAT3 signal pathway[J]. Applied Microbiology and Biotechnology, 2021, 105(14-15): 5973-5991. |
| 84 | DURMUSOGLU D, AL'ABRI I S, COLLINS S P, et al. In Situ Biomanufacturing of Small Molecules in the Mammalian Gut by Probiotic Saccharomyces boulardii[J]. ACS Synthetic Biology, 2021, 10(5): 1039-1052. |
| 85 | WANG Y, CHEN W, HAN Y, et al. Neuroprotective effect of engineered Clostridium butyricum-pMTL007-GLP-1 on Parkinson’s disease mice models via promoting mitophagy[J]. Bioengineering & Translational Medicine, 2023, 8(3): e10505. |
| 86 | ZHOU D, LI S, HU G, et al. Hypoglycemic effect of C. butyricum-pMTL007-GLP-1 engineered probiotics on type 2 diabetes mellitus[J]. Gut Microbes, 2025, 17(1): 2447814. |
| 87 | WANG W, PAN L, HE H, et al. Systematic Engineering for Efficient Uric Acid-Degrading Activity in Probiotic Yeast Saccharomyces boulardii[J]. ACS Synthetic Biology, 2025. |
| 88 | KWAK S, MAHMUD B, DANTAS G. A Tunable and Expandable Transactivation System in Probiotic Yeast Saccharomyces boulardii[J]. ACS Synthetic Biology, 2022, 11(1): 508-514. |
| 89 | DURMUSOGLU D, HALLER D J, AL'ABRI I S, et al. Programming Probiotics: Diet-Responsive Gene Expression and Colonization Control in Engineered S. boulardii[J]. ACS Synthetic Biology, 2024, 13(6): 1851-1865. |
| 90 | ZHOU X, WANG X, LUO H, et al. Exploiting heterologous and endogenous CRISPR-Cas systems for genome editing in the probiotic Clostridium butyricum[J]. Biotechnology and Bioengineering, 2021, 118(7): 2448-2459. |
| 91 | ZHENG L, TAN Y, HU Y, et al. CRISPR/Cas-Based Genome Editing for Human Gut Commensal Bacteroides Species[J]. ACS Synthetic Biology, 2022, 11(1): 464-472. |
| 92 | GROZDANOV L, RAASCH C, SCHULZE J, et al. Analysis of the Genome Structure of the Nonpathogenic Probiotic Escherichia coli Strain Nissle 1917[J]. Journal of Bacteriology, 2004, 186(16): 5432-5441. |
| 93 | BA F, ZHANG Y, JI X, et al. Expanding the toolbox of probiotic Escherichia coli Nissle 1917 for synthetic biology[J]. Biotechnology Journal, 2024, 19(1): 2300327. |
| 94 | FANG M, ZHANG R, WANG C, et al. Engineering probiotic Escherichia coli Nissle 1917 to block transfer of multiple antibiotic resistance genes by exploiting a type I CRISPR-Cas system[A]. 2024. |
| 95 | PRAVESCHOTINUNT P, DURAJ-THATTE A M, GELFAT I, et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut[J]. Nature Communications, 2019, 10(1): 5580. |
| 96 | DURRER K E, ALLEN M S, HUNT VON HERBING I. Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU[J]. PLOS ONE, 2017, 12(5): e0176286. |
| 97 | DUAN F F, LIU J H, MARCH J C. Engineered Commensal Bacteria Reprogram Intestinal Cells Into Glucose-Responsive Insulin-Secreting Cells for the Treatment of Diabetes[J]. Diabetes, 2015, 64(5): 1794-1803. |
| 98 | GOH Y J, BARRANGOU R. Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli[J]. Current Opinion in Biotechnology, 2019, 56: 163-171. |
| 99 | MU Y, ZHANG C, LI T, et al. Development and Applications of CRISPR/Cas9-Based Genome Editing in Lactobacillus[J]. International Journal of Molecular Sciences, 2022, 23(21): 12852. |
| 100 | PEI Z, LIU Y, YI Z, et al. Diversity within the species Clostridium butyricum: pan-genome, phylogeny, prophage, carbohydrate utilization, and antibiotic resistance[J]. Journal of Applied Microbiology, 2023, 134(7): lxad127. |
| 101 | SLUSARCZYK A L, LIN A, WEISS R. Foundations for the design and implementation of synthetic genetic circuits[J]. Nature Reviews Genetics, 2012, 13(6): 406-420. |
| 102 | XIA P F, LING H, FOO J L, et al. Synthetic genetic circuits for programmable biological functionalities[J]. Biotechnology Advances, 2019, 37(6): 107393. |
| 103 | HASTY J, MCMILLEN D, COLLINS J J. Engineered gene circuits[J]. Nature, 2002, 420(6912): 224-230. |
| 104 | SPRINZAK D, ELOWITZ M B. Reconstruction of genetic circuits[J]. Nature, 2005, 438(7067): 443-448. |
| 105 | ZENG W, GUO L, XU S, et al. High-Throughput Screening Technology in Industrial Biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 106 | ISHIBASHI N, YAMAZAKI S. Probiotics and safety[J]. The American Journal of Clinical Nutrition, 2001, 73(2): 465s-470s. |
| 107 | FULLER R. Probiotics in human medicine.[J]. Gut, 1991, 32(4): 439-442. |
| 108 | MA Y, MANNA A, MOON T S. Advances in engineering genetic circuits for microbial biocontainment[J]. Current Opinion in Systems Biology, 2023, 36: 100483. |
| 109 | WANG F, ZHANG W. Synthetic biology: Recent progress, biosafety and biosecurity concerns, and possible solutions[J]. Journal of Biosafety and Biosecurity, 2019, 1(1): 22-30. |
| 110 | ROTTINGHAUS A G, FERREIRO A, FISHBEIN S R S, et al. Genetically stable CRISPR-based kill switches for engineered microbes[J]. Nature Communications, 2022, 13(1): 672. |
| 111 | KARBALAEI-HEIDARI H R, BUDISA N. Advanced and Safe Synthetic Microbial Chassis with Orthogonal Translation System Integration[J]. ACS Synthetic Biology, 2024, 13(9): 2992-3002. |
| 112 | SHAO J, XUE S, YU G, et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice[J]. Science Translational Medicine, 2017, 9(387): eaal2298. |
| 113 | KRAWCZYK K, XUE S, BUCHMANN P, et al. Electrogenetic cellular insulin release for real-time glycemic control in type 1 diabetic mice[J]. Science, 2020, 368(6494): 993-1001. |
| 114 | CHENG A G, HO P Y, ARANDA-DÍAZ A, et al. Design, construction, and in vivo augmentation of a complex gut microbiome[J]. Cell, 2022, 185(19): 3617-3636.e19. |
| [1] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [2] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [3] | 鲁锦畅, 武耀康, 吕雪芹, 刘龙, 陈坚, 刘延峰. 神经酰胺类鞘脂的绿色生物制造[J]. 合成生物学, 2025, 6(2): 422-444. |
| [4] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [5] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [6] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [7] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [8] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [9] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [10] | 郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健. 乳酸菌的合成生物学工具及在合成益肤因子中的应用[J]. 合成生物学, 2025, 6(2): 320-333. |
| [11] | 张萍, 张维娇, 胥睿睿, 李江华, 陈坚, 康振. 防晒化合物类菌孢素氨基酸的生物合成[J]. 合成生物学, 2025, 6(2): 306-319. |
| [12] | 黄姝涵, 马赫, 罗云孜. 生物合成红景天苷的研究进展[J]. 合成生物学, 2025, 6(2): 391-407. |
| [13] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [14] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [15] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||