合成生物学 ›› 2025, Vol. 6 ›› Issue (5): 1058-1071.DOI: 10.12211/2096-8280.2025-075
构建根际合成微生物菌群促进作物养分高效吸收利用
郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福
- 南京农业大学资源与环境科学学院,江苏 南京 210095
-
收稿日期:2025-07-21修回日期:2025-09-09出版日期:2025-10-31发布日期:2025-11-05 -
通讯作者:张瑞福 -
作者简介:郑雷 (1995—),男,博士研究生。研究方向为根际微生物功能解析与植物养分吸收调控。E-mail:zzzl@stu.njau.edu.cn张瑞福 (1974—),男,教授,博士生导师。研究方向为根际微生物与生物肥料、农业有机废弃物微生物降解转化与有机肥料等。E-mail:rfzhang@njau.edu.cn -
基金资助:国家自然科学基金国际(地区)合作与交流项目(32361143785)
Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency
ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu
- The College of Resources and Environmental Sciences,Nanjing Agricultural University,Nanjing 210095,Jiangsu,China
-
Received:2025-07-21Revised:2025-09-09Online:2025-10-31Published:2025-11-05 -
Contact:ZHANG Ruifu
摘要:
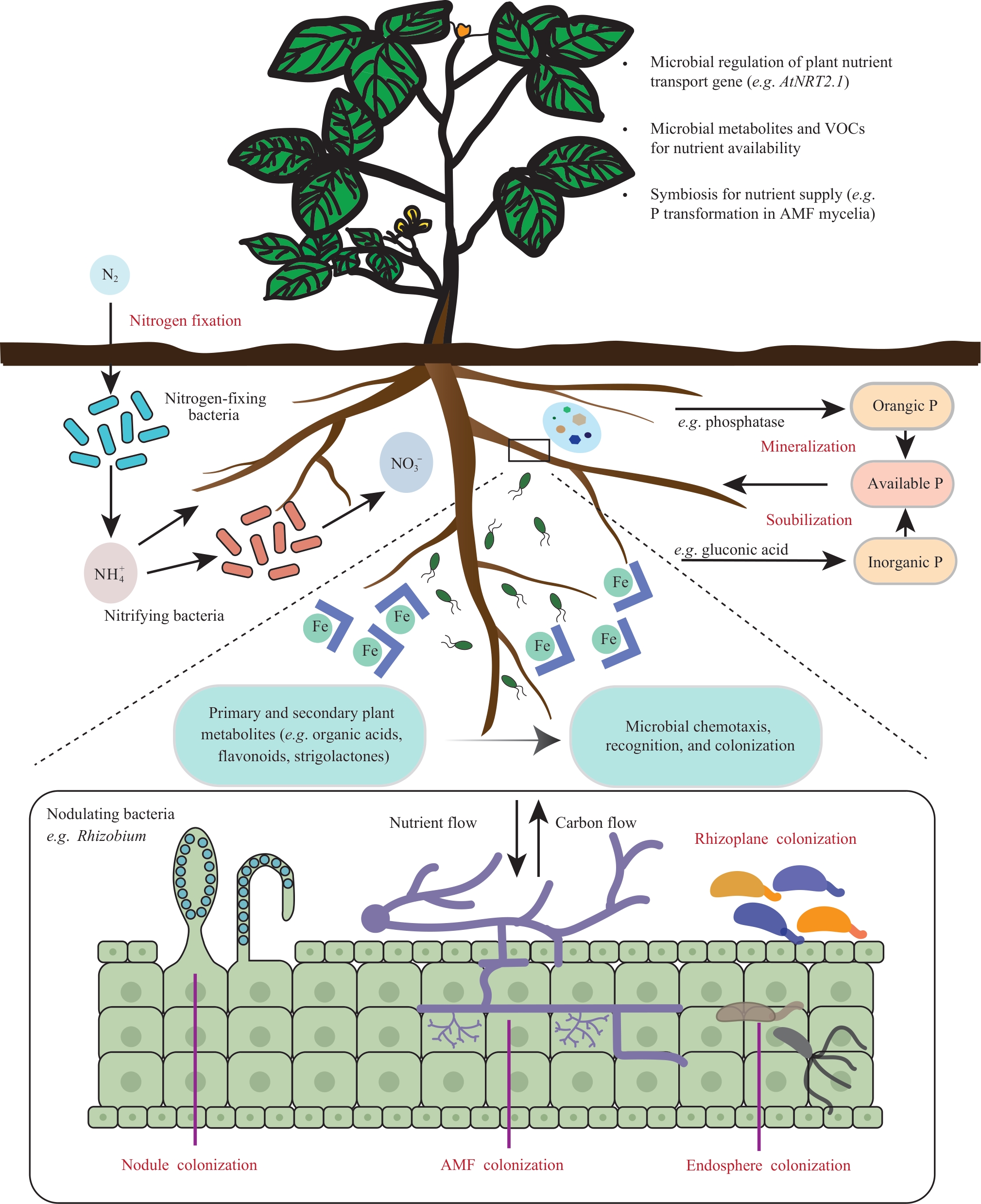
现代农业发展正面临养分利用效率低下和环境负担持续加剧的双重挑战。近年研究表明,根际微生物组(rhizosphere microbiome)作为植物的“第二基因组”,通过调控土壤氮、磷、铁等关键养分的生物地球化学循环,在植物高效获取养分过程中发挥核心驱动作用。合成生物学(synthetic biology)的快速发展为根际微生物组的精准解析与功能设计提供了创新性工具,通过模块化基因编辑、人工群落构建及宿主-微生物互作调控等策略显著提升植物养分利用效率,为突破传统农业依赖化肥、缓解资源浪费和环境压力提供全新的技术途径。本文系统综述了合成生物学驱动下根际微生物组工程在植物养分高效利用领域的研究进展,重点包括根际微生物组参与土壤养分循环的作用机制解析,合成生物学工具在单菌功能强化,群落协同调控、宿主-微生物互作优化等方面的关键作用以及当前技术发展中面临的微生物组复杂性限制、工程菌田间定植稳定性不足、跨作物普适性受限和潜在生态安全风险等诸多瓶颈。并展望了合成微生物组在可持续农业发展中的应用潜力,未来通过定向功能设计、智能响应系统构建及“植物-微生物-环境”协同调控,有望实现作物养分利用效率与可持续生产力的显著提升,从而为推动农业绿色转型提供关键科学技术支撑。
中图分类号:
引用本文
郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福. 构建根际合成微生物菌群促进作物养分高效吸收利用[J]. 合成生物学, 2025, 6(5): 1058-1071.
ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu. Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency[J]. Synthetic Biology Journal, 2025, 6(5): 1058-1071.
| [1] | LAMBERS H, RAVEN J A, SHAVER G R, et al. Plant nutrient-acquisition strategies change with soil age[J]. Trends in Ecology & Evolution, 2008, 23(2): 95-103. |
| [2] | SHI J C, WANG X L, WANG E T. Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems[J]. Annual Review of Plant Biology, 2023, 74: 569-607. |
| [3] | YANG J, LAN L Y, JIN Y, et al. Mechanisms underlying legume-rhizobium symbioses[J]. Journal of Integrative Plant Biology, 2022, 64(2): 244-267. |
| [4] | PANG Z Q, CHEN J, WANG T H, et al. Linking plant secondary metabolites and plant microbiomes: a review[J]. Frontiers in Plant Science, 2021, 12: 621276. |
| [5] | VENTURI V, KEEL C. Signaling in the rhizosphere[J]. Trends in Plant Science, 2016, 21(3): 187-198. |
| [6] | ZHOU X G, ZHANG J Y, KHASHI U RAHMAN M, et al. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes[J]. Molecular Plant, 2023, 16(5): 849-864. |
| [7] | YU P, HE X M, BAER M, et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation[J]. Nature Plants, 2021, 7(4): 481-499. |
| [8] | SINGH B K, HU H W, MACDONALD C A, et al. Microbiome-facilitated plant nutrient acquisition[J]. Cell Host & Microbe, 2025, 33(6): 869-881. |
| [9] | TRIVEDI P, LEACH J E, TRINGE S G, et al. Plant-microbiome interactions: from community assembly to plant health[J]. Nature Reviews Microbiology, 2020, 18(11): 607-621. |
| [10] | GRIFFIN C, OZ M T, DEMIRER G S. Engineering plant-microbe communication for plant nutrient use efficiency[J]. Current Opinion in Biotechnology, 2024, 88: 103150. |
| [11] | DAI R, ZHANG J Y, LIU F, et al. Crop root bacterial and viral genomes reveal unexplored species and microbiome patterns[J]. Cell, 2025, 188(9): 2521-2539.e22. |
| [12] | DONG Q Q, SU H J, SUN Y X, et al. Metagenomic insights into nitrogen cycling functional gene responses to nitrogen fixation and transfer in maize-peanut intercropping[J]. Plant, Cell & Environment, 2024, 47(12): 4557-4571. |
| [13] | ELIAS M, TANAKA M, SAKAI M, et al. C-terminal periplasmic domain of Escherichia coli quinoprotein glucose dehydrogenase transfers electrons to ubiquinone[J]. Journal of Biological Chemistry, 2001, 276(51): 48356-48361. |
| [14] | LIU Y, JIA B L, REN Y, et al. Bacterial social interactions in synthetic Bacillus consortia enhance plant growth[J]. iMeta, 2025, 4(4): e70053. |
| [15] | BERG G, KUSSTATSCHER P, ABDELFATTAH A, et al. Microbiome modulation-toward a better understanding of plant microbiome response to microbial inoculants[J]. Frontiers in Microbiology, 2021, 12: 650610. |
| [16] | ERLACHER A, CARDINALE M, GROSCH R, et al. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome[J]. Frontiers in Microbiology, 2014, 5: 175. |
| [17] | JIANG W Y, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| [18] | JOHNS N I, BLAZEJEWSKI T, GOMES A L, et al. Principles for designing synthetic microbial communities[J]. Current Opinion in Microbiology, 2016, 31: 146-153. |
| [19] | STENUIT B, AGATHOS S N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology[J]. Current Opinion in Biotechnology, 2015, 33: 305-317. |
| [20] | HU J, WEI Z, FRIMAN V P, et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression[J]. mBio, 2016, 7(6): e01790-16. |
| [21] | WANG J J, LI R C, ZHANG H, et al. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application[J]. BMC Microbiology, 2020, 20(1): 38. |
| [22] | WANG J, UWIRAGIYE Y, CAO M M, et al. Global land use change impacts on soil nitrogen availability and environmental losses[J]. Environmental Science & Technology, 2025, 59(33): 17595-17605. |
| [23] | HINSINGER P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review[J]. Plant and Soil, 2001, 237(2): 173-195. |
| [24] | GUERINOT M L, YI Y. Iron: nutritious, noxious, and not readily available[J]. Plant Physiology, 1994, 104(3): 815-820. |
| [25] | GALLOWAY J N, TOWNSEND A R, ERISMAN J W, et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions[J]. Science, 2008, 320(5878): 889-892. |
| [26] | YUE H, YUE W J, JIAO S, et al. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions[J]. Microbiome, 2023, 11(1): 70. |
| [27] | XU H R, LIU W D, HE Y H, et al. Plant-root microbiota interactions in nutrient utilization[J]. Frontiers of Agricultural Science and Engineering, 2025, 12(1): 16-26. |
| [28] | CRAINE J M, MORROW C, FIERER N. Microbial nitrogen limitation increases decomposition[J]. Ecology, 2007, 88(8): 2105-2113. |
| [29] | FONTAINE S, HENAULT C, AAMOR A, et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect[J]. Soil Biology and Biochemistry, 2011, 43(1): 86-96. |
| [30] | DAKORA F D, PHILLIPS D A. Root exudates as mediators of mineral acquisition in low-nutrient environments[J]. Plant and Soil, 2002, 245(1): 35-47. |
| [31] | LIU S B, HE F K, KUZYAKOV Y, et al. Nutrients in the rhizosphere: a meta-analysis of content, availability, and influencing factors[J]. Science of the Total Environment, 2022, 826: 153908. |
| [32] | SCHALK I J. Bacterial siderophores: diversity, uptake pathways and applications[J]. Nature Reviews Microbiology, 2025, 23(1): 24-40. |
| [33] | FAN X Y, GE A H, QI S S, et al. Root exudates and microbial metabolites: signals and nutrients in plant-microbe interactions[J]. Science China Life Sciences, 2025, 68(8): 2290-2302. |
| [34] | KUYPERS M M M, MARCHANT H K, KARTAL B. The microbial nitrogen-cycling network[J]. Nature Reviews Microbiology, 2018, 16(5): 263-276. |
| [35] | CHENG S S, GONG X, XUE W F, et al. Evolutionarily conserved core microbiota as an extended trait in nitrogen acquisition strategy of herbaceous species[J]. New Phytologist, 2024, 244(4): 1570-1584. |
| [36] | YANG N, NESME J, RØDER H L, et al. Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis [J]. NPJ Biofilms and Microbiomes, 2021, 7: 82. |
| [37] | PORTER S S, DUPIN S E, DENISON R F, et al. Host-imposed control mechanisms in legume-rhizobia symbiosis[J]. Nature Microbiology, 2024, 9(8): 1929-1939. |
| [38] | PRIYA H, DHAR D W, SINGH R, et al. Co-cultivation approach to decipher the influence of nitrogen-fixing Cyanobacterium on growth and N uptake in rice crop[J]. Current Microbiology, 2022, 79(2): 53. |
| [39] | HUREK T, REINHOLD-HUREK B. Azoarcus sp. strain BH72 as a model for nitrogen-fixing grass endophytes[J]. Journal of Biotechnology, 2003, 106(2-3): 169-178. |
| [40] | WALLER S, WILDER S L, SCHUELLER M J, et al. Examining the effects of the nitrogen environment on growth and N2-fixation of endophytic Herbaspirillum seropedicae in maize seedlings by applying 11C radiotracing[J]. Microorganisms, 2021, 9(8): 1582. |
| [41] | STEENHOUDT O, VANDERLEYDEN J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects[J]. FEMS Microbiology Reviews, 2000, 24(4): 487-506. |
| [42] | HASKETT T L, PARAMASIVAN P, MENDES M D, et al. Engineered plant control of associative nitrogen fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(16): e2117465119. |
| [43] | ZHANG L Y, ZHANG M L, HUANG S Y, et al. A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants[J]. Nature Communications, 2022, 13: 3361. |
| [44] | VAN DEYNZE A, ZAMORA P, DELAUX P M, et al. Nitrogen fixation in a Landrace of maize is supported by a mucilage-associated diazotrophic microbiota[J]. PLoS Biology, 2018, 16(8): e2006352. |
| [45] | CALVO P, ZEBELO S, MCNEAR D, et al. Plant growth-promoting rhizobacteria induce changes in Arabidopsis thaliana gene expression of nitrate and ammonium uptake genes[J]. Journal of Plant Interactions, 2019, 14(1): 224-231. |
| [46] | CHEN Y, LI Y C, FU Y S, et al. The beneficial rhizobacterium Bacillus velezensis SQR9 regulates plant nitrogen uptake via an endogenous signaling pathway[J]. Journal of Experimental Botany, 2024, 75(11): 3388-3400. |
| [47] | MCGRATH J W, CHIN J P, QUINN J P. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules[J]. Nature Reviews Microbiology, 2013, 11(6): 412-419. |
| [48] | GROSS A, LIN Y, WEBER P K, et al. The role of soil redox conditions in microbial phosphorus cycling in humid tropical forests[J]. Ecology, 2020, 101(2): e02928. |
| [49] | LIANG J L, LIU J, JIA P, et al. Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining[J]. The ISME Journal, 2020, 14(6): 1600-1613. |
| [50] | BILLAH M, KHAN M, BANO A, et al. Phosphorus and phosphate solubilizing bacteria: keys for sustainable agriculture[J]. Geomicrobiology Journal, 2019, 36(10): 904-916. |
| [51] | ZHAO B Y, JIA X Q, YU N, et al. Microbe-dependent and independent nitrogen and phosphate acquisition and regulation in plants[J]. New Phytologist, 2024, 242(4): 1507-1522. |
| [52] | DE ZUTTER N, AMEYE M, VERMEIR P, et al. Innovative rhizosphere-based enrichment under P-limitation selects for bacterial isolates with high-performance P-solubilizing traits[J]. Microbiology Spectrum, 2022, 10(6): e02052-22. |
| [53] | SINGH S K, WU X X, SHAO C Y, et al. Microbial enhancement of plant nutrient acquisition[J]. Stress Biology, 2022, 2(1): 3. |
| [54] | SHAO J H, MIAO Y Z, LIU K M, et al. Rhizosphere microbiome assembly involves seed-borne bacteria in compensatory phosphate solubilization[J]. Soil Biology and Biochemistry, 2021, 159: 108273. |
| [55] | PANG F, LI Q, SOLANKI M K, et al. Soil phosphorus transformation and plant uptake driven by phosphate-solubilizing microorganisms[J]. Frontiers in Microbiology, 2024, 15: 1383813. |
| [56] | LIU J P, XU W F, ZHANG Q, et al. OsPHR2-mediated recruitment of Pseudomonadaceae enhances rice phosphorus uptake[J]. Plant Communications, 2024, 5(8): 100930. |
| [57] | LIU C, BAI Z, LUO Y, et al. Multiomics dissection of Brassica napus L. lateral roots and endophytes interactions under phosphorus starvation[J]. Nature Communications, 2024, 15(1): 9732. |
| [58] | HARBORT C J, HASHIMOTO M, INOUE H, et al. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis [J]. Cell Host & Microbe, 2020, 28(6): 825-837.e6. |
| [59] | WANG N Q, WANG T Q, CHEN Y, et al. Microbiome convergence enables siderophore-secreting-rhizobacteria to improve iron nutrition and yield of peanut intercropped with maize[J]. Nature Communications, 2024, 15: 839. |
| [60] | ZAMIOUDIS C, KORTELAND J, VAN PELT J A, et al. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses[J]. The Plant Journal, 2015, 84(2): 309-322. |
| [61] | PRITY S A, SAJIB S A, DAS U, et al. Arbuscular mycorrhizal fungi mitigate Fe deficiency symptoms in sorghum through phytosiderophore-mediated Fe mobilization and restoration of redox status[J]. Protoplasma, 2020, 257(5): 1373-1385. |
| [62] | ZHANG P F, JIN T, KUMAR SAHU S, et al. The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis[J]. Molecules, 2019, 24(7): 1411. |
| [63] | NETT R S, MONTANARES M, MARCASSA A, et al. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution[J]. Nature Chemical Biology, 2017, 13(1): 69-74. |
| [64] | SALAZAR-CEREZO S, MARTÍNEZ-MONTIEL N, GARCÍA-SÁNCHEZ J, et al. Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria[J]. Microbiological Research, 2018, 208: 85-98. |
| [65] | HAYASHI S, GRESSHOFF P M, FERGUSON B J. Mechanistic action of gibberellins in legume nodulation[J]. Journal of Integrative Plant Biology, 2014, 56(10): 971-978. |
| [66] | GROßKINSKY D K, TAFNER R, MORENO M V, et al. Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis [J]. Scientific Reports, 2016, 6: 23310. |
| [67] | NUMAN M, BASHIR S, KHAN Y, et al. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review[J]. Microbiological Research, 2018, 209: 21-32. |
| [68] | CONWAY J M, WALTON W G, SALAS-GONZÁLEZ I, et al. Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome[J]. Nature Microbiology, 2022, 7(11): 1817-1833. |
| [69] | PACESA M, PELEA O, JINEK M. Past, present, and future of CRISPR genome editing technologies[J]. Cell, 2024, 187(5): 1076-1100. |
| [70] | CHEN L, ZHANG S, XUE N N, et al. Engineering a precise adenine base editor with minimal bystander editing[J]. Nature Chemical Biology, 2023, 19(1): 101-110. |
| [71] | XU P Y, SAITO M, FAURE G, et al. Structural insights into the diversity and DNA cleavage mechanism of Fanzor[J]. Cell, 2024, 187(19): 5238-5252.e20. |
| [72] | WEI Y H, GAO P F, PAN D, et al. Engineering eukaryotic transposon-encoded Fanzor2 system for genome editing in mammals[J]. Nature Chemical Biology, 2025. (2025-05-20)[2025-09-01]. . |
| [73] | HUA Y, TAY N E S, YE X J, et al. Protein editing using a coordinated transposition reaction[J]. Science, 2025, 388(6742): 68-74. |
| [74] | DIXON R A, POSTGATE J R. Genetic transfer of nitrogen fixation from Klebsiella pneumoniae to Escherichia coli [J]. Nature, 1972, 237(5350): 102-103. |
| [75] | TEMME K, ZHAO D H, VOIGT C A. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(18): 7085-7090. |
| [76] | WANG L Y, ZHANG L H, LIU Z Z, et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli [J]. PLoS Genetics, 2013, 9(10): e1003865. |
| [77] | LI X X, LIU Q, LIU X M, et al. Using synthetic biology to increase nitrogenase activity[J]. Microbial Cell Factories, 2016, 15: 43. |
| [78] | ZHANG T, YAN Y L, HE S, et al. Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501[J]. Research in Microbiology, 2012, 163(5): 332-339. |
| [79] | BAGESHWAR U K, SRIVASTAVA M, PARDHA-SARADHI P, et al. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield[J]. Applied and Environmental Microbiology, 2017, 83(15): e00590-17. |
| [80] | COMPANT S, CASSAN F, KOSTIĆ T, et al. Harnessing the plant microbiome for sustainable crop production[J]. Nature Reviews Microbiology, 2025, 23(1): 9-23. |
| [81] | WEN A, HAVENS K L, BLOCH S E, et al. Enabling biological nitrogen fixation for cereal crops in fertilized fields[J]. ACS Synthetic Biology, 2021, 10(12): 3264-3277. |
| [82] | RYU M H, ZHANG J, TOTH T, et al. Control of nitrogen fixation in bacteria that associate with cereals[J]. Nature Microbiology, 2020, 5(2): 314-330. |
| [83] | ALLEN R S, TILBROOK K, WARDEN A C, et al. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix[J]. Frontiers in Plant Science, 2017, 8: 287. |
| [84] | HE W S, BURÉN S, BAYSAL C, et al. Nitrogenase cofactor maturase NifB isolated from transgenic rice is active in FeMo-co synthesis[J]. ACS Synthetic Biology, 2022, 11(9): 3028-3036. |
| [85] | ALORI E T, GLICK B R, BABALOLA O O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture[J]. Frontiers in Microbiology, 2017, 8: 971. |
| [86] | BUCH A, ARCHANA G, NARESH KUMAR G. Heterologous expression of phosphoenolpyruvate carboxylase enhances the phosphate solubilizing ability of fluorescent pseudomonads by altering the glucose catabolism to improve biomass yield[J]. Bioresource Technology, 2010, 101(2): 679-687. |
| [87] | ADHIKARY H, SANGHAVI P B, MACWAN S R, et al. Artificial citrate operon confers mineral phosphate solubilization ability to diverse fluorescent pseudomonads[J]. PLoS One, 2014, 9(9): e107554. |
| [88] | SHULSE C N, CHOVATIA M, AGOSTO C, et al. Engineered root bacteria release plant-available phosphate from phytate[J]. Applied and Environmental Microbiology, 2019, 85(18): e01210-19. |
| [89] | LIU Y P, SHU X, CHEN L, et al. Plant commensal type Ⅶ secretion system causes iron leakage from roots to promote colonization[J]. Nature Microbiology, 2023, 8(8): 1434-1449. |
| [90] | CHEN J W, ZHANG X, KUANG M, et al. Endophytic Enterobacter sp. YG-14 mediated arsenic mobilization through siderophore and its role in enhancing phytostabilization[J]. Journal of Hazardous Materials, 2024, 465: 133206. |
| [91] | DE SOUZA R S C, ARMANHI J S L, ARRUDA P. From microbiome to traits: designing synthetic microbial communities for improved crop resiliency[J]. Frontiers in Plant Science, 2020, 11: 1179. |
| [92] | XI H C, NIE X Q, GAO F, et al. A bacterial spermidine biosynthetic pathway via carboxyaminopropylagmatine[J]. Science Advances, 2023, 9(43): eadj9075. |
| [93] | SHEN J Y, WANG M X, WANG E T. Exploitation of the microbiome for crop breeding[J]. Nature Plants, 2024, 10(4): 533-534. |
| [94] | VORHOLT J A, VOGEL C, CARLSTRÖM C I, et al. Establishing causality: opportunities of synthetic communities for plant microbiome research[J]. Cell Host & Microbe, 2017, 22(2): 142-155. |
| [95] | JING J Y, GARBEVA P, RAAIJMAKERS J M, et al. Strategies for tailoring functional microbial synthetic communities[J]. The ISME Journal, 2024, 18(1): wrae049. |
| [96] | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| [97] | WANG W, XIA Y W, ZHANG P P, et al. Narrow-spectrum resource-utilizing bacteria drive the stability of synthetic communities through enhancing metabolic interactions[J]. Nature Communications, 2025, 16: 6088. |
| [98] | KAUR S, EGIDI E, QIU Z G, et al. Synthetic community improves crop performance and alters rhizosphere microbial communities[J]. Journal of Sustainable Agriculture and Environment, 2022, 1(2): 118-131. |
| [99] | CHAI Y N, GE Y F, STOERGER V, et al. High-resolution phenotyping of sorghum genotypic and phenotypic responses to low nitrogen and synthetic microbial communities[J]. Plant, Cell & Environment, 2021, 44(5): 1611-1626. |
| [100] | NIU B, PAULSON J N, ZHENG X Q, et al. Simplified and representative bacterial community of maize roots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(12): E2450-E2459. |
| [101] | XUN W B, REN Y, YAN H, et al. Sustained inhibition of maize seed-borne Fusarium using a Bacillus-dominated rhizospheric stable core microbiota with unique cooperative patterns[J]. Advanced Science, 2023, 10(5): 2205215. |
| [102] | QIAO Y Z, WANG Z D, SUN H, et al. Synthetic community derived from grafted watermelon rhizosphere provides protection for ungrafted watermelon against Fusarium oxysporum via microbial synergistic effects[J]. Microbiome, 2024, 12(1): 101. |
| [103] | XU X M, DINESEN C, PIOPPI A, et al. Composing a microbial symphony: synthetic communities for promoting plant growth[J]. Trends in Microbiology, 2025, 33(7): 738-751. |
| [104] | NORTHEN T R, KLEINER M, TORRES M, et al. Community standards and future opportunities for synthetic communities in plant-microbiota research[J]. Nature Microbiology, 2024, 9(11): 2774-2784. |
| [105] | WANG C H, LI Y J, LI M J, et al. Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean[J]. Journal of Integrative Plant Biology, 2021, 63(6): 1021-1035. |
| [106] | LI Y J, LI R R, LIU R, et al. A simplified SynCom based on core-helper strain interactions enhances symbiotic nitrogen fixation in soybean[J]. Journal of Integrative Plant Biology, 2025, 67(6): 1582-1598. |
| [107] | LIU C Y, JIANG M T, YUAN M M, et al. Root microbiota confers rice resistance to aluminium toxicity and phosphorus deficiency in acidic soils[J]. Nature Food, 2023, 4(10): 912-924. |
| [108] | CASTRILLO G, TEIXEIRA P J P L, PAREDES S H, et al. Root microbiota drive direct integration of phosphate stress and immunity[J]. Nature, 2017, 543(7646): 513-518. |
| [109] | HASKETT T L, TKACZ A, POOLE P S. Engineering rhizobacteria for sustainable agriculture[J]. The ISME Journal, 2021, 15(4): 949-964. |
| [110] | OFEK M, VORONOV-GOLDMAN M, HADAR Y, et al. Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities[J]. Environmental Microbiology, 2014, 16(7): 2157-2167. |
| [111] | ZHONG X B, WANG J, SHI X L, et al. Genetically optimizing soybean nodulation improves yield and protein content[J]. Nature Plants, 2024, 10(5): 736-742. |
| [1] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2025, 6(5): 1203-1223. |
| [2] | 于文文, 吕雪芹, 李兆丰, 刘龙. 植物合成生物学与母乳低聚糖生物制造[J]. 合成生物学, 2025, 6(5): 992-997. |
| [3] | 颜钊涛, 周鹏飞, 汪阳忠, 张鑫, 谢雯燕, 田晨菲, 王勇. 植物合成生物学:植物细胞大规模培养的新机遇[J]. 合成生物学, 2025, 6(5): 1107-1125. |
| [4] | 孙扬, 陈立超, 石艳云, 王珂, 吕丹丹, 徐秀美, 张立新. 作物光合作用合成生物学的策略与展望[J]. 合成生物学, 2025, 6(5): 1025-1040. |
| [5] | 赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明. 天冬氨酸族饲用氨基酸微生物细胞工厂的创制[J]. 合成生物学, 2025, 6(5): 1184-1202. |
| [6] | 何杨昱, 杨凯, 王玮琳, 黄茜, 丘梓樱, 宋涛, 何流赏, 姚金鑫, 甘露, 何玉池. 国际基因工程机器大赛中植物合成生物学主题的设计与实践[J]. 合成生物学, 2025, 6(5): 1243-1254. |
| [7] | 张学博, 朱成姝, 陈睿雲, 金庆姿, 刘晓, 熊燕, 陈大明. 农业合成生物学:政策规划与产业发展协同推进[J]. 合成生物学, 2025, 6(5): 1224-1242. |
| [8] | 刘婕, 郜钰, 马永硕, 尚轶. 合成生物学在农业中的进展及挑战[J]. 合成生物学, 2025, 6(5): 998-1024. |
| [9] | 李超, 张焕, 杨军, 王二涛. 固氮合成生物学研究进展[J]. 合成生物学, 2025, 6(5): 1041-1057. |
| [10] | 魏家秀, 嵇佩云, 节庆雨, 黄秋燕, 叶浩, 戴俊彪. 植物人工染色体的构建与应用[J]. 合成生物学, 2025, 6(5): 1093-1106. |
| [11] | 方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898. |
| [12] | 朱欣悦, 陈恬恬, 邵恒煊, 唐曼玉, 华威, 程艳玲. 益生菌辅助防治恶性肿瘤的研究进展[J]. 合成生物学, 2025, 6(4): 899-919. |
| [13] | 吴晓燕, 宋琪, 许睿, 丁陈君, 陈方, 郭勍, 张波. 合成生物学研发竞争态势对比分析[J]. 合成生物学, 2025, 6(4): 940-955. |
| [14] | 张建康, 王文君, 郭洪菊, 白北辰, 张亚飞, 袁征, 李彦辉, 李航. 基于机器视觉的高通量微生物克隆挑选工作站研制及应用[J]. 合成生物学, 2025, 6(4): 956-971. |
| [15] | 李全飞, 陈乾, 刘浩, 贺坤东, 潘亮, 雷鹏, 谷益安, 孙良, 李莎, 邱益彬, 王瑞, 徐虹. 高黏性蛋白材料的合成生物学及应用[J]. 合成生物学, 2025, 6(4): 806-828. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||