合成生物学 ›› 2025, Vol. 6 ›› Issue (5): 998-1024.DOI: 10.12211/2096-8280.2025-065
合成生物学在农业中的进展及挑战
刘婕1, 郜钰1, 马永硕1,2, 尚轶1
- 1.云南省马铃薯生物学重点实验室,马铃薯科学研究院,云南师范大学,云南 昆明 650500
2.麻省理工学院化学工程系,美国 马萨诸塞州 剑桥市 02142
-
收稿日期:2025-06-23修回日期:2025-09-07出版日期:2025-10-31发布日期:2025-11-05 -
通讯作者:马永硕,尚轶 -
作者简介:刘婕 (1996—),女,博士研究生。研究方向为植物天然产物在植物以及微生物的高效合成。E-mail:liujie19960901@163.com马永硕 (1986—),男,研究员,博士生导师。研究方向为合成生物学与代谢工程。E-mail:mayongshuo2000@163.com尚轶 (1982—),男,研究员,博士生导师。研究方向为作物营养与风味品质,合成生物学。E-mail:shangyi@ynnu.edu.cn -
基金资助:国家自然科学基金委联合基金重点项目(U2202206);云南省“兴滇英才支持计划”云岭学者专项(XDYC-YLXZ-2022-0019)
Progress and challenges of synthetic biology in agriculture
LIU Jie1, GAO Yu1, MA Yongshuo1,2, SHANG Yi1
- 1.Key Laboratory for Potato Biology of Yunnan Province,The CAAS-YNNU-YINMORE Joint Academy of Potato Science,Yunnan Normal University,Kunming 650500,Yunnan,China
2.Department of Chemical Engineering,Massachusetts Institute of Technology,Cambridge 02142,Massachusetts,USA
-
Received:2025-06-23Revised:2025-09-07Online:2025-10-31Published:2025-11-05 -
Contact:MA Yongshuo, SHANG Yi
摘要:
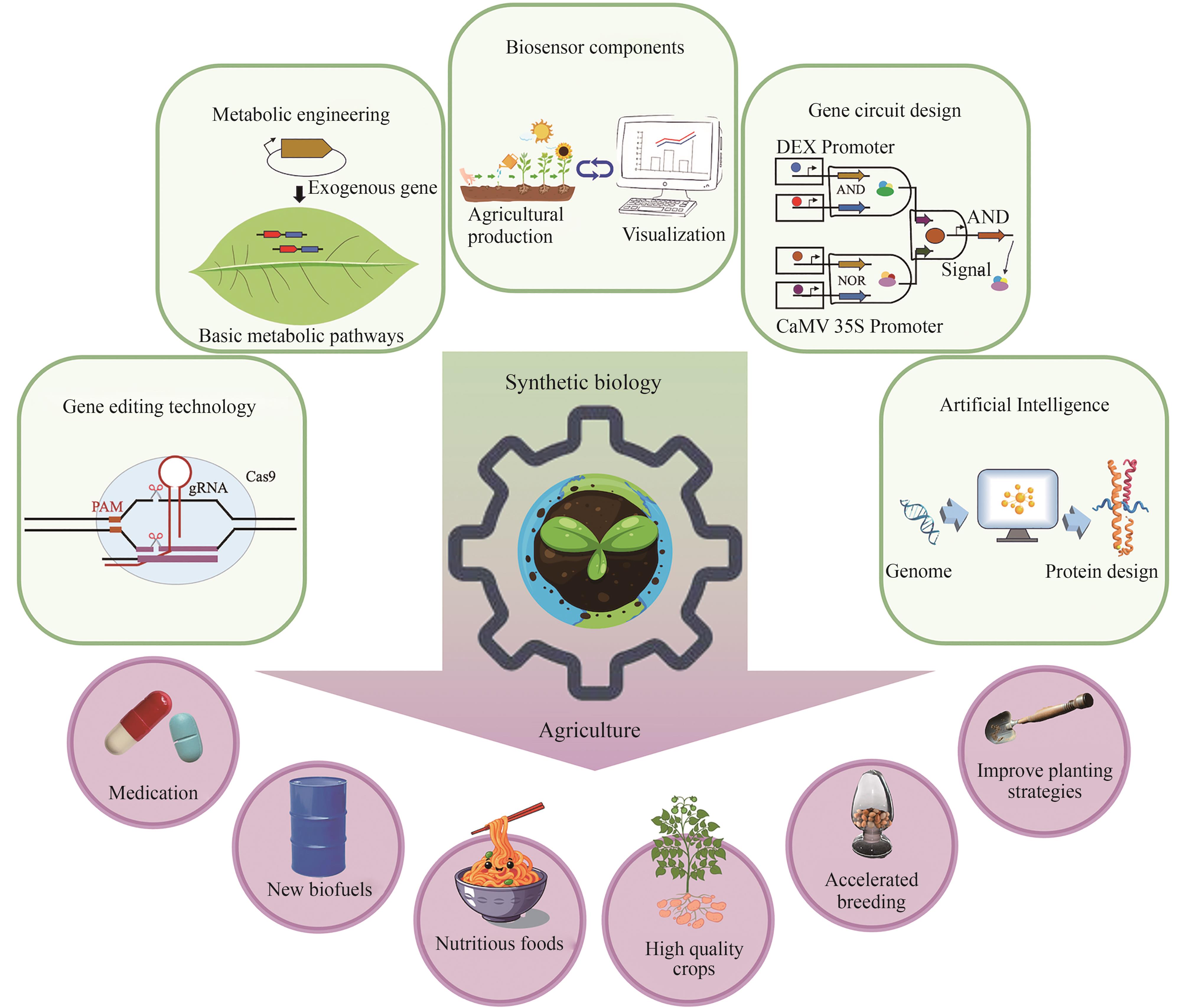
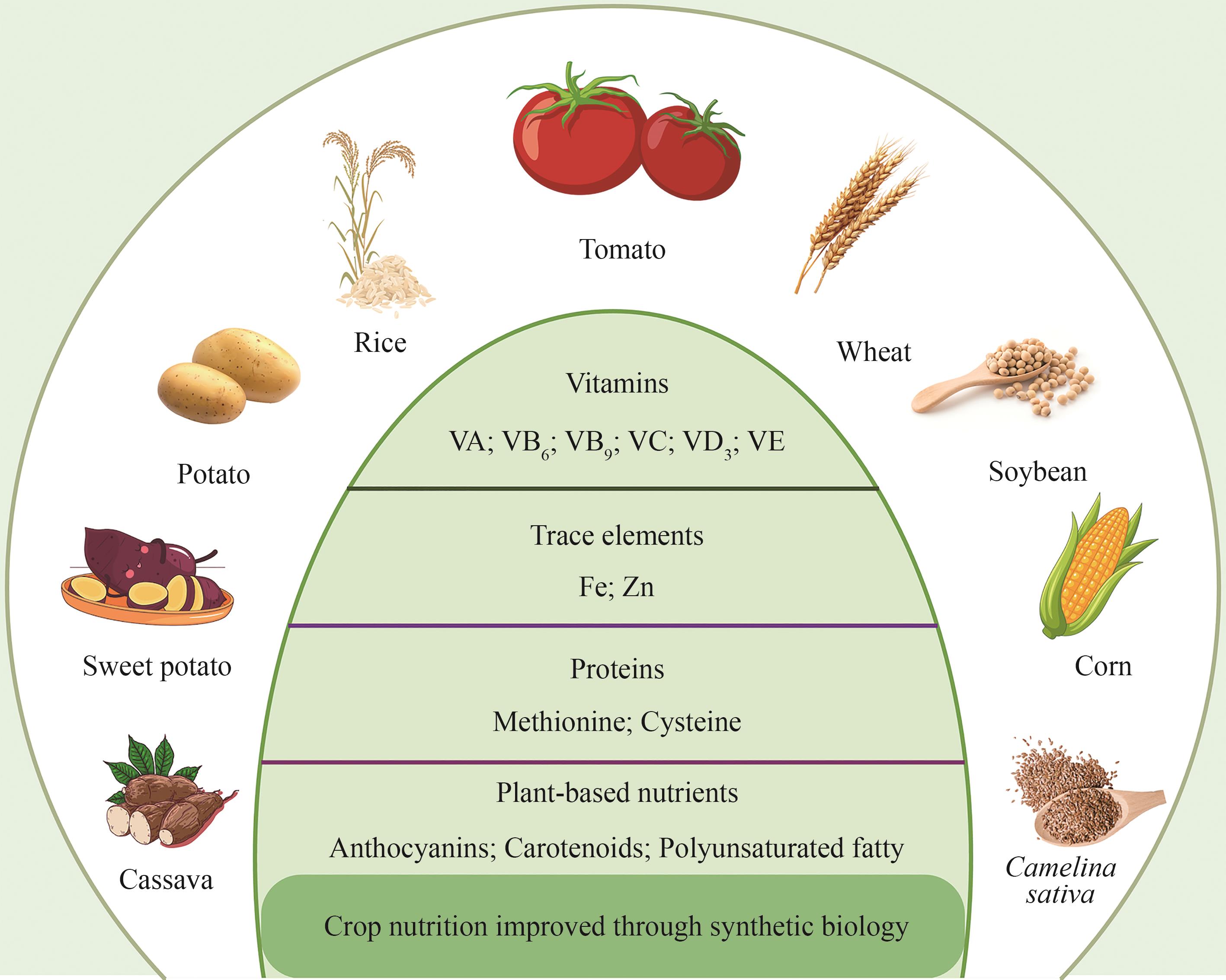
合成生物学通过工程化设计与新生命系统构建,为农业带来了革命性的突破。与传统农业技术相比,合成生物学汇聚农业科技领域的高新技术,可以更高效、更广泛地解决光合作用、生物固氮、作物抗逆、农业生态可持续性等世界性农业难题。合成生物技术不仅可以提高作物产量和优化营养品质,还可以利用生物质副产物产生健康的肥料和土壤,实现废弃物资源化循环的新模式,是应对人口增加和气候变化、促进生物经济可持续发展的战略制高点。本文回顾了农业合成生物学的发展历程,综述了基因编辑技术、代谢工程策略、生物传感器元件开发、基因回路设计、人工智能等在农业中广泛应用的合成生物技术的最新研究进展。阐述了合成生物学在农业中的核心应用,包括提高作物产量和资源利用率、增强抗逆性、作物营养强化以及改善微生物互作等方面。合成生物学在农业领域的多维应用,将有效保障粮食安全并助力未来农业可持续发展。
中图分类号:
引用本文
刘婕, 郜钰, 马永硕, 尚轶. 合成生物学在农业中的进展及挑战[J]. 合成生物学, 2025, 6(5): 998-1024.
LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture[J]. Synthetic Biology Journal, 2025, 6(5): 998-1024.
| Year | Findings | References |
|---|---|---|
| 2005 | 二代黄金大米的总胡萝卜素得到23倍的增加 | [ |
| 2011 | 植物传感器开始发展 | [ |
| 2012 | 番茄中抗坏血酸含量的增加 | [ |
| 2014 | 谷类作物中固氮途径的引入,增加作物固氮量 | [ |
| 2014 | 油料作物种子中高效合成ω-3不饱和脂肪酸 | [ |
| 2016 | 烟草叶片中青蒿素的合成 | [ |
| 2017 | 番茄果实中GABA的大量积累 | [ |
| 2017 | 大麦中引入固氮系统,提高氮利用效率 | [ |
| 2018 | 水稻胚乳中虾青素的生物合成 | [ |
| 2019 | 烟草中引入光呼吸途径,增加C3作物的产量 | [ |
| 2019 | 水稻中引入光呼吸旁路,增加光合效率 | [ |
| 2020 | 初级编辑器在作物中的应用 | [ |
| 2021 | C3作物中C4高光效特征的模拟 | [ |
| 2022 | 烟草中马钱子碱的生物合成途径重构 | [ |
| 2024 | 烟草中疫苗佐剂QS-21的生物合成途径重构 | [ |
表1 合成生物学在农业领域的关键成果
Table1 Key achievements of synthetic biology in agriculture
| Year | Findings | References |
|---|---|---|
| 2005 | 二代黄金大米的总胡萝卜素得到23倍的增加 | [ |
| 2011 | 植物传感器开始发展 | [ |
| 2012 | 番茄中抗坏血酸含量的增加 | [ |
| 2014 | 谷类作物中固氮途径的引入,增加作物固氮量 | [ |
| 2014 | 油料作物种子中高效合成ω-3不饱和脂肪酸 | [ |
| 2016 | 烟草叶片中青蒿素的合成 | [ |
| 2017 | 番茄果实中GABA的大量积累 | [ |
| 2017 | 大麦中引入固氮系统,提高氮利用效率 | [ |
| 2018 | 水稻胚乳中虾青素的生物合成 | [ |
| 2019 | 烟草中引入光呼吸途径,增加C3作物的产量 | [ |
| 2019 | 水稻中引入光呼吸旁路,增加光合效率 | [ |
| 2020 | 初级编辑器在作物中的应用 | [ |
| 2021 | C3作物中C4高光效特征的模拟 | [ |
| 2022 | 烟草中马钱子碱的生物合成途径重构 | [ |
| 2024 | 烟草中疫苗佐剂QS-21的生物合成途径重构 | [ |
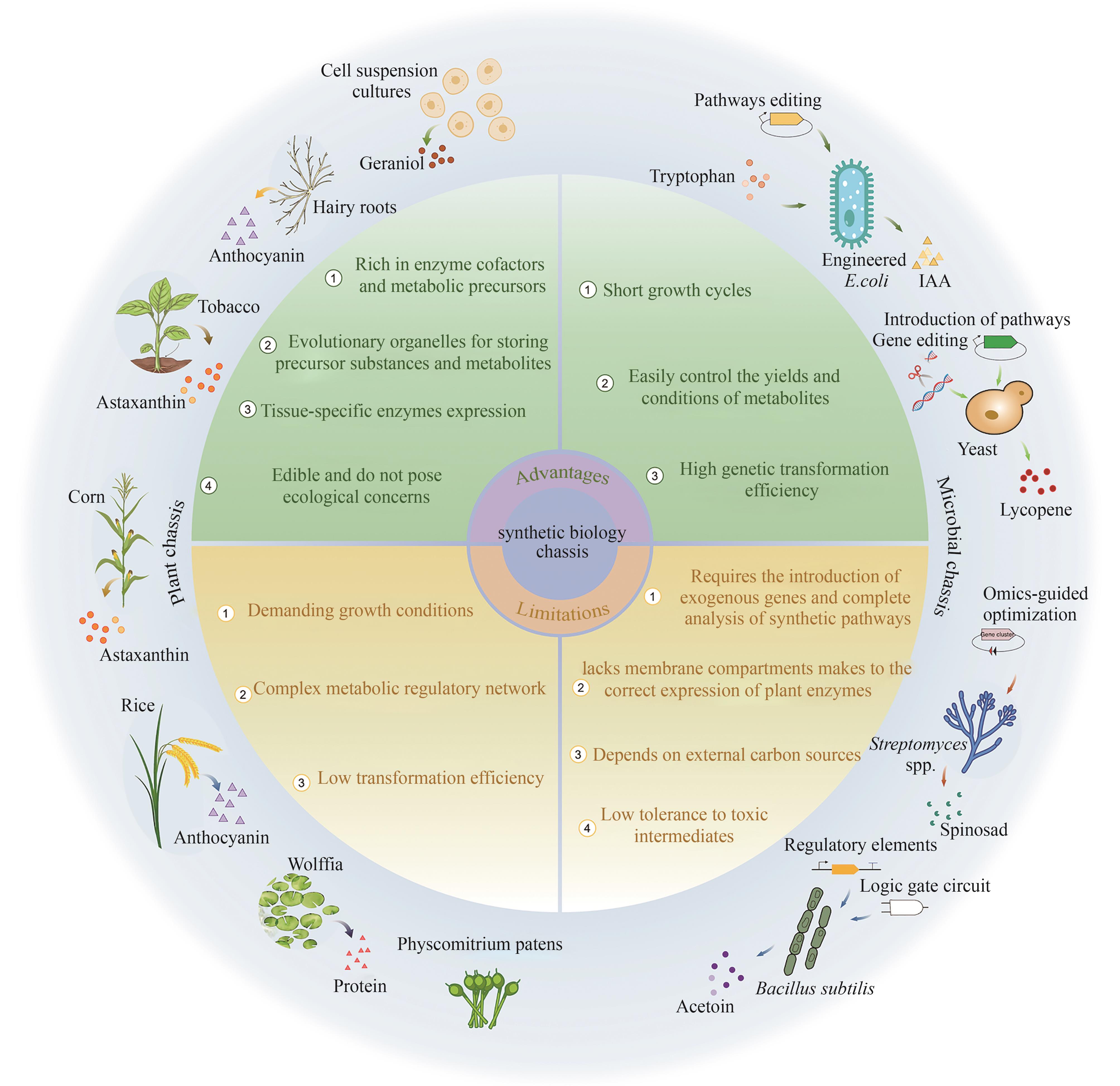
| 优/缺点 | 植物底盘 | 微生物底盘 |
|---|---|---|
| 优点 | 富含辅酶因子和前体物质 高度进化的细胞器可以储存前体物质和代谢物 组织特异性的酶表达模式 可以直接食用 | 生长周期短 培养条件和产量可控性强 基因转化效率高 |
| 缺点 | 生长环境要求高 代谢调控网络复杂 转化效率低 | 需要外源基因引入且依赖完整的途径解析 缺乏植物来源的酶表达所需的隔膜系统 依赖外部碳源 对有毒中间产物耐受性低 |
表2 植物和微生物底盘的优缺点
Table2 Advantages and disadvantages of plant chassis and microbial chassis
| 优/缺点 | 植物底盘 | 微生物底盘 |
|---|---|---|
| 优点 | 富含辅酶因子和前体物质 高度进化的细胞器可以储存前体物质和代谢物 组织特异性的酶表达模式 可以直接食用 | 生长周期短 培养条件和产量可控性强 基因转化效率高 |
| 缺点 | 生长环境要求高 代谢调控网络复杂 转化效率低 | 需要外源基因引入且依赖完整的途径解析 缺乏植物来源的酶表达所需的隔膜系统 依赖外部碳源 对有毒中间产物耐受性低 |
| [106] | WU R, DUAN L N, PRUNEDA-PAZ J L, et al. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation[J]. Plant Physiology, 2018, 177(4): 1650-1665. |
| [107] | MÜLLER B, SHEEN J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis[J]. Nature, 2008, 453(7198): 1094-1097. |
| [108] | RIZZA A, WALIA A, TANG B J, et al. Visualizing cellular gibberellin levels using the nlsGPS1 Förster resonance energy transfer (FRET) biosensor[J]. Journal of Visualized Experiments, 2019(143): e58739. |
| [109] | LARRIEU A, CHAMPION A, LEGRAND J, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants[J]. Nature Communications, 2015, 6: 6043. |
| [110] | POUVREAU B, VANHERCKE T, SINGH S. From plant metabolic engineering to plant synthetic biology: the evolution of the design/build/test/learn cycle[J]. Plant Science, 2018, 273: 3-12. |
| [111] | WONG M H, GIRALDO J P, KWAK S Y, et al. Nitroaromatic detection and infrared communication from wild-type plants using plant nanobionics[J]. Nature Materials, 2017, 16(2): 264-272. |
| [112] | PENG Y H, ALLEN S, MILLWOOD R J, et al. ‘Fukusensor’: a genetically engineered plant for reporting DNA damage in response to gamma radiation[J]. Plant Biotechnology Journal, 2014, 12(9): 1329-1332. |
| [113] | JEZ J M, LEE S G, SHERP A M. The next green movement: Plant biology for the environment and sustainability[J]. Science, 2016, 353(6305): 1241-1244. |
| [114] | FICHMAN Y, MILLER G, MITTLER R. Whole-plant live imaging of reactive oxygen species[J]. Molecular Plant, 2019, 12(9): 1203-1210. |
| [115] | MCADAM E L, REID J B, FOO E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation[J]. Journal of Experimental Botany, 2018, 69(8): 2117-2130. |
| [116] | CRUZ A P Z, FERREIRA V, PIANZZOLA M J, et al. A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain[J]. Molecular Plant-Microbe Interactions, 2014, 27(3): 277-285. |
| [117] | MOHAMMAD-RAZDARI A, ROUSSEAU D, BAKHSHIPOUR A, et al. Recent advances in E-monitoring of plant diseases[J]. Biosensors and Bioelectronics, 2022, 201: 113953. |
| [118] | JUGDER B E, ERTAN H, BOHL S, et al. Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation[J]. Frontiers in Microbiology, 2016, 7: 249. |
| [119] | SAHA G, SHAHRIN F, KHAN F H, et al. Smart IoT-driven precision agriculture: land mapping, crop prediction, and irrigation system[J]. PLoS One, 2025, 20(3): e0319268. |
| [120] | CUZICK A, MAGUIRE K, HAMMOND-KOSACK K E. Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers[J]. New Phytologist, 2009, 181(4): 901-912. |
| [121] | JEONG H J, JUNG K H. Rice tissue-specific promoters and condition-dependent promoters for effective translational application[J]. Journal of Integrative Plant Biology, 2015, 57(11): 913-924. |
| [122] | JUSIAK B, CLETO S, PEREZ-PIÑERA P, et al. Engineering synthetic gene circuits in living cells with CRISPR technology[J]. Trends in Biotechnology, 2016, 34(7): 535-547. |
| [123] | LLOYD J P B, LY F, GONG P, et al. Synthetic memory circuits for stable cell reprogramming in plants[J]. Nature Biotechnology, 2022, 40(12): 1862-1872. |
| [124] | KHAN M A, HERRING G, ZHU J Y, et al. CRISPRi-based circuits to control gene expression in plants[J]. Nature Biotechnology, 2025, 43(3): 416-430. |
| [125] | WEINBERG B H, HANG PHAM N T, CARABALLO L D, et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells[J]. Nature Biotechnology, 2017, 35(5): 453-462. |
| [126] | GUIZIOU S, MARANAS C J, CHU J C, et al. An integrase toolbox to record gene-expression during plant development[J]. Nature Communications, 2023, 14: 1844. |
| [127] | LI S S, LI Z L, TAN G Y, et al. In vitro allosteric transcription factor-based biosensing[J]. Trends in Biotechnology, 2023, 41(8): 1080-1095. |
| [128] | FERREIRA S S, ANTUNES M S. Genetically encoded Boolean logic operators to sense and integrate phenylpropanoid metabolite levels in plants[J]. New Phytologist, 2024, 243(2): 674-687. |
| [129] | LIANG Y, RICHARDSON S, YAN J W, et al. Endoribonuclease-based two-component repressor systems for tight gene expression control in plants[J]. ACS Synthetic Biology, 2017, 6(5): 806-816. |
| [1] | LUCIDO A, BASALLO O, MARIN-SANGUINO A, et al. Multiscale mathematical modeling in systems biology: a framework to boost plant synthetic biology[J]. Plants, 2025, 14(3): 470. |
| [2] | KE J, WANG B, YOSHIKUNI Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture[J]. Trends in Biotechnology, 2021, 39(3): 244-261. |
| [3] | 张博, 马永硕, 尚轶, 等. 植物合成生物学研究进展[J]. 合成生物学, 2020, 1(2): 121-140. |
| ZHANG B, MA Y S, SHANG Y, et al. Recent advances in plant synthetic biology[J]. Synthetic Biology Journal, 2020, 1(2): 121-140. | |
| [4] | BROPHY J A N, MAGALLON K J, DUAN L N, et al. Synthetic genetic circuits as a means of reprogramming plant roots[J]. Science, 2022, 377(6607): 747-751. |
| [5] | ZHANG X N, LIU C L, DAI J B, et al. Enabling technology and core theory of synthetic biology[J]. Science China Life Sciences, 2023, 66(8): 1742-1785. |
| [6] | DOUDNA J A, CHARPENTIER E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| [7] | TIAN Y S, WANG B, PENG R H, et al. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering[J]. Plant Biotechnology Journal, 2019, 17(5): 849-851. |
| [8] | XIONG D L. Perspectives of improving rice photosynthesis for higher grain yield[J]. Crop and Environment, 2024, 3(3): 123-137. |
| [9] | 燕永亮, 田长富, 杨建国, 等. 人工高效生物固氮体系创建及其农业应用[J]. 生命科学, 2021, 33(12): 1532-1543. |
| YAN Y L, TIAN C F, YANG J G, et al. Establishment of artificial efficiency biological nitrogen fixation system and its agricultural application[J]. Chinese Bulletin of Life Sciences, 2021, 33(12): 1532-1543. | |
| [10] | 潘明慧, 杨雪, 杜国忠, 等. 合成生物学助力链霉菌天然产物农药创制与产业化[J]. 植物保护, 2023, 49(5): 371-389. |
| [130] | KASENIIT K E, KATZ N, KOLBER N S, et al. Modular, programmable RNA sensing using ADAR editing in living cells[J]. Nature Biotechnology, 2023, 41(4): 482-487. |
| [131] | KONERMANN S, LOTFY P, BRIDEAU N J, et al. Transcriptome engineering with RNA-targeting type Ⅵ-D CRISPR effectors[J]. Cell, 2018, 173(3): 665-676.e14. |
| [132] | MARAND A P, CHEN Z L, GALLAVOTTI A, et al. A cis-regulatory atlas in maize at single-cell resolution[J]. Cell, 2021, 184(11): 3041-3055.e21. |
| [133] | HE Z H, LUO Y T, ZHOU X K, et al. scPlantDB: a comprehensive database for exploring cell types and markers of plant cell atlases[J]. Nucleic Acids Research, 2024, 52(D1): D1629-D1638. |
| [134] | RAI K, WANG Y D, O’CONNELL R W, et al. Using machine learning to enhance and accelerate synthetic biology[J]. Current Opinion in Biomedical Engineering, 2024, 31: 100553. |
| [135] | LAM H Y I, ONG X E, MUTWIL M. Large language models in plant biology[J]. Trends in Plant Science, 2024, 29(10): 1145-1155. |
| [136] | ZHU W C, HAN R, SHANG X Y, et al. The CropGPT project: Call for a global, coordinated effort in precision design breeding driven by AI using biological big data[J]. Molecular Plant, 2024, 17(2): 215-218. |
| [137] | JI Y R, ZHOU Z H, LIU H, et al. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome[J]. Bioinformatics, 2021, 37(15): 2112-2120. |
| [138] | ABRAMSON J, ADLER J, DUNGER J, et al. Addendum: accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 636(8042): E4. |
| [139] | CUI H T, WANG C, MAAN H, et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI[J]. Nature Methods, 2024, 21(8): 1470-1480. |
| [140] | WANG J, ZHANG L, WANG S D, et al. AlphaFold-guided bespoke gene editing enhances field-grown soybean oil contents[J]. Advanced Science, 2025, 12(23): 2500290. |
| [141] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [10] | PAN M H, YANG X, DU G Z, et al. The exploitation and bio-manufacture of natural product pesticides from Streptomyces by synthetic biology[J]. Plant Protection, 2023, 49(5): 371-389. |
| [11] | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| [12] | YE X, AL-BABILI S, KLÖTI A, et al. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm[J]. Science, 2000, 287(5451): 303-305. |
| [13] | BHULLAR S, CHAKRAVARTHY S, ADVANI S, et al. Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping[J]. Plant Physiology, 2003, 132(2): 988-998. |
| [14] | ENGLER C, KANDZIA R, MARILLONNET S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647. |
| [15] | ENGLER C, GRUETZNER R, KANDZIA R, et al. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes[J]. PLoS One, 2009, 4(5): e5553. |
| [16] | AMOS M. Population-based microbial computing: a third wave of synthetic biology?[J]. International Journal of General Systems, 2014, 43(7): 770-782. |
| [17] | FISHER A K, FREEDMAN B G, BEVAN D R, et al. A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories[J]. Computational and Structural Biotechnology Journal, 2014, 11(18): 91-99. |
| [18] | LU X F, VORA H, KHOSLA C. Overproduction of free fatty acids in E. coli: implications for biodiesel production[J]. Metabolic Engineering, 2008, 10(6): 333-339. |
| [19] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [20] | SHAN Q W, WANG Y P, LI J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system[J]. Nature Biotechnology, 2013, 31(8): 686-688. |
| [21] | CHAN A N, WANG L L, ZHU Y J, et al. Identification through fine mapping and verification using CRISPR/Cas9-targeted mutagenesis for a minor QTL controlling grain weight in rice[J]. Theoretical and Applied Genetics, 2021, 134(1): 327-337. |
| [141] | SONG C Z, LIN Y H. AI-enabled directed evolution for protein engineering and optimization[J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [142] | SHARMA A, JAIN A, GUPTA P, et al. Machine learning applications for precision agriculture: a comprehensive review[J]. IEEE Access, 2021, 9: 4843-4873. |
| [143] | LIAKOS K G, BUSATO P, MOSHOU D, et al. Machine learning in agriculture: a review[J]. Sensors, 2018, 18(8): 2674. |
| [144] | ESLAMI M, ADLER A, CACERES R S, et al. Artificial intelligence for synthetic biology[J]. Communications of the ACM, 2022, 65(5): 88-97. |
| [145] | YANG J S, REYNA-LLORENS I. Plant synthetic biology: exploring the frontiers of sustainable agriculture and fundamental plant biology[J]. Journal of Experimental Botany, 2023, 74(13): 3787-3790. |
| [146] | ORR D J, ALCÂNTARA A, KAPRALOV M V, et al. Surveying Rubisco diversity and temperature response to improve crop photosynthetic efficiency[J]. Plant Physiology, 2016, 172(2): 707-717. |
| [147] | LIN M T, OCCHIALINI A, ANDRALOJC P J, et al. A faster Rubisco with potential to increase photosynthesis in crops[J]. Nature, 2014, 513(7519): 547-550. |
| [148] | MANNING T, BIRCH R, STEVENSON T, et al. Bacterial Form Ⅱ Rubisco can support wild-type growth and productivity in Solanum tuberosum cv. Desiree (potato) under elevated CO2 [J]. PNAS Nexus, 2023, 2(2): pgac305. |
| [149] | GUNN L H, MARTIN AVILA E, BIRCH R, et al. The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(41): 25890-25896. |
| [150] | MATSUMURA H, SHIOMI K, YAMAMOTO A, et al. Hybrid rubisco with complete replacement of rice Rubisco small subunits by sorghum counterparts confers C4 plant-like high catalytic activity[J]. Molecular Plant, 2020, 13(11): 1570-1581. |
| [151] | MAO Y W, CATHERALL E, DÍAZ-RAMOS A, et al. The small subunit of Rubisco and its potential as an engineering target[J]. Journal of Experimental Botany, 2023, 74(2): 543-561. |
| [152] | ADLER L, DÍAZ-RAMOS A, MAO Y W, et al. New horizons for building pyrenoid-based CO2-concentrating mechanisms in plants to improve yields[J]. Plant Physiology, 2022, 190(3): 1609-1627. |
| [22] | JARVIS D E, HO Y S, LIGHTFOOT D J, et al. The genome of Chenopodium quinoa [J]. Nature, 2017, 542(7641): 307-312. |
| [23] | ZIMIN A V, PUIU D, HALL R, et al. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum [J]. GigaScience, 2017, 6(11): 1-7. |
| [24] | YU W C, YAU Y Y, BIRCHLER J A. Plant artificial chromosome technology and its potential application in genetic engineering[J]. Plant Biotechnology Journal, 2016, 14(5): 1175-1182. |
| [25] | XU C H, BIRCHLER J A. Editorial: Plant artificial chromosomes: progress and perspectives[J]. Frontiers in Plant Science, 2023, 14: 1290386. |
| [26] | RAVIKUMAR S, BAYLON M G, PARK S J, et al. Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery[J]. Microbial Cell Factories, 2017, 16(1): 62. |
| [27] | ARACIC S, MANNA S, PETROVSKI S, et al. Innovative biological approaches for monitoring and improving water quality[J]. Frontiers in Microbiology, 2015, 6: 826. |
| [28] | NEETHIRAJAN S, RAGAVAN V, WENG X, et al. Biosensors for sustainable food engineering: challenges and perspectives[J]. Biosensors, 2018, 8(1): 23. |
| [29] | 刘华梅, 许国建. 微生物农药苏云金杆菌G033A[J]. 农药科学与管理, 2018, 39(4): 59-60. |
| LIU H M, XU G J. Microbial pesticide Bacillus thuringiensis G033A[J]. Pesticide Science and Administration, 2018, 39(4): 59-60. | |
| [30] | 王晓梅, 李辉尚, 杨小薇. 全球农业合成生物学发展现状及对中国的启示[J]. 农业展望, 2023, 19(4): 71-76. |
| WANG X M, LI H S, YANG X W. Development status of global agricultural synthetic biology and its enlightenment to China[J]. Agricultural Outlook, 2023, 19(4): 71-76. | |
| [31] | WALTZ E. GABA-enriched tomato is first CRISPR-edited food to enter market[J]. Nature Biotechnology, 2022, 40(1): 9-11. |
| [153] | GONG H Y, LI Y, FANG G, et al. Transgenic rice expressing ictb and FBP/sbpase derived from cyanobacteria exhibits enhanced photosynthesis and mesophyll conductance to CO2 [J]. PLoS One, 2015, 10(10): e0140928. |
| [154] | LONG B M, HEE W Y, SHARWOOD R E, et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts[J]. Nature Communications, 2018, 9: 3570. |
| [155] | CHEN T Y, HOJKA M, DAVEY P, et al. Engineering α-carboxysomes into plant chloroplasts to support autotrophic photosynthesis[J]. Nature Communications, 2023, 14: 2118. |
| [156] | FURBANK R T. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types?[J]. Journal of Experimental Botany, 2011, 62(9): 3103-3108. |
| [157] | FURBANK R, KELLY S, VON CAEMMERER S. Photosynthesis and food security: the evolving story of C4 rice[J]. Photosynthesis Research, 2023, 158(2): 121-130. |
| [158] | ERMAKOVA M, DANILA F R, FURBANK R T, et al. On the road to C4 rice: advances and perspectives[J]. The Plant Journal, 2020, 101(4): 940-950. |
| [159] | ERMAKOVA M, ARRIVAULT S, GIULIANI R, et al. Installation of C4 photosynthetic pathway enzymes in rice using a single construct[J]. Plant Biotechnology Journal, 2021, 19(3): 575-588. |
| [160] | SMITH E N, VAN AALST M, TOSENS T, et al. Improving photosynthetic efficiency toward food security: strategies, advances, and perspectives[J]. Molecular Plant, 2023, 16(10): 1547-1563. |
| [161] | LIN X L, LONG Y M, YAO Z, et al. Synthetic photorespiratory bypass more stably increases potato yield per plant by improving photosynthesis[J]. Plant Biotechnology Journal, 2025, 23(7): 2526-2536. |
| [162] | MO B Q, CHEN X F, YANG J J, et al. Engineering of photorespiration-dependent glycine betaine biosynthesis improves photosynthetic carbon fixation and panicle architecture in rice[J]. Journal of Integrative Plant Biology, 2025, 67(4): 979-992. |
| [163] | WALTZ E. A new crop of microbe startups raises big bucks, takes on the establishment [J]. Nature Biotechnology, 2017, 35(12): 1120-1122. |
| [164] | ALLEN R S, TILBROOK K, WARDEN A C, et al. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix[J]. Frontiers in Plant Science, 2017, 8: 287. |
| [32] | PAINE J A, SHIPTON C A, CHAGGAR S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content[J]. Nature Biotechnology, 2005, 23(4): 482-487. |
| [33] | ANTUNES M S, MOREY K J, SMITH J J, et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway[J]. PLoS One, 2011, 6(1): e16292. |
| [34] | BULLEY S, WRIGHT M, ROMMENS C, et al. Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase[J]. Plant Biotechnology Journal, 2012, 10(4): 390-397. |
| [35] | ROGERS C, OLDROYD G E D. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals[J]. Journal of Experimental Botany, 2014, 65(8): 1939-1946. |
| [36] | RUIZ-LOPEZ N, HASLAM R P, NAPIER J A, et al. Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop[J]. The Plant Journal, 2014, 77(2): 198-208. |
| [37] | WANG B, KASHKOOLI A B, SALLETS A, et al. Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua [J]. Metabolic Engineering, 2016, 38: 159-169. |
| [38] | NONAKA S, ARAI C, TAKAYAMA M, et al. Efficient increase of γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis[J]. Scientific Reports, 2017, 7: 7057. |
| [39] | PERCHLIK M, TEGEDER M. Improving plant nitrogen use efficiency through alteration of amino acid transport processes[J]. Plant Physiology, 2017, 175(1): 235-247. |
| [40] | ZHU Q L, ZENG D C, YU S Z, et al. From golden rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm[J]. Molecular Plant, 2018, 11(12): 1440-1448. |
| [41] | SOUTH P F, CAVANAGH A P, LIU H W, et al. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field[J]. Science, 2019, 363(6422): eaat9077. |
| [42] | SHEN B R, WANG L M, LIN X L, et al. Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice[J]. Molecular Plant, 2019, 12(2): 199-214. |
| [43] | LIN Q P, ZONG Y, XUE C X, et al. Prime genome editing in rice and wheat[J]. Nature Biotechnology, 2020, 38(5): 582-585. |
| [165] | BIRCHLER J A, SWYERS N C. Engineered minichromosomes in plants [J]. Experimental Cell Research, 2020, 388(2): 111852. |
| [166] | OLDROYD G E, DIXON R. Biotechnological solutions to the nitrogen problem[J]. Current Opinion in Biotechnology, 2014, 26: 19-24. |
| [167] | BAGESHWAR U K, SRIVASTAVA M, PARDHA-SARADHI P, et al. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield[J]. Applied and Environmental Microbiology, 2017, 83(15): e00590-17. |
| [168] | GEDDES B A, PARAMASIVAN P, JOFFRIN A, et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria[J]. Nature Communications, 2019, 10: 3430. |
| [169] | BLOCH S E, CLARK R, GOTTLIEB S S, et al. Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph[J]. Journal of Experimental Botany, 2020, 71(15): 4591-4603. |
| [170] | CHRISTIAENS O, TARDAJOS M G, MARTINEZ REYNA Z L, et al. Increased RNAi efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers[J]. Frontiers in Physiology, 2018, 9: 316. |
| [171] | HUAN Y C, KONG Q, MOU H J, et al. Antimicrobial peptides: classification, design, application and research progress in multiple fields[J]. Frontiers in Microbiology, 2020, 11: 582779. |
| [172] | MARCOS J F, MUÑOZ A, PÉREZ-PAYÁ E, et al. Identification and rational design of novel antimicrobial peptides for plant protection[J]. Annual Review of Phytopathology, 2008, 46: 273-301. |
| [173] | MONTESINOS E. Functional peptides for plant disease control[J]. Annual Review of Phytopathology, 2023, 61: 301-324. |
| [174] | ZEITLER B, BERNHARD A, MEYER H, et al. Production of a de-novo designed antimicrobial peptide in Nicotiana benthamiana [J]. Plant Molecular Biology, 2013, 81(3): 259-272. |
| [175] | HOLÁSKOVÁ E, GALUSZKA P, MIČÚCHOVÁ A, et al. Molecular farming in barley: development of a novel production platform to produce human antimicrobial peptide LL-37[J]. Biotechnology Journal, 2018, 13(6): e1700628. |
| [176] | MIRZAEE M, HOLÁSKOVÁ E, MIČÚCHOVÁ A, et al. Long-lasting stable expression of human LL-37 antimicrobial peptide in transgenic barley plants[J]. Antibiotics, 2021, 10(8): 898. |
| [44] | ROELL M S, SCHADA VON BORZYKOWSKI L, WESTHOFF P, et al. A synthetic C4 shuttle via the β-hydroxyaspartate cycle in C3 plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(21): e2022307118. |
| [45] | HONG B K, GRZECH D, CAPUTI L, et al. Biosynthesis of strychnine[J]. Nature, 2022, 607(7919): 617-622. |
| [46] | MARTIN L B B, KIKUCHI S, REJZEK M, et al. Complete biosynthesis of the potent vaccine adjuvant QS-21[J]. Nature Chemical Biology, 2024, 20(4): 493-502. |
| [47] | WU H X, YANG J H, SHEN P J, et al. High-level production of indole-3-acetic acid in the metabolically engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2021, 69(6): 1916-1924. |
| [48] | ZHANG Y, CHIU T Y, ZHANG J T, et al. Systematical engineering of synthetic yeast for enhanced production of lycopene[J]. Bioengineering, 2021, 8(1): 14. |
| [49] | ZHANG Y, YUAN M D, WU X X, et al. The construction and optimization of engineered yeast chassis for efficient biosynthesis of 8-hydroxygeraniol[J]. mLife, 2023, 2(4): 438-449. |
| [50] | ZHU Y, LI J X, PENG L Y, et al. High-yield production of protopanaxadiol from sugarcane molasses by metabolically engineered Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2022, 21(1): 230. |
| [51] | TAN G Y, DENG K H, LIU X H, et al. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces [J]. ACS Synthetic Biology, 2017, 6(6): 995-1005. |
| [52] | WANG Q, BAO T, HU M K, et al. Efficient acetoin production in Bacillus subtilis by multivariate modular metabolic engineering with spatiotemporal modulation[J]. ACS Sustainable Chemistry & Engineering, 2025, 13(5): 1927-1936. |
| [53] | SABATE R, DE GROOT N S, VENTURA S. Protein folding and aggregation in bacteria[J]. Cellular and Molecular Life Sciences, 2010, 67(16): 2695-2715. |
| [54] | TIAN Y, KONG L Z, LI Q, et al. Structural diversity, evolutionary origin, and metabolic engineering of plant specialized benzylisoquinoline alkaloids[J]. Natural Product Reports, 2024, 41(11): 1787-1810. |
| [55] | SUN Q Y, DING L W, LOMONOSSOFF G P, et al. Improved expression and purification of recombinant human serum albumin from transgenic tobacco suspension culture[J]. Journal of Biotechnology, 2011, 155(2): 164-172. |
| [177] | BUNDÓ M, SHI X Q, VERNET M, et al. Rice seeds as biofactories of rationally designed and cell-penetrating antifungal PAF peptides[J]. Frontiers in Plant Science, 2019, 10: 731. |
| [178] | KOCH A, KUMAR N, WEBER L, et al. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(48): 19324-19329. |
| [179] | SÚNICO V, HIGUERA J J, MOLINA-HIDALGO F J, et al. The intragenesis and synthetic biology approach towards accelerating genetic gains on strawberry: development of new tools to improve fruit quality and resistance to pathogens[J]. Plants, 2022, 11(1): 57. |
| [180] | MOORE B D, ANDREW R L, KÜLHEIM C, et al. Explaining intraspecific diversity in plant secondary metabolites in an ecological context[J]. New Phytologist, 2014, 201(3): 733-750. |
| [181] | AHUJA I, KISSEN R, BONES A M. Phytoalexins in defense against pathogens[J]. Trends in Plant Science, 2012, 17(2): 73-90. |
| [182] | HU H, LI J J, DELATTE T, et al. Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids[J]. Plant Biotechnology Journal, 2018, 16(8): 1434-1445. |
| [183] | MITTLER R, BLUMWALD E. Genetic engineering for modern agriculture: challenges and perspectives[J]. Annual Review of Plant Biology, 2010, 61: 443-462. |
| [184] | SHI H T, YE T T, CHAN Z L. Comparative proteomic responses of two bermudagrass (Cynodon dactylon (L). Pers.) varieties contrasting in drought stress resistance[J]. Plant Physiology and Biochemistry, 2014, 82: 218-228. |
| [185] | HU H H, XIONG L Z. Genetic engineering and breeding of drought-resistant crops[J]. Annual Review of Plant Biology, 2014, 65: 715-741. |
| [186] | CHEN S J, XU K, KONG D Y, et al. Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2;1[J]. Plant Biotechnology Journal, 2022, 20(9): 1743-1755. |
| [187] | DING L, MILHIET T, PARENT B, et al. The plasma membrane aquaporin ZmPIP2;5 enhances the sensitivity of stomatal closure to water deficit[J]. Plant, Cell & Environment, 2022, 45(4): 1146-1156. |
| [188] | XU Y, HU W, LIU J H, et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.)[J]. Plant Physiology and Biochemistry, 2020, 147: 66-76. |
| [56] | VASILEV N, SCHMITZ C, DONG L M, et al. Comparison of plant-based expression platforms for the heterologous production of geraniol[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2014, 117(3): 373-380. |
| [57] | YOUSEFIAN S, LOHRASEBI T, FARHADPOUR M, et al. Effect of methyl jasmonate on phenolic acids accumulation and the expression profile of their biosynthesis-related genes in Mentha spicata hairy root cultures[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2020, 142(2): 285-297. |
| [58] | WU S J, XIE X G, FENG K M, et al. Transcriptome sequencing and signal transduction for the enhanced tanshinone production in Salvia miltiorrhiza hairy roots induced by Trichoderma atroviride D16 polysaccharide fraction[J]. Bioscience, Biotechnology, and Biochemistry, 2022, 86(8): 1049-1059. |
| [59] | KUŹMA Ł, KISIEL W, KRÓLICKA A, et al. Genetic transformation of Salvia austriaca by Agrobacterium rhizogenes and diterpenoid isolation[J]. Die Pharmazie, 2011, 66(11): 904-907. |
| [60] | LIU J, ZHAO Y X, ZHANG J M, et al. Production of species-specific anthocyanins through an inducible system in plant hairy roots[J]. Metabolic Engineering, 2024, 81: 182-196. |
| [61] | NOGUEIRA M, ENFISSI E M A, WELSCH R, et al. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: a new tool for engineering ketocarotenoids[J]. Metabolic Engineering, 2019, 52: 243-252. |
| [62] | BEYRAGHDAR KASHKOOLI A, VAN DER KROL A R, RABE P, et al. Substrate promiscuity of enzymes from the sesquiterpene biosynthetic pathways from Artemisia annua and Tanacetum parthenium allows for novel combinatorial sesquiterpene production[J]. Metabolic Engineering, 2019, 54: 12-23. |
| [63] | FORESTIER E C F, CZECHOWSKI T, CORDING A C, et al. Developing a Nicotiana benthamiana transgenic platform for high-value diterpene production and candidate gene evaluation[J]. Plant Biotechnology Journal, 2021, 19(8): 1614-1623. |
| [64] | ZHU Q L, YU S Z, ZENG D C, et al. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system[J]. Molecular Plant, 2017, 10(7): 918-929. |
| [65] | LIU X Q, MA X H, WANG H, et al. Metabolic engineering of astaxanthin-rich maize and its use in the production of biofortified eggs[J]. Plant Biotechnology Journal, 2021, 19(9): 1812-1823. |
| [66] | LAM E, MICHAEL T P. Wolffia, a minimalist plant and synthetic biology chassis[J]. Trends in Plant Science, 2022, 27(5): 430-439. |
| [67] | BI G Q, ZHAO S J, YAO J W, et al. Near telomere-to-telomere genome of the model plant Physcomitrium patens [J]. Nature Plants, 2024, 10(2): 327-343. |
| [189] | SAJA-GARBARZ D, LIBIK-KONIECZNY M, FELLNER M, et al. Silicon-induced alterations in the expression of aquaporins and antioxidant system activity in well-watered and drought-stressed oilseed rape[J]. Plant Physiology and Biochemistry, 2022, 174: 73-86. |
| [190] | HOSSEINIFARD M, STEFANIAK S, GHORBANI JAVID M, et al. Contribution of exogenous proline to abiotic stresses tolerance in plants: a review[J]. International Journal of Molecular Sciences, 2022, 23(9): 5186. |
| [191] | ARAÚJO W L, NUNES-NESI A, OSORIO S, et al. Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture[J]. The Plant Cell, 2011, 23(2): 600-627. |
| [192] | MOVAHEDI A, DZINYELA R, AGHAEI-DARGIRI S, et al. Advanced study of drought-responsive protein pathways in plants[J]. Agronomy, 2023, 13(3): 849. |
| [193] | EDWARDS R A, NG X Y, TUCKER M R, et al. Plant synthetic biology as a tool to help eliminate hidden hunger[J]. Current Opinion in Biotechnology, 2024, 88: 103168. |
| [194] | BEYENE G, SOLOMON F R, CHAUHAN R D, et al. Provitamin A biofortification of cassava enhances shelf life but reduces dry matter content of storage roots due to altered carbon partitioning into starch[J]. Plant Biotechnology Journal, 2018, 16(6): 1186-1200. |
| [195] | LOW J W, MWANGA R O M, ANDRADE M, et al. Tackling vitamin A deficiency with biofortified sweetpotato in sub-Saharan Africa[J]. Global Food Security, 2017, 14: 23-30. |
| [196] | LI K T, MOULIN M, MANGEL N, et al. Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency[J]. Nature Biotechnology, 2015, 33(10): 1029-1032. |
| [197] | DE LEPELEIRE J, STROBBE S, VERSTRAETE J, et al. Folate biofortification of potato by Tuber-specific expression of four folate biosynthesis genes[J]. Molecular Plant, 2018, 11(1): 175-188. |
| [198] | BULLEY S, LAING W. The regulation of ascorbate biosynthesis[J]. Current Opinion in Plant Biology, 2016, 33: 15-22. |
| [199] | WANG L Y, MENG X, YANG D Y, et al. Overexpression of tomato GDP-L-galactose phosphorylase gene in tobacco improves tolerance to chilling stress[J]. Plant Cell Reports, 2014, 33(9): 1441-1451. |
| [200] | ALI B, PANTHA S, ACHARYA R, et al. Enhanced ascorbate level improves multi-stress tolerance in a widely grown indica rice variety without compromising its agronomic characteristics[J]. Journal of Plant Physiology, 2019, 240: 152998. |
| [201] | MA L C, WANG Y R, LIU W X, et al. Overexpression of an alfalfa GDP-mannose 3,5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation[J]. Biotechnology Letters, 2014, 36(11): 2331-2341. |
| [202] | LI X J, YE J, MUNIR S, et al. Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato[J]. International Journal of Molecular Sciences, 2019, 20(7): 1558. |
| [203] | ZHANG G Y, LIU R R, ZHANG C Q, et al. Manipulation of the rice L-galactose pathway: evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance[J]. PLoS One, 2015, 10(5): e0125870. |
| [204] | LI J, SCARANO A, GONZALEZ N M, et al. Biofortified tomatoes provide a new route to vitamin D sufficiency[J]. Nature Plants, 2022, 8(6): 611-616. |
| [205] | UPADHYAYA D C, BAGRI D S, UPADHYAYA C P, et al. Genetic engineering of potato (Solanum tuberosum L.) for enhanced α-tocopherols and abiotic stress tolerance[J]. Physiologia Plantarum, 2021, 173(1): 116-128. |
| [206] | BOONYAVES K, WU T Y, GRUISSEM W, et al. Enhanced grain iron levels in rice expressing an IRON-REGULATED METAL TRANSPORTER, NICOTIANAMINE SYNTHASE, and FERRITIN gene cassette[J]. Frontiers in Plant Science, 2017, 8: 130. |
| [207] | NARAYANAN N, BEYENE G, CHAUHAN R D, et al. Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin[J]. Nature Biotechnology, 2019, 37(2): 144-151. |
| [208] | HARRINGTON S A, CONNORTON J M, NYANGOMA N I M, et al. A two-gene strategy increases iron and zinc concentrations in wheat flour, improving mineral bioaccessibility[J]. Plant Physiology, 2023, 191(1): 528-541. |
| [209] | NAGESH C R, PRASHAT G R, GOSWAMI S, et al. Sulfate transport and metabolism: strategies to improve the seed protein quality[J]. Molecular Biology Reports, 2024, 51(1): 242. |
| [210] | KIM W S, SUN-HYUNG J, OEHRLE N W, et al. Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman-Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds[J]. Scientific Reports, 2020, 10: 14989. |
| [211] | HUANG Y C, WANG H H, ZHU Y D, et al. THP9 enhances seed protein content and nitrogen-use efficiency in maize[J]. Nature, 2022, 612(7939): 292-300. |
| [212] | LEE S, PARK J, LEE J, et al. OsASN1 overexpression in rice increases grain protein content and yield under nitrogen-limiting conditions[J]. Plant & Cell Physiology, 2020, 61(7): 1309-1320. |
| [213] | LIU X Q, LI S Z, YANG W Z, et al. Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich maize[J]. Plant & Cell Physiology, 2018, 59(10): 1942-1955. |
| [214] | GONZALI S, PERATA P. Anthocyanins from purple tomatoes as novel antioxidants to promote human health[J]. Antioxidants, 2020, 9(10): 1017. |
| [215] | BEYER P, AL-BABILI S, YE X D, et al. Golden Rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency[J]. The Journal of Nutrition, 2002, 132(3): 506S-510S. |
| [216] | HAN L H, SILVESTRE S, SAYANOVA O, et al. Using field evaluation and systematic iteration to rationalize the accumulation of omega-3 long-chain polyunsaturated fatty acids in transgenic Camelina sativa [J]. Plant Biotechnology Journal, 2022, 20(9): 1833-1852. |
| [217] | RYU M H, ZHANG J, TOTH T, et al. Control of nitrogen fixation in bacteria that associate with cereals[J]. Nature Microbiology, 2020, 5(2): 314-330. |
| [218] | SHULSE C N, CHOVATIA M, AGOSTO C, et al. Engineered root bacteria release plant-available phosphate from phytate[J]. Applied and Environmental Microbiology, 2019, 85(18): e01210-19. |
| [219] | SHAO J H, LI S Q, ZHANG N, et al. Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9[J]. Microbial Cell Factories, 2015, 14: 130. |
| [220] | ZÚÑIGA A, DE LA FUENTE F, FEDERICI F, et al. An engineered device for indoleacetic acid production under quorum sensing signals enables Cupriavidus pinatubonensis JMP134 to stimulate plant growth[J]. ACS Synthetic Biology, 2018, 7(6): 1519-1527. |
| [221] | TRDÁ L, BAREŠOVÁ M, ŠAŠEK V, et al. Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase[J]. Frontiers in Microbiology, 2017, 8: 1374. |
| [222] | SALOMON M V, BOTTINI R, DE SOUZA FILHO G A, et al. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine[J]. Physiologia Plantarum, 2014, 151(4): 359-374. |
| [223] | MULLINS A J, MURRAY J A H, BULL M J, et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria [J]. Nature Microbiology, 2019, 4(6): 996-1005. |
| [224] | LI Z L, HUANG P J, WANG M Y, et al. Stepwise increase of thaxtomins production in Streptomyces albidoflavus J1074 through combinatorial metabolic engineering[J]. Metabolic Engineering, 2021, 68: 187-198. |
| [225] | LIU Y P, ZHU A P, TAN H M, et al. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana[J]. Microbiome, 2019, 7(1): 74. |
| [226] | SANATI NEZHAD A. Microfluidic platforms for plant cells studies[J]. Lab on a Chip, 2014, 14(17): 3262-3274. |
| [227] | MASSALHA H, KORENBLUM E, MALITSKY S, et al. Live imaging of root-bacteria interactions in a microfluidics setup[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(17): 4549-4554. |
| [228] | KEHE J, KULESA A, ORTIZ A, et al. Massively parallel screening of synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(26): 12804-12809. |
| [229] | ZENGLER K, HOFMOCKEL K, BALIGA N S, et al. EcoFABs: advancing microbiome science through standardized fabricated ecosystems[J]. Nature Methods, 2019, 16(7): 567-571. |
| [230] | LIU P, PANDA K, EDWARDS S A, et al. Transposase-assisted target-site integration for efficient plant genome engineering[J]. Nature, 2024, 631(8021): 593-600. |
| [231] | 邵洁, 刘海利, 王勇. 植物合成生物学的现在与未来[J]. 合成生物学, 2020, 1(4): 395-412. |
| SHAO J, LIU H L, WANG Y. Present and future of plant synthetic biology[J]. Synthetic Biology Journal, 2020, 1(4): 395-412. | |
| [232] | SANDHYA D, JOGAM P, ALLINI V R, et al. The present and potential future methods for delivering CRISPR/Cas9 components in plants[J]. Journal of Genetic Engineering and Biotechnology, 2020, 18(1): 25. |
| [233] | CHEN L, LIU G Q, ZHANG T. Integrating machine learning and genome editing for crop improvement[J]. aBIOTECH, 2024, 5(2): 262-277. |
| [234] | BAUER-PANSKUS A, MIYAZAKI J, KAWALL K, et al. Risk assessment of genetically engineered plants that can persist and propagate in the environment[J]. Environmental Sciences Europe, 2020, 32(1): 32. |
| [235] | LI J, ZHAO H M, ZHENG L X, et al. Advances in synthetic biology and biosafety governance[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 598087. |
| [236] | 王盼娣, 熊小娟, 付萍, 等. 《生物安全法》实施背景下对合成生物学的监管[J]. 华中农业大学学报, 2021, 40(6): 231-245. |
| WANG P D, XIONG X J, FU P, et al. Regulation of synthetic biology under background of implementing Biosafety Law[J]. Journal of Huazhong Agricultural University, 2021, 40(6): 231-245. | |
| [237] | DE SOUZA A P, BURGESS S J, DORAN L, et al. Soybean photosynthesis and crop yield are improved by accelerating recovery from photoprotection[J]. Science, 2022, 377(6608): 851-854. |
| [238] | SPRONCKEN C C M, LIU P, MONNEY J, et al. Large-area, self-healing block copolymer membranes for energy conversion[J]. Nature, 2024, 630(8018): 866-871. |
| [239] | HE Y, NING T T, XIE T T, et al. Large-scale production of functional human serum albumin from transgenic rice seeds[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(47): 19078-19083. |
| [68] | LI Y Z, LIANG J, DENG B F, et al. Applications and prospects of CRISPR/Cas9-mediated base editing in plant breeding[J]. Current Issues in Molecular Biology, 2023, 45(2): 918-935. |
| [69] | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| [70] | LI C, ZHANG R, MENG X B, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors[J]. Nature Biotechnology, 2020, 38(7): 875-882. |
| [71] | MIKAMI M, TOKI S, ENDO M. In planta processing of the SpCas9-gRNA complex[J]. Plant & Cell Physiology, 2017, 58(11): 1857-1867. |
| [72] | CAMPA C C, WEISBACH N R, SANTINHA A J, et al. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts[J]. Nature Methods, 2019, 16(9): 887-893. |
| [73] | LI C, ZONG Y, JIN S, et al. SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds[J]. Genome Biology, 2020, 21(1): 141. |
| [74] | LIU H J, JIAN L M, XU J T, et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize[J]. The Plant Cell, 2020, 32(5): 1397-1413. |
| [75] | BAI M Y, YUAN J H, KUANG H Q, et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean[J]. Plant Biotechnology Journal, 2020, 18(3): 721-731. |
| [76] | JACOBS T B, ZHANG N, PATEL D, et al. Generation of a collection of mutant tomato lines using pooled CRISPR libraries[J]. Plant Physiology, 2017, 174(4): 2023-2037. |
| [77] | KUANG Y J, LI S F, REN B, et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms[J]. Molecular Plant, 2020, 13(4): 565-572. |
| [78] | HU N, TIAN H L, LI Y H, et al. pHNRhCas9NG, single expression cassette-based dual-component dual-transcription unit CRISPR/Cas9 system for plant genome editing[J]. Trends in Biotechnology, 2025, 43(7): 1788-1808. |
| [79] | TATSIS E C, O’CONNOR S E. New developments in engineering plant metabolic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 126-132. |
| [80] | WU Q Y, HUANG Z Y, WANG J Y, et al. Construction of an Escherichia coli cell factory to synthesize taxadien-5α-ol, the key precursor of anti-cancer drug paclitaxel[J]. Bioresources and Bioprocessing, 2022, 9(1): 82. |
| [81] | IGNEA C, ATHANASAKOGLOU A, IOANNOU E, et al. Carnosic acid biosynthesis elucidated by a synthetic biology platform[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(13): 3681-3686. |
| [82] | TAN H X, CHEN X H, LIANG N, et al. Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast[J]. Journal of Experimental Botany, 2019, 70(18): 4819-4834. |
| [83] | ZHANG J, HANSEN L G, GUDICH O, et al. A microbial supply chain for production of the anti-cancer drug vinblastine[J]. Nature, 2022, 609(7926): 341-347. |
| [84] | LIU J Q, WANG X, DAI G Z, et al. Microbial chassis engineering drives heterologous production of complex secondary metabolites[J]. Biotechnology Advances, 2022, 59: 107966. |
| [85] | KWAN B D, SELIGMANN B, NGUYEN T D, et al. Leveraging synthetic biology and metabolic engineering to overcome obstacles in plant pathway elucidation[J]. Current Opinion in Plant Biology, 2023, 71: 102330. |
| [86] | BUTELLI E, TITTA L, GIORGIO M, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors[J]. Nature Biotechnology, 2008, 26(11): 1301-1308. |
| [87] | MALHOTRA K, SUBRAMANIYAN M, RAWAT K, et al. Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells[J]. Molecular Plant, 2016, 9(11): 1464-1477. |
| [88] | POLTURAK G, BREITEL D, GROSSMAN N, et al. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants[J]. New Phytologist, 2016, 210(1): 269-283. |
| [89] | MAJER E, LLORENTE B, RODRÍGUEZ-CONCEPCIÓN M, et al. Rewiring carotenoid biosynthesis in plants using a viral vector[J]. Scientific Reports, 2017, 7: 41645. |
| [90] | FUCHS L K, HOLLAND A H, LUDLOW R A, et al. Genetic manipulation of biosynthetic pathways in mint[J]. Frontiers in Plant Science, 2022, 13: 928178. |
| [91] | D’ANDREA L, SIMON-MOYA M, LLORENTE B, et al. Interference with Clp protease impairs carotenoid accumulation during tomato fruit ripening[J]. Journal of Experimental Botany, 2018, 69(7): 1557-1568. |
| [92] | PETRIE J R, SHRESTHA P, BELIDE S, et al. Metabolic engineering Camelina sativa with fish oil-like levels of DHA[J]. PLoS One, 2014, 9(1): e85061. |
| [93] | KIM J Y, KIM J H, JANG Y H, et al. Transcriptome and metabolite profiling of tomato SGR-knockout null lines using the CRISPR/Cas9 system[J]. International Journal of Molecular Sciences, 2023, 24(1): 109. |
| [94] | TU M X, FANG J H, ZHAO R K, et al. CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera)[J]. Horticulture Research, 2022, 9: uhac022. |
| [95] | WEN D, WU L, WANG M Y, et al. CRISPR/Cas9-mediated targeted mutagenesis of FtMYB45 promotes flavonoid biosynthesis in Tartary buckwheat (Fagopyrum tataricum)[J]. Frontiers in Plant Science, 2022, 13: 879390. |
| [96] | DEVIREDDY A R, ZANDALINAS S I, FICHMAN Y, et al. Integration of reactive oxygen species and hormone signaling during abiotic stress[J]. The Plant Journal, 2021, 105(2): 459-476. |
| [97] | MIYAWAKI A, LLOPIS J, HEIM R, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin[J]. Nature, 1997, 388(6645): 882-887. |
| [98] | BRUNOUD G, WELLS D M, OLIVA M, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution[J]. Nature, 2012, 482(7383): 103-106. |
| [99] | LIAO C Y, SMET W, BRUNOUD G, et al. Reporters for sensitive and quantitative measurement of auxin response[J]. Nature Methods, 2015, 12(3): 207-210. |
| [100] | JONES A M. A new look at stress: abscisic acid patterns and dynamics at high-resolution[J]. New Phytologist, 2016, 210(1): 38-44. |
| [101] | FERNANDEZ-MORENO J P, STEPANOVA A N. Monitoring ethylene in plants: genetically encoded reporters and biosensors[J]. Small Methods, 2020, 4(8): 1900260. |
| [102] | RIZZA A, JONES A M. The makings of a gradient: spatiotemporal distribution of gibberellins in plant development[J]. Current Opinion in Plant Biology, 2019, 47: 9-15. |
| [103] | ORTEGA-VILLASANTE C, BURÉN S, BARÓN-SOLA Á, et al. In vivo ROS and redox potential fluorescent detection in plants: present approaches and future perspectives[J]. Methods, 2016, 109: 92-104. |
| [104] | ZÜRCHER E, TAVOR-DESLEX D, LITUIEV D, et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta[J]. Plant Physiology, 2013, 161(3): 1066-1075. |
| [105] | STEINER E, LIVNE S, KOBINSON-KATZ T, et al. The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class Ⅰ TCP proteolysis to promote sensitivity to cytokinin[J]. Plant Physiology, 2016, 171(2): 1485-1494. |
| [1] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2025, 6(5): 1203-1223. |
| [2] | 赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明. 天冬氨酸族饲用氨基酸微生物细胞工厂的创制[J]. 合成生物学, 2025, 6(5): 1184-1202. |
| [3] | 何杨昱, 杨凯, 王玮琳, 黄茜, 丘梓樱, 宋涛, 何流赏, 姚金鑫, 甘露, 何玉池. 国际基因工程机器大赛中植物合成生物学主题的设计与实践[J]. 合成生物学, 2025, 6(5): 1243-1254. |
| [4] | 郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福. 构建根际合成微生物菌群促进作物养分高效吸收利用[J]. 合成生物学, 2025, 6(5): 1058-1071. |
| [5] | 方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898. |
| [6] | 李永珠, 陈禹. 酵母基因组规模模型进展及发展趋势[J]. 合成生物学, 2025, 6(3): 585-602. |
| [7] | 夏辰亮, 张泽成, 管星悦, 唐乾元. 统计物理与人工智能驱动的蛋白质结构生物信息学[J]. 合成生物学, 2025, 6(3): 547-565. |
| [8] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| [9] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [10] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [11] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [12] | 黄姝涵, 马赫, 罗云孜. 生物合成红景天苷的研究进展[J]. 合成生物学, 2025, 6(2): 391-407. |
| [13] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [14] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [15] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||