合成生物学 ›› 2025, Vol. 6 ›› Issue (4): 873-898.DOI: 10.12211/2096-8280.2025-006
微生物合成高级醇的发展趋势与挑战
方馨仪1, 孙丽超2,3, 霍毅欣2,3, 王颖4, 岳海涛1
- 1.新疆大学,新疆 乌鲁木齐 830000
2.北京理工大学生命学院,分子医学与生物诊疗重点实验室,北京 100081
3.北京理工大学唐山研究院,河北 唐山 063000
4.北京理工大学化学与化工学院,生物化工研究所,医药分子科学与制剂工程工信部重点实验室,北京 100081
-
收稿日期:2025-01-21修回日期:2025-04-18出版日期:2025-08-31发布日期:2025-09-03 -
通讯作者:王颖,岳海涛 -
作者简介:方馨仪 (2000—),女,硕士研究生。研究方向为合成生物学,生物信息,大语言模型。E-mail:107552301711@stu.xju.edu.cn王颖 (1987—),女,特别研究员,博士生导师。研究方向为合成生物学与代谢工程。E-mail:wy2015@bit.edu.cn岳海涛 (1980—),男,教授,博士生导师。研究方向为抗逆元件挖掘设计与高版本底盘构建等。E-mail:yuehaitao@tsinghua.org.cn -
基金资助:河北省自然科学基金面上项目“基于氨基酸资源分配的代谢调控策略及其在异丁醇生产中的应用”(C2023105022);唐山市科技计划项目(23130228E);新疆维吾尔自治区重点研发任务专项(2023B02034);新疆维吾尔自治区重点研发任务专项(2023B02034-2);新疆维吾尔自治区青年拔尖人才-基础研究人才(20243122875);国家自然科学基金(U2003305)
Trends and challenges in microbial synthesis of higher alcohols
FANG Xinyi1, SUN Lichao2,3, HUO Yixin2,3, WANG Ying4, YUE Haitao1
- 1.Xinjiang University,Urumqi 830000,Xinjiang,China
2.Key Laboratory of Molecular Medicine and Biotherapy,School of Life Science,Beijing Institute of Technology,Beijing 100081,China
3.Tangshan Research Institute,BIT,Tangshan 063000,Hebei,China
4.Key Laboratory of Medicinal Molecule Science and Pharmaceutical Engineering,Ministry of Industry and Information Technology,Institute of Biochemical Engineering,School of Chemistry and Chemical Engineering,Beijing Institute of Technology,Beijing 100081,China
-
Received:2025-01-21Revised:2025-04-18Online:2025-08-31Published:2025-09-03 -
Contact:WANG Ying, YUE Haitao
摘要:
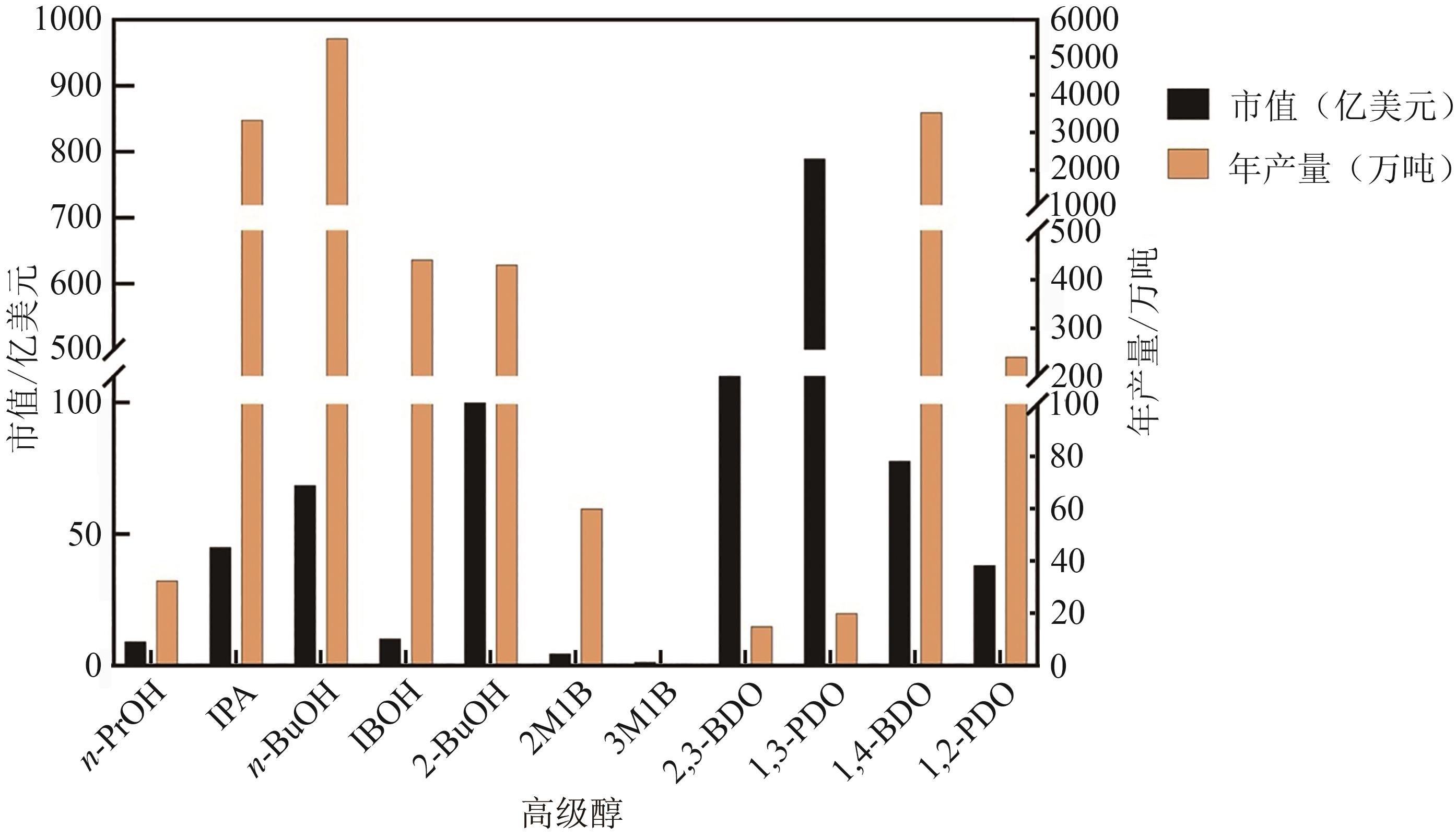
高级醇是指含有三个或以上碳原子的醇类。传统高级醇的生产主要依赖于石化资源,然而其不可再生性限制了相关产业的发展,因此开发可持续的生物基高级醇生产技术成为研究热点。本文综述了高级醇的市场规模、主要应用领域及其经济价值,并重点分析了异丁醇、1,3-丁二醇和2,3-丁二醇的市场表现。进一步探讨了高级醇的生物合成路径,包括乙酰辅酶A依赖途径、支链氨基酸合成途径和脂肪酸链延长途径,同时总结了代谢工程优化策略,如辅因子平衡调节、竞争途径敲除、酶优化及高产菌株筛选。此外,本文还综述了基于新技术的多维度优化策略,未来有望通过生物传感器、高效基因编辑和计算机辅助代谢工程等技术的结合,进一步优化微生物细胞工厂的设计,有助于提高高级醇的工业化生产效率,为可再生能源和绿色化学工业的发展提供重要支持。
中图分类号:
引用本文
方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898.
FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols[J]. Synthetic Biology Journal, 2025, 6(4): 873-898.
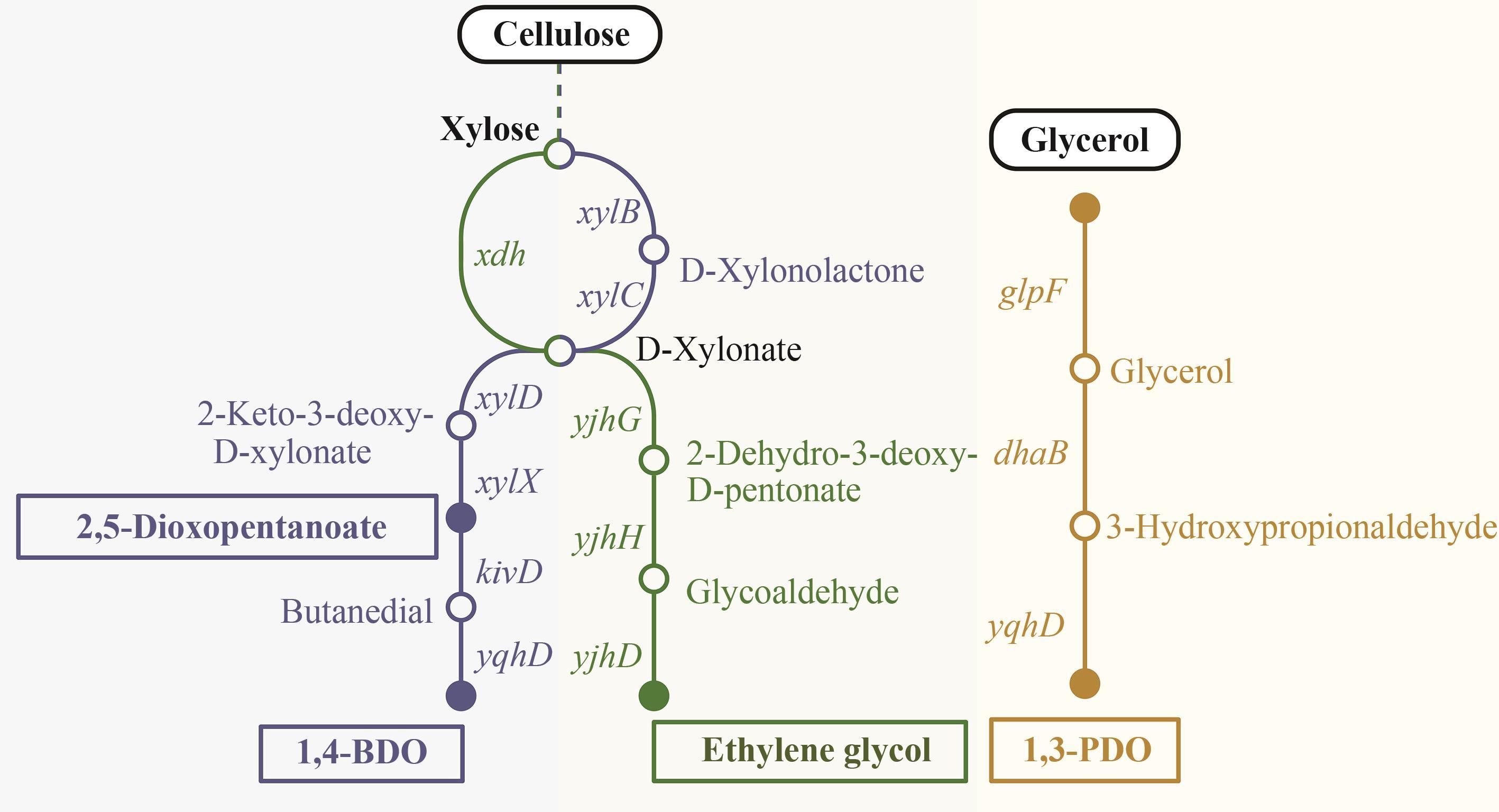
图2 纤维素代谢合成高级醇的途径(1,3-PDO—1,3-丙二醇;1,4-BDO—1,4-丁二醇;glpF—甘油通道蛋白编码基因;dhaB—甘油脱氢酶编码基因;kivD—α-酮异戊酸脱羧酶编码基因;yqhD—NADPH依赖的醛还原酶YqhD编码基因;xylB—木酮糖激酶编码基因;xylC—木糖内酯酶XylC编码基因;xdh—黄嘌呤脱氢酶编码基因;yjhH—2-脱氢-3-脱氧-D-戊酮酸醛缩酶编码基因;yjhG—D-木糖酸脱水酶编码基因;xylX—甲苯甲酸1,2-双加氧酶α亚基编码基因)
Fig. 2 Metabolic pathway of higher alcohol synthesis from cellulose(1,3-PDO—1,3-Propanediol; 1,4-BDO—1,4-Butanediol; glpF—glycerol facilitator gene; dhaB—glycerol dehydratase gene; kivD—alpha-ketoisovalerate decarboxylase gene; yqhD—NADPH-dependent aldehyde reductase YqhD gene; xylB—xylulokinase gene; xylC—xylonolactonase XylC gene; xdh—xanthine dehydrogenase gene; yjhH—putative 2-dehydro-3-deoxy-D-pentonate aldolase gene; yjhG—D-xylonate dehydratase gene; xylX—toluate 1,2-dioxygenase subunit alpha gene)
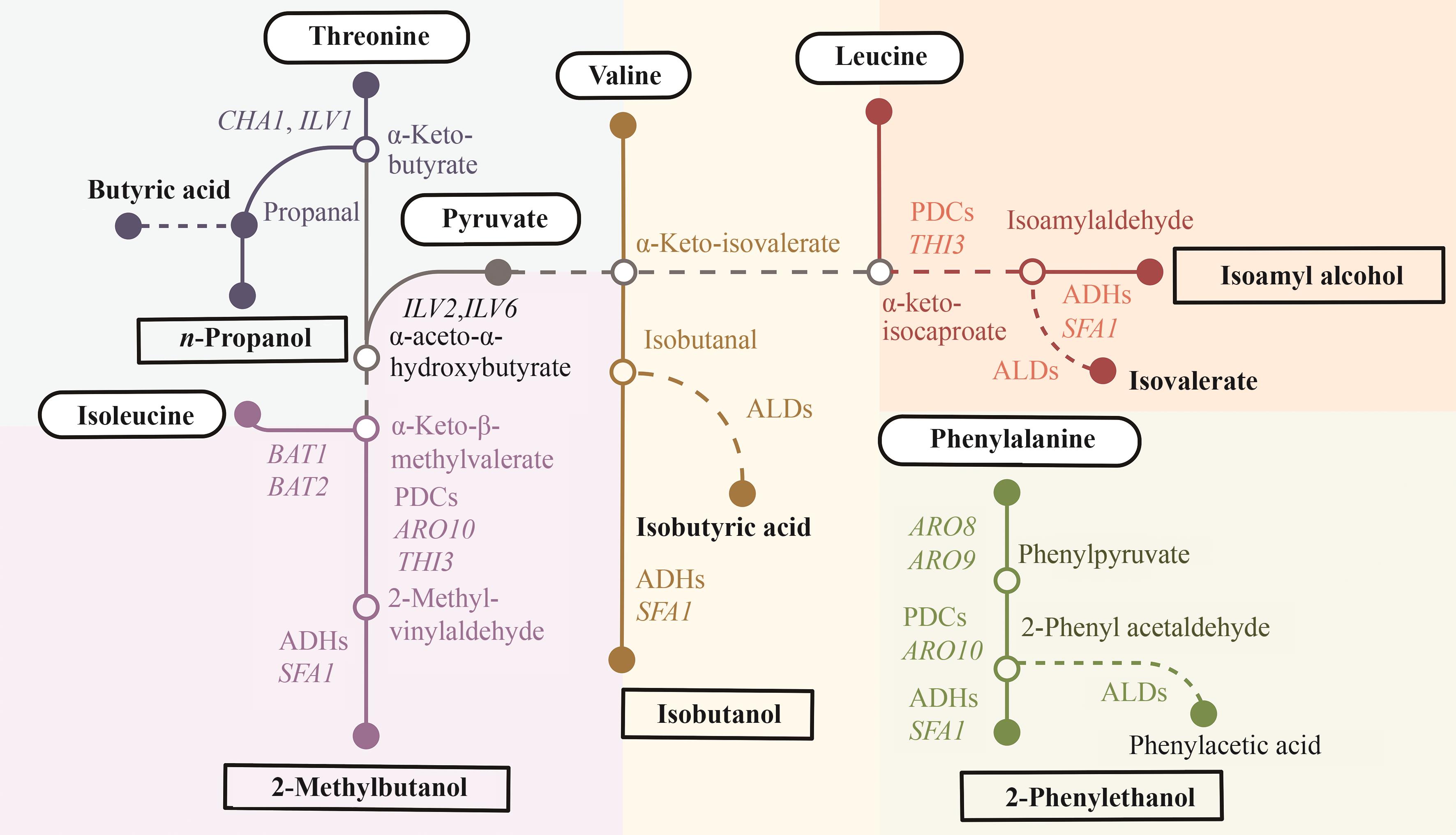
图3 基于蛋白质代谢的高级醇生物合成途径(CHA1—L-丝氨酸/L-苏氨酸氨裂解酶CHA1编码基因;ILV1—苏氨酸氨裂解酶ILV1编码基因;BAT1—支链氨基酸转氨酶BAT1编码基因;BAT2—支链氨基酸转氨酶 BAT2编码基因;PDCs—丙酮酸脱羧酶;THI3—支链-2-氧酸脱羧酶THI3编码基因;ADHs—醇脱氢酶;SFA1—双功能乙醇脱氢酶/S-(羟甲基)编码基因;ARO10—苯基丙酮酸脱羧酶ARO10编码基因;ARO8—双功能2-氨基己二酸转氨酶/芳香族氨基酸:2-酮戊二酸转氨酶编码基因;ARO9—芳香族氨基酸:2-氧代戊二酸转氨酶编码基因;ALDs—醛脱氢酶;ILV2—乙酰乳酸合成酶催化亚基编码基因; ILV6—乙酰乳酸合成酶调节亚基编码基因)
Fig. 3 Biosynthetic pathways of higher alcohols based on protein metabolism(CHA1—L-serine/L-threonine ammonia-lyase CHA1 gene; ILV1—threonine ammonia-lyase ILV1 gene; BAT1—branched-chain-amino-acid transaminase BAT1 gene; BAT2—branched-chain-amino-acid transaminase BAT2 gene; PDCs—pyruvate decarboxylases; THI3—branched-chain-2-oxoacid decarboxylase THI3 gene; ADHs—alcohol dehydrogenases; SFA1—bifunctional alcohol dehydrogenase/S-(hydroxymethyl) glutathione dehydrogenase gene; ARO10—phenylpyruvate decarboxylase ARO10 gene; ARO8—bifunctional 2-aminoadipate transaminase/aromatic-amino-acid:2-oxoglutarate transaminase gene; ARO9—aromatic-amino-acid:2-oxoglutarate transaminase gene; ALDs—aldehyde dehydrogenases; ILV2—acetolactate synthase catalytic subunit gene; ILV6—acelolactate synthase regulatory subunit gene)
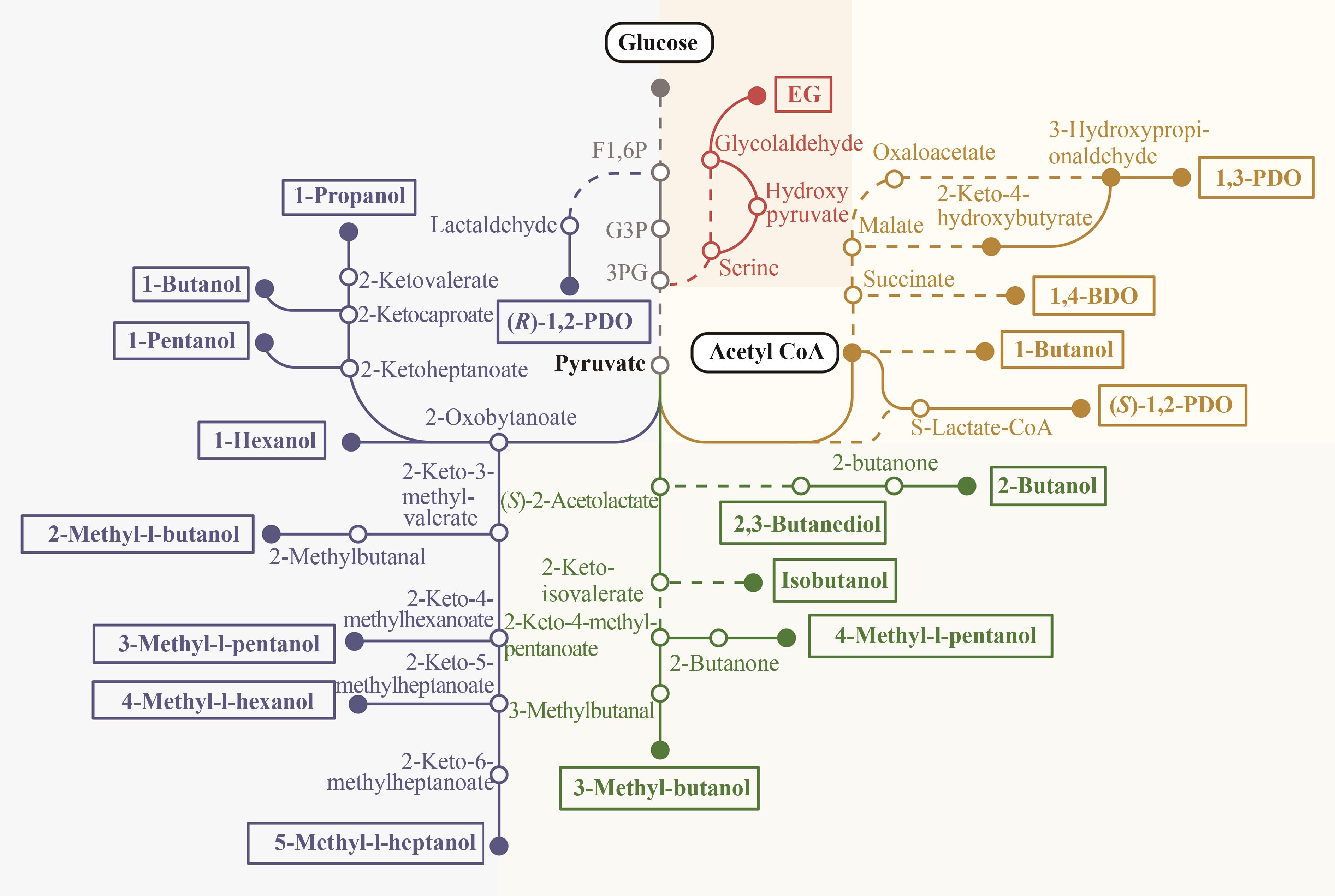
图4 葡萄糖、乙酰辅酶A合成高级醇[EG—乙二醇;1,3-PDO—1,3-丙二醇;1,4-BDO—1,4-丁二醇;(R)-1,2-PDO—R-1,2-丙二醇;(S)-1,2-PDO—S-1,2-丙二醇;Acetyl-CoA—乙酰辅酶A;S-Lactate-CoA—琥珀酰-乳酸辅酶A]
Fig. 4 Synthesis of higher alcohols from glucose and acetyl-CoA[EG—Ethylene glycol; 1,3-PDO—1,3-Propanediol; 1,4-BDO—1,4-Butanediol; (R)-1,2-PDO—(R)-1,2-Propanediol; (S)-1,2-PDO—(S)-1,2-Propanediol; Acetyl CoA—Acetyl coenzyme A; S-Lactate-CoA—Succinyl-lactate coenzyme A]
产物 Products | 宿主 Hosts | 基因类型(敲除;过表达) Genotypes (knockout; overexpression) | 底物 Substrates | 发酵条件 Fermentation conditions | 产量/(g/L) Titer/(g/L) | 参考文献 References |
|---|---|---|---|---|---|---|
| n-PrOH | Escherichia coli | ΔilvA ΔilvB; kivD adh2 cimAmut leuABCD | Glucose | Shake flask | 2.78 | [ |
| E. coli | ΔlacI ΔlysA ΔmetA ΔtdhA ΔiclR ΔilvIH ΔilvBNΔrpoS thrAC1034T lysCC1005T | Glycerol | Bioreactor (Fed-batch) | 10.3 | [ | |
| IPA | E. coli | None; lacIq thl atoDA adc adhB-593 | Glucose | Stirred flask | 143 | [ |
| E. coli | None; thl atoDA adc adhB-593 bgl-blc | Cellobiose | Shake flask | 4.1 | [ | |
| E. coli | None; thl ctfAB adc adhB-593 | Glucose | Shake baffled flask | 13.6 | [ | |
| Alcohol mixture | Clostridium acetobutylicum | Δbuk:: ermC; adc ctfAB adhB-593 | Glucose | Bioreactor | 35 | [ |
| C. acetobutylicum | Δbuk ΔC1502; adc ctfAB adhB-593 | Glucose | Bioreactor | 20.4 | [ | |
| n-BuOH | Synechococcus elongatus | None; atoB, hbd, crt, ter, AdhE2 | CO2 | Flask and bioreactor | 0.015 | [ |
| S. elongatus | None; nphT7 phaB aphaJ ter bld yqhD | CO2 | Static capped flask | 0.029 9 | [ | |
| Clostridium tyrobutyricum | Δpta Δbuk; adhED485G | Glucose | Bioreactor | 130 | [ | |
| C. tyrobutyricum | None; adhE2 | Mannitol | Bioreactor; anaerobic | 20.5 | [ | |
| S. cerevisiae | None; thl, hbd, crt, bcd, etfAB, adhE2 | Galactose | Shake flask | 0.002 5 | [ | |
| Pseudomonas putida | None; thl, hbd, crt, bcd, etfAB, adhE2 | Glycerol | Shake flask | 0.024 | [ | |
| P. putida | None; thl, hbd, crt, bcd, etfAB, adhE1 | Glycerol | Shake flask | 0.122 | [ | |
| Lactobacillus brevis | None; thl, hbd, crt, bcd, etfAB | Glucose | Vial | 0.3 | [ | |
| Clostridium ljungdahlii | None; thlA, hbd, crt, bcd, adhE, and bdhA | Syngas | Shake flask | 0.148 | [ | |
| E. coli | ΔldhA ΔadhE ΔfrdBC Δpta; atoB hbd crt adhE2 fdh ter | Glucose | Bioreactor; anaerobic | 30 | [ | |
| IBOH | E. coli | ΔadhE ΔldhA ΔfrdBC Δfnr Δpta ΔpflB; alsS ilvCD kivD adh2 | Glucose | Static capped flask | 22 | [ |
| E. coli | None; alsS ilvCD kivD adhA | Glucose | Bioreactor; | 56 | [ | |
| S. elongatus | None; alsS, ilvCD, kivd, yqhD | CO2 | Bottle | 0.450 | [ | |
| Corynebacterium crenatum | None; ILV2, ILV3, ILV5, kivD, ADH6 | Lignocellulose | Bottle | 5.61 | [ | |
| Bacillus megaterium | None; kivd, yqhD | Glucose | Test tube | 0.3 | [ | |
| Bacillus subtilis | Δldh; ilvCD alsS kivD adh2 | Glucose | Bioreactor | 3.83 | [ | |
| Clostridium cellulolyticum | None; kivD yqhD alsS ilvCD | Cellulose | Not specified | 0.66 | [ | |
| Corynebacterium glutamicum | ΔaceE Δpqo ΔilvE ΔldhA Δmdh; ilvBNCD pntAB kivD adhA | Glucose | Bioreactor | 13 | [ | |
| Ralstonia eutropha | ΔphaB2C2 ΔphaC1AB1;alsS ilvCD kivD yqhD | CO2 | Bioreactor with electrodes | 0.09 | [ | |
| R. eutropha | ΔphaCAB ΔilvE ΔbkdAB ΔaceE; adh ilvBHCD kivD | Fructose | Shake flask | 0.27 | [ | |
| S. cerevisiae | Δlpd; kivD ADH6 ILV2 ILV5c ILV3c ILV2C MAE1 | Glucose | Shake flask | 1.62 | [ | |
| Zymomonas mobilis | None; kdcA Pgap als ilvC ilvD | Glucose | Shake flask | 4 | [ | |
| Clostridium thermocellum | Δhpt; kivd, alsS, ilvBN, ilvCD, ilvD | Cellulose | Consolidated bioprocessing | 5.4 | [ | |
| C. glutamicum | ΔilvE; ilvBNC, ilvD, adhA, kivd | Glucose | Shake flask | 20.8 | [ | |
| 2-BuOH | Lactobacillus diolivorans | None; pduQ, PDO-DH (NAD(P)H) | meso-2,3-butanediol/glycerol | Bioreactor | 13.4 | [ |
| Klebsiella pneumoniae | ΔldhA; pduCDEGHQ337AorF375I, adh | Glucose | Shake flask | 1.03 | [ | |
| L. brevis | None; None | meso-2,3-butanediol | Test tube | 0.88 | [ | |
| Lactobacillus buchneri | None; None | butaNone | Test tube | 0.04 | [ | |
| S. cerevisiae | None; pduCDEGH, adh | meso-2,3‐butanediol | Shake flask | 0.004 | [ | |
| Lactobacillus spp. | None; None | meso-2,3‐butanediol | Shake flask | 0.41 | [ | |
| 2M1B | E. coli | ΔmetA Δtdh; ilvGM ilvCD ilvA kivDadh2 thrABC | Glucose | Shake baffled flask | 1.25 | [ |
| Brevibacterium flavum | None; ilvC-ilvD-alsS, kivd-ADH2 | Glucose; duckweed | Shake flash | 19.5/17.5 | [ | |
| C. crenatum | None; Cgl1271, Cgl1273, Cgl1268,kivd; adh2 | Glucose | Shake flash | 5.26 | [ | |
| 3M1B | E. coli | None; alsS ilvCD kivD adh2leuAG462D leuBCD | Glucose | Shake flask | 9.5 | [ |
| B. flavum | None; ilvC-ilvD-alsS, kivd-ADH2 | Glucose; duckweed | Shake flask | 0.79/0.78 | [ | |
| C. crenatum | None; Cgl1271, Cgl1273, Cgl1268,kivd; adh2 | Glucose | Shake flask | 3.78 | [ | |
| 2,3-BDO | Serratia marcescens | swrI, swrR, slaA, slaB, slaC, slaR; swrR | Glucose | Shake flask | 42.5 | [ |
| S. marcescens | ΔswrW; swrW | Sucrose | Fed-batch | 152 | [ | |
| Z. mobilis | ΔPDC; LlkivD, ARO10, THI3, ADH6, ADH7, PpIlv2, PpIlv6, PpIlv5,PpIlv3, ScATF1 | Glucose | Shake flask | 13.3 | [ | |
| E. coli | None; budB, budA, budC | Glucose | Shake flask | meso-BDO,17.7 | [ | |
| E. coli | None; alsS, alsD, bdhA from K. pneumoniae | Glucose | Shake flask | (R,R)-BDO,5.8 | [ | |
| E. coli | None; alsS, alsD, adh from C. beijerinckii | Glucose | Shake flask | (R,R)-BDO,5.1 | [ | |
| E. coli | None; alsS, alsD, adh from T. brockii | Glucose | Shake flask | (R,R)-BDO,6.1 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Shake flask | (S,S)-BDO,2.2 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Shake flask | 0.66 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Bioreactor | (R,R)-BDO,30.5 | [ | |
| E. coli | ΔldhA, ΔpflB, ΔadhE, ΔlpdA::K.p.lpd E354 K, Δmdh, ΔarcA; gltAR164L, ilvBN, aldB, bdh1 | Glucose | Bioreactor | 88 | [ | |
| E. coli | None; budA, budB, budC | Glucose | Bioreactor High oxygen | 52.1 | [ | |
| E. coli | ΔldhA, ΔadhE, Δpta, ΔfrdA; budA,budB, budC | Glucose | Bioreactor Low oxygen | 68.1 | [ | |
| E. coli | ΔldhA, Δpta, ΔadhE, ΔpoxB; alaS, alsD, budC | Glucose | Shake flask | meso-BDO,14.5 | [ | |
| E. coli | None; bdh, fdh | Diacetyl | Bioreactor | (S,S)-BDO,31.7 | [ | |
| E. coli | None; budC, bdh | Diacetyl | Shake flask | (S,S)-BDO,2.2 | [ | |
| E. coli | None; budA, budB, budC | Sugar beet molasses | Fed-batch | 56.2 | [ | |
| E. coli | ΔfrdABCD, ΔldhA, ΔadhE, ΔlpdA, Δpta; budB, budA, budC | Algal hydrolysate | Shake flask | meso-(S,S)-BDO,14.1 | [ | |
| E. coli | ΔpoxB, ΔldhA, ΔackA, Δpta; alsS, alsD, budC, ced3A | Cellodextrin | Shake flask | meso-BDO,5.5 | [ | |
| 1,3-PDO | E. coli | ΔgldA, ΔglpK, ΔaldA, ΔaldB, ΔmgsA, ΔptsHI, replacing gapA promoter with the synthetic short 1.5 GI promoter (SEQ ID NO:28); galP | Glucose | Bioreactor | 112 | [ |
| E. coli | None; gpd1-gpp2 fusion gene,dha operon, rpoS | Glucose | Bioreactor | 12.1 | [ | |
| E. coli | None; dhaB, yqhD, gdrA, gdrB, fdh1, gapN, galP, glk | Glycerol | Shake flask | 13.47 | [ | |
| E. coli | None; dhaB1, dhaB2, yqhD | Glycerol | Bioreactor | 104.4 | [ | |
| E. coli | None; dhaB1, dhaB2, dhaT | Glycerol and glucose | Bioreactor | 41.7 | [ | |
| Corynebacterium glutamicum | Δald, Δpyk, Δadh, ΔpoxB, ΔldhA, Δppc, Δzwf; hdpA-gldA, gpd1, gpp2, yqhD, pduCEDGHDownregulation: gapA | Glucose and xylose | Fed-batch | 110.4 | [ | |
| Vibrio natriegens | Δpta-ackA, ΔarcA, ΔadhE, ΔaldB, Δldh, Δpfl, ΔsthA, ΔglpR, ΔaldA, ΔfrdABCD; pntAB, phaP | Glycerol | Fed-batch | 69.5 | [ | |
| E. coli | ΔthrB;yqhD, lysC, serCR42W/R77W, metL, pdc | Glucose | Fed-batch | 3.03 | [ | |
| E. coli | None; mcrC, pduP, mcrN, yqhD, prpE | Glucose; Xylose, Glycerol, Acetate | Shake flask; Fed-batch | 2.93; 7.98 | [ | |
| E. coli | ΔlysC, ΔgltA; ppc | Glucose | Shake flask; Fed-batch | 6.41; 11.21 | [ | |
| E. coli | ΔglpK, ΔptsG; yqhD, pntAB, galP, glk | Isoprene | Shake flask | 2.5 | [ | |
| 1,3-BDO | E. coli | None; phaAB, bld | Glucose | Fed-batch | 15.76 | [ |
| E. coli | Δldh, Δpta, ΔackA, ΔadhE; bldL273T, yqhD, phaAB, pntAB | Glucose | Optimized fermentation | 13.40 | [ | |
| E. coli | Δldh, Δpta, ΔackA, ΔadhE; phaAB, yqhD, pntAB, car, sfp | Glucose | Optimized fermentation | 0.40 | [ | |
| E. coli | Δzwf, Δedd, ΔpfkA, ΔpfkB; pk, glpX, thl, hbd, tesB, car | Glucose | Fed-batch | 22.66 | [ | |
| E. coli | ΔadhE, ΔpoxB, ΔldhA, Δpta-ackA, ΔatoB, ΔtesB, ΔyciA; phaAB, bld, yqhD | Glucose | Fed-batch | 23.13 | [ | |
| E. coli | ΔadhE, ΔpoxB, ΔldhA, ΔyciA, ΔpdhR, Δpgi, ΔgntR; phaAB, bld, yjgB, zwf | Glucose | Fed-batch | 71.1 | [ | |
| E. coli | Δpta, ΔyjgB, ΔadhE, ΔldhA, ΔpflB, ΔadhP, ΔyqhD, ΔeutG, ΔilvB, ΔpoxB; AKR, DERA, PDC | Acetaldehyd; 3-Hydroxybutanal (3-HB) | Biotransformation | 2.4 | [ | |
| 1,4-BDO | E. coli | Δsad::cat2-bld-bdh, ΔlacZ::cat1-sucD 4hbd, ΔllpdA::K.p.lpdA D354KΔpflB, ΔarcA, Δmdh, ΔadhE, ΔldhA, knock down tesB; gabD, ybgC, gltAR163L | Glucose | Bioreactor | 1.8 | [ |
| E. coli | ΔadhE, ΔldhA, ΔpflB, Δmdh, ΔarcA, lpdA::K.p.lpdD354K; gltAR163L, sucA, 4hbd, cat2, ald, adh | Glucose | Bioreactor | 18 | [ | |
| E. coli | ΔaraA, Δicd; araC, araD, araA, araB, araE, kivd, yqhd | L-Arabinose | Bioreactor | 1.51 | [ | |
| E. coli | None; None | Glucose | Commercial-scale fermentation | >125 | [ | |
| E. coli | None; gadB, gabT, yqhD,car, ppc, gltAR163L | Amino acids (AAs), Glucose | General metabolic platform | 1.41 | [ | |
| E. coli | ΔyagE, ΔxylA, ΔyjhH; KvidV461I,xylBCDX, yqhD | Glucose, Xylose | D-xylose, L-arabinose, D-galacturonate | 12 | [ | |
| E. coli | ΔuxaC, ΔgarL, Δicd; udh, garD, ycbC, xylA(CC), kivd, yqhd | D-Galacturonate | Bioreactor | 16.5 | [ | |
| E. coli | ΔxylA, ΔyjhH, ΔyagE, Δicd; xylB, xylC, xylD, xylX, xylA(CC), kivdV461I, yqhd | Xylose | Bioreactor | 12 | [ | |
| 1,2-PDO | E. coli | ΔpoxB, ΔfrdA, ΔmgsA, ΔadhE::pdcD; gldA, mgs | Glucose | Shake flask | 0.7 | [ |
| E. coli | ΔlldD::mmsB, ΔackA-pta::pct, ΔldhA::Lldh; pct, pdcD, mmsB | Glucose | Shake flask | 1.04 | [ | |
| E. coli | ΔldhA::KanR;mgs, gldA, fucO | Glucose | Bioreactor | 4.5 | [ | |
| E. coli | Δzwf, ΔtpiA, ΔgloA, ΔldhA, ΔadhE,; mgsA, gldA, fucO | Glucose | Shake flask | 5.13 | [ | |
| E. coli | ΔlldD, Δdld, ΔldhA, ΔadhE; pct,pduP, yahK | D-/L-Lactate | Shake flask | (R)-1,2-PDO,1.5; (S)-1,2-PDO,1.7 | [ | |
| E. coli | Δac kA-pta, ΔldhA, ΔdhaK; dhaKL, gldA, mgsA, yqhD | Glycerol | Bioreactor | 5.6 | [ | |
| BT | E. coli | ΔyiaE, ΔycdW, ΔxylA KivD(Lactococcus lactis); None | Xylose | Fed-batch, carbon flux optimization | 10.03 | [ |
| E. coli | None; YqhD, YjhG | Xylose | Optimized cultivation | 5.1 | [ | |
| E. coli | ΔxylA, ΔxylB, ΔyjhE, ΔyagH, ΔycdW;xdh, mdlC | Xylose | Fed-batch | 2.38 | [ | |
| E. coli | None; XylD, KdcA (Lactococcus lactis), AdhP (E. coli) | Xylose | Optimized cultivation | 5.1 | [ | |
| E. coli | ΔfadE; XylD, fadD | Xylose, free fatty acids | Fed-batch | 1.1 (BT esters) | [ | |
| S. cerevisiae | None; XylB, XylD | Xylose, rice straw hydrolysate | Aerobic fermentation | 1.7 | [ |
表1 微生物生产高级醇的进展
Table 1 Advances in the microbial production of higher alcohols
产物 Products | 宿主 Hosts | 基因类型(敲除;过表达) Genotypes (knockout; overexpression) | 底物 Substrates | 发酵条件 Fermentation conditions | 产量/(g/L) Titer/(g/L) | 参考文献 References |
|---|---|---|---|---|---|---|
| n-PrOH | Escherichia coli | ΔilvA ΔilvB; kivD adh2 cimAmut leuABCD | Glucose | Shake flask | 2.78 | [ |
| E. coli | ΔlacI ΔlysA ΔmetA ΔtdhA ΔiclR ΔilvIH ΔilvBNΔrpoS thrAC1034T lysCC1005T | Glycerol | Bioreactor (Fed-batch) | 10.3 | [ | |
| IPA | E. coli | None; lacIq thl atoDA adc adhB-593 | Glucose | Stirred flask | 143 | [ |
| E. coli | None; thl atoDA adc adhB-593 bgl-blc | Cellobiose | Shake flask | 4.1 | [ | |
| E. coli | None; thl ctfAB adc adhB-593 | Glucose | Shake baffled flask | 13.6 | [ | |
| Alcohol mixture | Clostridium acetobutylicum | Δbuk:: ermC; adc ctfAB adhB-593 | Glucose | Bioreactor | 35 | [ |
| C. acetobutylicum | Δbuk ΔC1502; adc ctfAB adhB-593 | Glucose | Bioreactor | 20.4 | [ | |
| n-BuOH | Synechococcus elongatus | None; atoB, hbd, crt, ter, AdhE2 | CO2 | Flask and bioreactor | 0.015 | [ |
| S. elongatus | None; nphT7 phaB aphaJ ter bld yqhD | CO2 | Static capped flask | 0.029 9 | [ | |
| Clostridium tyrobutyricum | Δpta Δbuk; adhED485G | Glucose | Bioreactor | 130 | [ | |
| C. tyrobutyricum | None; adhE2 | Mannitol | Bioreactor; anaerobic | 20.5 | [ | |
| S. cerevisiae | None; thl, hbd, crt, bcd, etfAB, adhE2 | Galactose | Shake flask | 0.002 5 | [ | |
| Pseudomonas putida | None; thl, hbd, crt, bcd, etfAB, adhE2 | Glycerol | Shake flask | 0.024 | [ | |
| P. putida | None; thl, hbd, crt, bcd, etfAB, adhE1 | Glycerol | Shake flask | 0.122 | [ | |
| Lactobacillus brevis | None; thl, hbd, crt, bcd, etfAB | Glucose | Vial | 0.3 | [ | |
| Clostridium ljungdahlii | None; thlA, hbd, crt, bcd, adhE, and bdhA | Syngas | Shake flask | 0.148 | [ | |
| E. coli | ΔldhA ΔadhE ΔfrdBC Δpta; atoB hbd crt adhE2 fdh ter | Glucose | Bioreactor; anaerobic | 30 | [ | |
| IBOH | E. coli | ΔadhE ΔldhA ΔfrdBC Δfnr Δpta ΔpflB; alsS ilvCD kivD adh2 | Glucose | Static capped flask | 22 | [ |
| E. coli | None; alsS ilvCD kivD adhA | Glucose | Bioreactor; | 56 | [ | |
| S. elongatus | None; alsS, ilvCD, kivd, yqhD | CO2 | Bottle | 0.450 | [ | |
| Corynebacterium crenatum | None; ILV2, ILV3, ILV5, kivD, ADH6 | Lignocellulose | Bottle | 5.61 | [ | |
| Bacillus megaterium | None; kivd, yqhD | Glucose | Test tube | 0.3 | [ | |
| Bacillus subtilis | Δldh; ilvCD alsS kivD adh2 | Glucose | Bioreactor | 3.83 | [ | |
| Clostridium cellulolyticum | None; kivD yqhD alsS ilvCD | Cellulose | Not specified | 0.66 | [ | |
| Corynebacterium glutamicum | ΔaceE Δpqo ΔilvE ΔldhA Δmdh; ilvBNCD pntAB kivD adhA | Glucose | Bioreactor | 13 | [ | |
| Ralstonia eutropha | ΔphaB2C2 ΔphaC1AB1;alsS ilvCD kivD yqhD | CO2 | Bioreactor with electrodes | 0.09 | [ | |
| R. eutropha | ΔphaCAB ΔilvE ΔbkdAB ΔaceE; adh ilvBHCD kivD | Fructose | Shake flask | 0.27 | [ | |
| S. cerevisiae | Δlpd; kivD ADH6 ILV2 ILV5c ILV3c ILV2C MAE1 | Glucose | Shake flask | 1.62 | [ | |
| Zymomonas mobilis | None; kdcA Pgap als ilvC ilvD | Glucose | Shake flask | 4 | [ | |
| Clostridium thermocellum | Δhpt; kivd, alsS, ilvBN, ilvCD, ilvD | Cellulose | Consolidated bioprocessing | 5.4 | [ | |
| C. glutamicum | ΔilvE; ilvBNC, ilvD, adhA, kivd | Glucose | Shake flask | 20.8 | [ | |
| 2-BuOH | Lactobacillus diolivorans | None; pduQ, PDO-DH (NAD(P)H) | meso-2,3-butanediol/glycerol | Bioreactor | 13.4 | [ |
| Klebsiella pneumoniae | ΔldhA; pduCDEGHQ337AorF375I, adh | Glucose | Shake flask | 1.03 | [ | |
| L. brevis | None; None | meso-2,3-butanediol | Test tube | 0.88 | [ | |
| Lactobacillus buchneri | None; None | butaNone | Test tube | 0.04 | [ | |
| S. cerevisiae | None; pduCDEGH, adh | meso-2,3‐butanediol | Shake flask | 0.004 | [ | |
| Lactobacillus spp. | None; None | meso-2,3‐butanediol | Shake flask | 0.41 | [ | |
| 2M1B | E. coli | ΔmetA Δtdh; ilvGM ilvCD ilvA kivDadh2 thrABC | Glucose | Shake baffled flask | 1.25 | [ |
| Brevibacterium flavum | None; ilvC-ilvD-alsS, kivd-ADH2 | Glucose; duckweed | Shake flash | 19.5/17.5 | [ | |
| C. crenatum | None; Cgl1271, Cgl1273, Cgl1268,kivd; adh2 | Glucose | Shake flash | 5.26 | [ | |
| 3M1B | E. coli | None; alsS ilvCD kivD adh2leuAG462D leuBCD | Glucose | Shake flask | 9.5 | [ |
| B. flavum | None; ilvC-ilvD-alsS, kivd-ADH2 | Glucose; duckweed | Shake flask | 0.79/0.78 | [ | |
| C. crenatum | None; Cgl1271, Cgl1273, Cgl1268,kivd; adh2 | Glucose | Shake flask | 3.78 | [ | |
| 2,3-BDO | Serratia marcescens | swrI, swrR, slaA, slaB, slaC, slaR; swrR | Glucose | Shake flask | 42.5 | [ |
| S. marcescens | ΔswrW; swrW | Sucrose | Fed-batch | 152 | [ | |
| Z. mobilis | ΔPDC; LlkivD, ARO10, THI3, ADH6, ADH7, PpIlv2, PpIlv6, PpIlv5,PpIlv3, ScATF1 | Glucose | Shake flask | 13.3 | [ | |
| E. coli | None; budB, budA, budC | Glucose | Shake flask | meso-BDO,17.7 | [ | |
| E. coli | None; alsS, alsD, bdhA from K. pneumoniae | Glucose | Shake flask | (R,R)-BDO,5.8 | [ | |
| E. coli | None; alsS, alsD, adh from C. beijerinckii | Glucose | Shake flask | (R,R)-BDO,5.1 | [ | |
| E. coli | None; alsS, alsD, adh from T. brockii | Glucose | Shake flask | (R,R)-BDO,6.1 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Shake flask | (S,S)-BDO,2.2 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Shake flask | 0.66 | [ | |
| E. coli | None; budA, budB, ydjL | Glucose | Bioreactor | (R,R)-BDO,30.5 | [ | |
| E. coli | ΔldhA, ΔpflB, ΔadhE, ΔlpdA::K.p.lpd E354 K, Δmdh, ΔarcA; gltAR164L, ilvBN, aldB, bdh1 | Glucose | Bioreactor | 88 | [ | |
| E. coli | None; budA, budB, budC | Glucose | Bioreactor High oxygen | 52.1 | [ | |
| E. coli | ΔldhA, ΔadhE, Δpta, ΔfrdA; budA,budB, budC | Glucose | Bioreactor Low oxygen | 68.1 | [ | |
| E. coli | ΔldhA, Δpta, ΔadhE, ΔpoxB; alaS, alsD, budC | Glucose | Shake flask | meso-BDO,14.5 | [ | |
| E. coli | None; bdh, fdh | Diacetyl | Bioreactor | (S,S)-BDO,31.7 | [ | |
| E. coli | None; budC, bdh | Diacetyl | Shake flask | (S,S)-BDO,2.2 | [ | |
| E. coli | None; budA, budB, budC | Sugar beet molasses | Fed-batch | 56.2 | [ | |
| E. coli | ΔfrdABCD, ΔldhA, ΔadhE, ΔlpdA, Δpta; budB, budA, budC | Algal hydrolysate | Shake flask | meso-(S,S)-BDO,14.1 | [ | |
| E. coli | ΔpoxB, ΔldhA, ΔackA, Δpta; alsS, alsD, budC, ced3A | Cellodextrin | Shake flask | meso-BDO,5.5 | [ | |
| 1,3-PDO | E. coli | ΔgldA, ΔglpK, ΔaldA, ΔaldB, ΔmgsA, ΔptsHI, replacing gapA promoter with the synthetic short 1.5 GI promoter (SEQ ID NO:28); galP | Glucose | Bioreactor | 112 | [ |
| E. coli | None; gpd1-gpp2 fusion gene,dha operon, rpoS | Glucose | Bioreactor | 12.1 | [ | |
| E. coli | None; dhaB, yqhD, gdrA, gdrB, fdh1, gapN, galP, glk | Glycerol | Shake flask | 13.47 | [ | |
| E. coli | None; dhaB1, dhaB2, yqhD | Glycerol | Bioreactor | 104.4 | [ | |
| E. coli | None; dhaB1, dhaB2, dhaT | Glycerol and glucose | Bioreactor | 41.7 | [ | |
| Corynebacterium glutamicum | Δald, Δpyk, Δadh, ΔpoxB, ΔldhA, Δppc, Δzwf; hdpA-gldA, gpd1, gpp2, yqhD, pduCEDGHDownregulation: gapA | Glucose and xylose | Fed-batch | 110.4 | [ | |
| Vibrio natriegens | Δpta-ackA, ΔarcA, ΔadhE, ΔaldB, Δldh, Δpfl, ΔsthA, ΔglpR, ΔaldA, ΔfrdABCD; pntAB, phaP | Glycerol | Fed-batch | 69.5 | [ | |
| E. coli | ΔthrB;yqhD, lysC, serCR42W/R77W, metL, pdc | Glucose | Fed-batch | 3.03 | [ | |
| E. coli | None; mcrC, pduP, mcrN, yqhD, prpE | Glucose; Xylose, Glycerol, Acetate | Shake flask; Fed-batch | 2.93; 7.98 | [ | |
| E. coli | ΔlysC, ΔgltA; ppc | Glucose | Shake flask; Fed-batch | 6.41; 11.21 | [ | |
| E. coli | ΔglpK, ΔptsG; yqhD, pntAB, galP, glk | Isoprene | Shake flask | 2.5 | [ | |
| 1,3-BDO | E. coli | None; phaAB, bld | Glucose | Fed-batch | 15.76 | [ |
| E. coli | Δldh, Δpta, ΔackA, ΔadhE; bldL273T, yqhD, phaAB, pntAB | Glucose | Optimized fermentation | 13.40 | [ | |
| E. coli | Δldh, Δpta, ΔackA, ΔadhE; phaAB, yqhD, pntAB, car, sfp | Glucose | Optimized fermentation | 0.40 | [ | |
| E. coli | Δzwf, Δedd, ΔpfkA, ΔpfkB; pk, glpX, thl, hbd, tesB, car | Glucose | Fed-batch | 22.66 | [ | |
| E. coli | ΔadhE, ΔpoxB, ΔldhA, Δpta-ackA, ΔatoB, ΔtesB, ΔyciA; phaAB, bld, yqhD | Glucose | Fed-batch | 23.13 | [ | |
| E. coli | ΔadhE, ΔpoxB, ΔldhA, ΔyciA, ΔpdhR, Δpgi, ΔgntR; phaAB, bld, yjgB, zwf | Glucose | Fed-batch | 71.1 | [ | |
| E. coli | Δpta, ΔyjgB, ΔadhE, ΔldhA, ΔpflB, ΔadhP, ΔyqhD, ΔeutG, ΔilvB, ΔpoxB; AKR, DERA, PDC | Acetaldehyd; 3-Hydroxybutanal (3-HB) | Biotransformation | 2.4 | [ | |
| 1,4-BDO | E. coli | Δsad::cat2-bld-bdh, ΔlacZ::cat1-sucD 4hbd, ΔllpdA::K.p.lpdA D354KΔpflB, ΔarcA, Δmdh, ΔadhE, ΔldhA, knock down tesB; gabD, ybgC, gltAR163L | Glucose | Bioreactor | 1.8 | [ |
| E. coli | ΔadhE, ΔldhA, ΔpflB, Δmdh, ΔarcA, lpdA::K.p.lpdD354K; gltAR163L, sucA, 4hbd, cat2, ald, adh | Glucose | Bioreactor | 18 | [ | |
| E. coli | ΔaraA, Δicd; araC, araD, araA, araB, araE, kivd, yqhd | L-Arabinose | Bioreactor | 1.51 | [ | |
| E. coli | None; None | Glucose | Commercial-scale fermentation | >125 | [ | |
| E. coli | None; gadB, gabT, yqhD,car, ppc, gltAR163L | Amino acids (AAs), Glucose | General metabolic platform | 1.41 | [ | |
| E. coli | ΔyagE, ΔxylA, ΔyjhH; KvidV461I,xylBCDX, yqhD | Glucose, Xylose | D-xylose, L-arabinose, D-galacturonate | 12 | [ | |
| E. coli | ΔuxaC, ΔgarL, Δicd; udh, garD, ycbC, xylA(CC), kivd, yqhd | D-Galacturonate | Bioreactor | 16.5 | [ | |
| E. coli | ΔxylA, ΔyjhH, ΔyagE, Δicd; xylB, xylC, xylD, xylX, xylA(CC), kivdV461I, yqhd | Xylose | Bioreactor | 12 | [ | |
| 1,2-PDO | E. coli | ΔpoxB, ΔfrdA, ΔmgsA, ΔadhE::pdcD; gldA, mgs | Glucose | Shake flask | 0.7 | [ |
| E. coli | ΔlldD::mmsB, ΔackA-pta::pct, ΔldhA::Lldh; pct, pdcD, mmsB | Glucose | Shake flask | 1.04 | [ | |
| E. coli | ΔldhA::KanR;mgs, gldA, fucO | Glucose | Bioreactor | 4.5 | [ | |
| E. coli | Δzwf, ΔtpiA, ΔgloA, ΔldhA, ΔadhE,; mgsA, gldA, fucO | Glucose | Shake flask | 5.13 | [ | |
| E. coli | ΔlldD, Δdld, ΔldhA, ΔadhE; pct,pduP, yahK | D-/L-Lactate | Shake flask | (R)-1,2-PDO,1.5; (S)-1,2-PDO,1.7 | [ | |
| E. coli | Δac kA-pta, ΔldhA, ΔdhaK; dhaKL, gldA, mgsA, yqhD | Glycerol | Bioreactor | 5.6 | [ | |
| BT | E. coli | ΔyiaE, ΔycdW, ΔxylA KivD(Lactococcus lactis); None | Xylose | Fed-batch, carbon flux optimization | 10.03 | [ |
| E. coli | None; YqhD, YjhG | Xylose | Optimized cultivation | 5.1 | [ | |
| E. coli | ΔxylA, ΔxylB, ΔyjhE, ΔyagH, ΔycdW;xdh, mdlC | Xylose | Fed-batch | 2.38 | [ | |
| E. coli | None; XylD, KdcA (Lactococcus lactis), AdhP (E. coli) | Xylose | Optimized cultivation | 5.1 | [ | |
| E. coli | ΔfadE; XylD, fadD | Xylose, free fatty acids | Fed-batch | 1.1 (BT esters) | [ | |
| S. cerevisiae | None; XylB, XylD | Xylose, rice straw hydrolysate | Aerobic fermentation | 1.7 | [ |
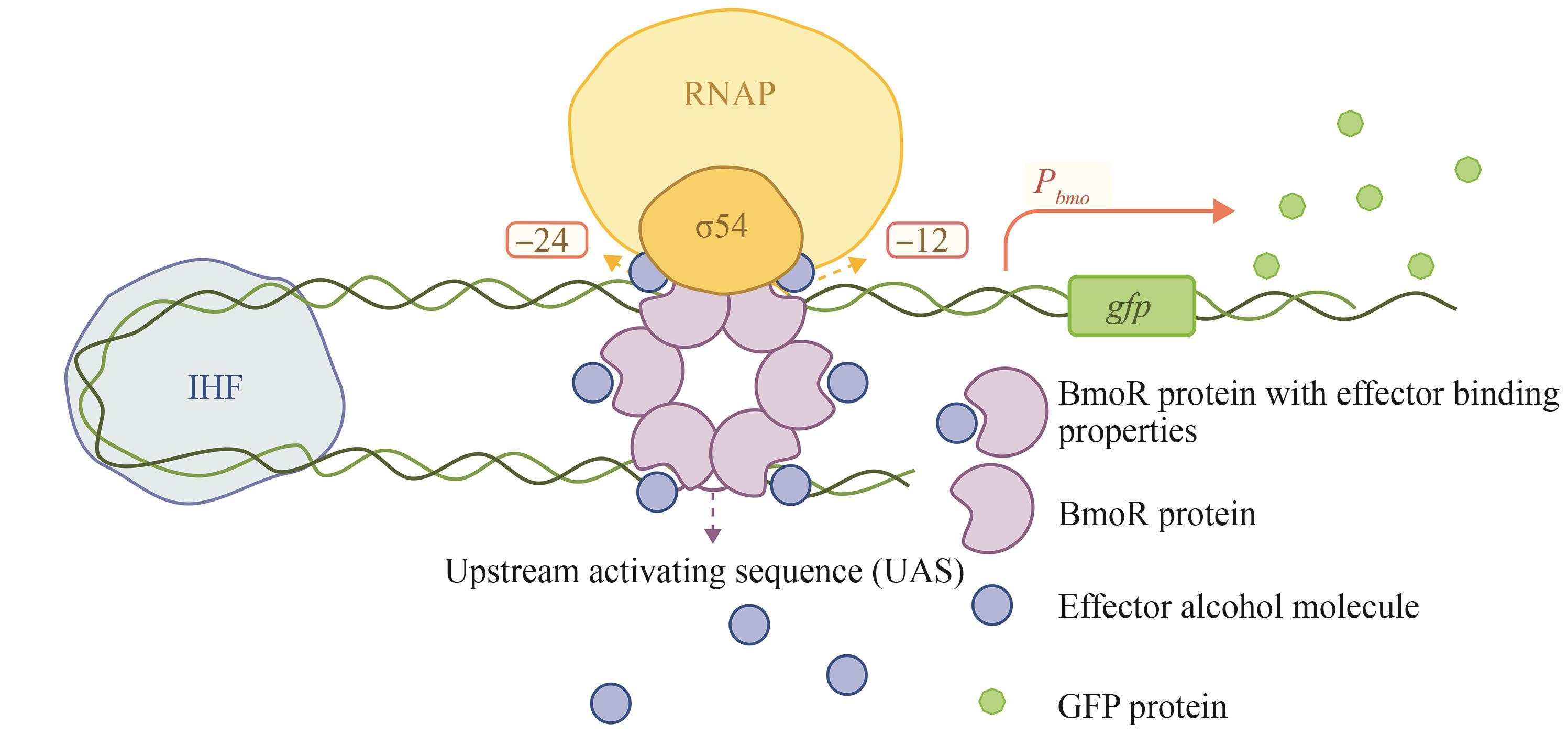
图5 BmoR传感器作用机制[σ54依赖型启动子Pbmo 受醇响应蛋白BmoR的调控。当BmoR蛋白感应到醇分子的存在时,会形成多聚体,并结合至启动子Pbmo 上游的激活序列(UAS)。该激活序列位于σ⁵⁴-RNA聚合酶结合位点(图中所示为-24和-12区域)上游。由于σ54-RNA聚合酶与BmoR之间存在一定距离,宿主整合因子(IHF)蛋白通过促使DNA形成环状结构,实现远距离的转录调控]
Fig. 5 Mechanism of action for the BmoR sensor[The σ54-dependent promoter Pbmo is regulated by the BmoR protein, which responds to alcohol. When alcohol is present, BmoR forms a multimer (depicted as a hexamer in the figure, a presumed form) and attaches to the upstream activating sequence (UAS) of Pbmo, located upstream of the σ54-RNA polymerase binding sites (shown in the figure as the -24 and -12 regions). Due to the distance between the σ54-RNA polymerase and BmoR, DNA looping facilitated by the integration host factor (IHF) protein achieves long-distance regulation.]
| [87] | BOOCK J T, FREEDMAN A J E, TOMPSETT G A, et al. Engineered microbial biofuel production and recovery under supercritical carbon dioxide[J]. Nature Communications, 2019, 10: 587. |
| [88] | LI S S, JIA X Q, WEN J P. Improved 2-methyl-1-propanol production in an engineered Bacillus subtilis by constructing inducible pathways[J]. Biotechnology Letters, 2012, 34(12): 2253-2258. |
| [89] | HIGASHIDE W, LI Y C, YANG Y F, et al. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose[J]. Applied and Environmental Microbiology, 2011, 77(8): 2727-2733. |
| [90] | BLOMBACH B, RIESTER T, WIESCHALKA S, et al. Corynebacterium glutamicum tailored for efficient isobutanol production[J]. Applied and Environmental Microbiology, 2011, 77(10): 3300-3310. |
| [91] | LI H, OPGENORTH P H, WERNICK D G, et al. Integrated electromicrobial conversion of CO2 to higher alcohols[J]. Science, 2012, 335(6076): 1596. |
| [92] | LU J N, BRIGHAM C J, GAI C S, et al. Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha [J]. Applied Microbiology and Biotechnology, 2012, 96(1): 283-297. |
| [93] | MATSUDA F, ISHII J, KONDO T, et al. Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance[J]. Microbial Cell Factories, 2013, 12: 119. |
| [94] | QIU M Y, SHEN W, YAN X Y, et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production[J]. Biotechnology for Biofuels, 2020, 13: 15. |
| [95] | LIN P P, MI L, MORIOKA A H, et al. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum [J]. Metabolic Engineering, 2015, 31: 44-52. |
| [96] | HASEGAWA S, JOJIMA T, SUDA M, et al. Isobutanol production in Corynebacterium glutamicum: suppressed succinate by-production by pckA inactivation and enhanced productivity via the Entner-Doudoroff pathway[J]. Metabolic Engineering, 2020, 59: 24-35. |
| [97] | RUSSMAYER H, MARX H, SAUER M. Microbial 2-butanol production with Lactobacillus diolivorans [J]. Biotechnology for Biofuels, 2019, 12: 262. |
| [98] | CHEN Z, WU Y, HUANG J H, et al. Metabolic engineering of Klebsiella pneumoniae for the de novo production of 2-butanol as a potential biofuel[J]. Bioresource Technology, 2015, 197: 260-265. |
| [99] | GHIACI P, LAMEIRAS F, NORBECK J, et al. Production of 2-butanol through meso-2,3-butanediol consumption in lactic acid bacteria[J]. FEMS Microbiology Letters, 2014, 360(1): 70-75. |
| [100] | GHIACI P, NORBECK J, LARSSON C. 2-Butanol and butanone production in Saccharomyces cerevisiae through combination of a B12 dependent dehydratase and a secondary alcohol dehydrogenase using a TEV-based expression system[J]. PLoS One, 2014, 9(7): e102774. |
| [101] | SPERANZA G, CORTI S, FONTANA G, et al. Conversion of meso-2,3-butanediol into 2-butanol by Lactobacilli. Stereochemical and enzymatic aspects[J]. Journal of agricultural and food chemistry, 1997, 45(9): 3476-3480. |
| [102] | SU H F, LIN J F, WANG Y H, et al. Engineering Brevibacterium flavum for the production of renewable bioenergy: C4–C5 advanced alcohols[J]. Biotechnology and Bioengineering, 2017, 114(9): 1946-1958. |
| [103] | SU H F, CHEN H, LIN J F. Enriching the production of 2-methyl-1-butanol in fermentation process using Corynebacterium crenatum [J]. Current Microbiology, 2020, 77(8): 1699-1706. |
| [104] | CONNOR M R, CANN A F, LIAO J C. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation[J]. Applied Microbiology and Biotechnology, 2010, 86(4): 1155-1164. |
| [105] | RAO B, ZHANG L Y, SUN J A, et al. Characterization and regulation of the 2,3-butanediol pathway in Serratia marcescens [J]. Applied Microbiology and Biotechnology, 2012, 93(5): 2147-2159. |
| [106] | ZHANG L Y, SUN J A, HAO Y L, et al. Microbial production of 2,3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30[J]. Journal of Industrial Microbiology & Biotechnology, 2010, 37(8): 857-862. |
| [107] | YANG S H, MOHAGHEGHI A, FRANDEN M A, et al. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars[J]. Biotechnology for Biofuels, 2016, 9(1): 189. |
| [108] | UI S, OKAJIMA Y, MIMURA A, et al. Molecular generation of an Escherichia coli strain producing only the meso-isomer of 2,3-butanediol[J]. Journal of Fermentation and Bioengineering, 1997, 84(3): 185-189. |
| [109] | YAN Y J, LEE C C, LIAO J C. Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases[J]. Organic & Biomolecular Chemistry, 2009, 7(19): 3914-3917. |
| [110] | CHU H P, XIN B, LIU P H, et al. Metabolic engineering of Escherichia coli for production of (2S,3S)-butane-2,3-diol from glucose[J]. Biotechnology for Biofuels, 2015, 8: 143. |
| [111] | RESHAMWALA S M S, DEB S S, LALI A M. A shortened, two-enzyme pathway for 2,3-butanediol production in Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(9): 1273-1277. |
| [112] | TONG Y J, JI X J, SHEN M Q, et al. Constructing a synthetic constitutive metabolic pathway in Escherichia coli for (R,R)-2,3-butanediol production[J]. Applied Microbiology and Biotechnology, 2016, 100(2): 637-647. |
| [113] | HWANG H J, LEE S Y, LEE P C. Engineering and application of synthetic nar promoter for fine-tuning the expression of metabolic pathway genes in Escherichia coli [J]. Biotechnology for Biofuels, 2018, 11: 103. |
| [114] | ERIAN A M, GIBISCH M, PFLÜGL S. Engineered E. coli W enables efficient 2,3-butanediol production from glucose and sugar beet molasses using defined minimal medium as economic basis[J]. Microbial Cell Factories, 2018, 17(1): 190. |
| [115] | LI Z J, JIAN J, WEI X X, et al. Microbial production of meso-2,3-butanediol by metabolically engineered Escherichia coli under low oxygen condition[J]. Applied Microbiology and Biotechnology, 2010, 87(6): 2001-2009. |
| [116] | WANG Y, LI L X, MA C Q, et al. Engineering of cofactor regeneration enhances (2S,3S)-2,3-butanediol production from diacetyl[J]. Scientific Reports, 2013, 3: 2643. |
| [117] | UI S, TAKUSAGAWA Y, SATO T, et al. Production of L-2,3-butanediol by a new pathway constructed in Escherichia coli [J]. Letters in Applied Microbiology, 2004, 39(6): 533-537. |
| [118] | MAZUMDAR S, LEE J, OH M K. Microbial production of 2,3-butanediol from seaweed hydrolysate using metabolically engineered Escherichia coli [J]. Bioresource Technology, 2013, 136: 329-336. |
| [119] | SHIN H D, YOON S H, WU J R, et al. High-yield production of meso-2,3-butanediol from cellodextrin by engineered E. coli biocatalysts[J]. Bioresource Technology, 2012, 118: 367-373. |
| [120] | JAIN R, SUN X X, YUAN Q P, et al. Systematically engineering Escherichia coli for enhanced production of 1,2-propanediol and 1-propanol[J]. ACS Synthetic Biology, 2015, 4(6): 746-756. |
| [121] | LIANG Q F, ZHANG H J, LI S N, et al. Construction of stress-induced metabolic pathway from glucose to 1,3-propanediol in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2011, 89(1): 57-62. |
| [122] | YANG B, LIANG S X, LIU H H, et al. Metabolic engineering of Escherichia coli for 1,3-propanediol biosynthesis from glycerol[J]. Bioresource Technology, 2018, 267: 599-607. |
| [123] | TANG X M, TAN Y S, ZHU H, et al. Microbial conversion of glycerol to 1,3-propanediol by an engineered strain of Escherichia coli [J]. Applied and Environmental Microbiology, 2009, 75(6): 1628-1634. |
| [124] | YUN J H, ZABED H M, ZHANG Y F, et al. Co-fermentation of glycerol and glucose by a co-culture system of engineered Escherichia coli strains for 1,3-propanediol production without vitamin B12 supplementation[J]. Bioresource Technology, 2021, 319: 124218. |
| [125] | LI Z H, DONG Y F, LIU Y, et al. Systems metabolic engineering of Corynebacterium glutamicum for high-level production of 1,3-propanediol from glucose and xylose[J]. Metabolic Engineering, 2022, 70: 79-88. |
| [126] | ZHANG Y, SUN Q, LIU Y, et al. Development of a plasmid stabilization system in Vibrio natriegens for the high production of 1,3-propanediol and 3-hydroxypropionate[J]. Bioresources and Bioprocessing, 2021, 8(1): 125. |
| [127] | ZHANG Y J, MA C W, DISCHERT W, et al. Engineering of phosphoserine aminotransferase increases the conversion of L-homoserine to 4-hydroxy-2-ketobutyrate in a glycerol-independent pathway of 1,3-propanediol production from glucose[J]. Biotechnology Journal, 2019, 14(9): 1900003. |
| [128] | LI M D, ZHANG Y, LI J C, et al. Biosynthesis of 1,3-propanediol via a new pathway from glucose in Escherichia coli [J]. ACS Synthetic Biology, 2023, 12(7): 2083-2093. |
| [129] | GUO J, CAO Y J, LIU H, et al. Improving the production of isoprene and 1,3-propanediol by metabolically engineered Escherichia coli through recycling redox cofactor between the dual pathways[J]. Applied Microbiology and Biotechnology, 2019, 103(6): 2597-2608. |
| [130] | KATAOKA N, VANGNAI A S, UEDA H, et al. Enhancement of (R)-1,3-butanediol production by engineered Escherichia coli using a bioreactor system with strict regulation of overall oxygen transfer coefficient and pH[J]. Bioscience, Biotechnology, and Biochemistry, 2014, 78(4): 695-700. |
| [131] | LIU Y, CEN X C, LIU D H, et al. Metabolic engineering of Escherichia coli for high-yield production of (R)-1,3-butanediol[J]. ACS Synthetic Biology, 2021, 10(8): 1946-1955. |
| [132] | WANG J, ZHANG R H, ZHANG J L, et al. Tunable hybrid carbon metabolism coordination for the carbon-efficient biosynthesis of 1,3-butanediol in Escherichia coli [J]. Green Chemistry, 2021, 23(21): 8694-8706. |
| [133] | ISLAM T, NGUYEN-VO T P, GAUR V K, et al. Metabolic engineering of Escherichia coli for biological production of 1,3-butanediol[J]. Bioresource Technology, 2023, 376: 128911. |
| [134] | ISLAM T, NGUYEN-VO T P, CHO S, et al. Metabolic engineering of Escherichia coli for enhanced production of 1,3-butanediol from glucose[J]. Bioresource Technology, 2023, 389: 129814. |
| [1] | PRZYSTAŁOWSKA H, ZEYLAND J, SZYMANOWSKA-POWAŁOWSKA D, et al. 1,3-Propanediol production by new recombinant Escherichia coli containing genes from pathogenic bacteria[J]. Microbiological Research, 2015, 171: 1-7. |
| [2] | ABDERRAZAK H B, FILDIER A, ROMDHANE H B, et al. Synthesis of new poly(ether ketone)s derived from biobased diols[J]. Macromolecular Chemistry and Physics, 2013, 214(13): 1423-1433. |
| [3] | GARCÍA-HERNÁNDEZ A E, SEGOVIA-HERNÁNDEZ J G, SÁNCHEZ-RAMÍREZ E, et al. Sustainable aviation fuel from butanol: a study in optimizing economic and environmental impact through process intensification[J]. Chemical Engineering and Processing-Process Intensification, 2024, 200: 109769. |
| [4] | ZHU L F, GUAN X C, XIE N Z, et al. Fermentative production of enantiomerically pure S-1,2-propanediol from glucose by engineered E. coli strain[J]. Applied Microbiology and Biotechnology, 2016, 100(3): 1241-1251. |
| [5] | SALUSJÄRVI L, HAVUKAINEN S, KOIVISTOINEN O, et al. Biotechnological production of glycolic acid and ethylene glycol: current state and perspectives[J]. Applied Microbiology and Biotechnology, 2019, 103(6): 2525-2535. |
| [6] | YU C, CAO Y J, ZOU H B, et al. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols[J]. Applied Microbiology and Biotechnology, 2011, 89(3): 573-583. |
| [7] | CANN A F, LIAO J C. Production of 2-methyl-1-butanol in engineered Escherichia coli [J]. Applied Microbiology and Biotechnology, 2008, 81(1): 89-98. |
| [8] | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels[J]. Nature, 2008, 451(7174): 86-89. |
| [9] | RADDADI N, CHERIF A, DAFFONCHIO D, et al. Biotechnological applications of extremophiles, extremozymes and extremolytes[J]. Applied Microbiology and Biotechnology, 2015, 99(19): 7907-7913. |
| [10] | CHEN G Q, JIANG X R. Next generation industrial biotechnology based on extremophilic bacteria[J]. Current Opinion in Biotechnology, 2018, 50: 94-100. |
| [11] | YIN J, CHEN J C, WU Q, et al. Halophiles, coming stars for industrial biotechnology[J]. Biotechnology Advances, 2015, 33(7): 1433-1442. |
| [12] | Oxo alcohol market size, share & trends analysis report by application, outlook regional, strategies competitive, and forecasts segment, 2019 To 2025[R/OL]. 2019[2024-12-01]. . |
| [135] | KIM T H, FLICK R, BRUNZELLE J, et al. Novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol: structural and biochemical studies[J]. Applied and Environmental Microbiology, 2017, 83(7): e03172-16. |
| [136] | WU M Y, SUNG L Y, LI H, et al. Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1,4-BDO biosynthesis[J]. ACS Synthetic Biology, 2017, 6(12): 2350-2361. |
| [137] | TAI Y S, XIONG M Y, JAMBUNATHAN P, et al. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products[J]. Nature Chemical Biology, 2016, 12(4): 247-253. |
| [138] | BURGARD A, BURK M J, OSTERHOUT R, et al. Development of a commercial scale process for production of 1,4-butanediol from sugar[J]. Current Opinion in Biotechnology, 2016, 42: 118-125. |
| [139] | WANG J, LI C Y, ZOU Y S, et al. Bacterial synthesis of C3-C5 diols via extending amino acid catabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(32): 19159-19167. |
| [140] | ALTARAS N E, CAMERON D C. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli [J]. Applied and Environmental Microbiology, 1999, 65(3): 1180-1185. |
| [141] | ALTARAS N E, CAMERON D C. Enhanced production of (R)-1,2-propanediol by metabolically engineered Escherichia coli [J]. Biotechnology Progress, 2000, 16(6): 940-946. |
| [142] | NIU W, GUO J T. Stereospecific microbial conversion of lactic acid into 1,2-propanediol[J]. ACS Synthetic Biology, 2015, 4(4): 378-382. |
| [143] | CLOMBURG J M, GONZALEZ R. Metabolic engineering of Escherichia coli for the production of 1,2-propanediol from glycerol[J]. Biotechnology and Bioengineering, 2011, 108(4): 867-879. |
| [144] | JING P Y, CAO X, LU X Y, et al. Modification of an engineered Escherichia coli by a combined strategy of deleting branch pathway, fine-tuning xylose isomerase expression, and substituting decarboxylase to improve 1,2,4-butanetriol production[J]. Journal of Bioscience and Bioengineering, 2018, 126(5): 547-552. |
| [145] | ZHANG N N, WANG J B, ZHANG Y, et al. Metabolic pathway optimization for biosynthesis of 1,2,4-butanetriol from xylose by engineered Escherichia coli [J]. Enzyme and Microbial Technology, 2016, 93-94: 51-58. |
| [146] | 孙雷, 杨帆, 朱泰承, 等. 大肠杆菌合成1,2,4-丁三醇的途径优化[J]. 生物工程学报, 2016, 32(1): 51-63. |
| [13] | Tridecyl alcohol market report 2025 (Global edition) [R/OL]. [2024-12-01]. . |
| [14] | JAISWAL C. Polyols market research report information by product (polyether, polyester), by application (flexible foam, foamrigid, coatings, adhesives & sealants), and by region (North America, Europe, Asia-Pacific, and rest of the world)-market forecast till 2032[R/OL]. Market Research Future, 2025[2025-04-18]. . |
| [15] | New frontiers: the implication of the revised energy performance of buildings directive in the energy-efficient building market and renewable energy market[EB/OL]. [2025-02-01]. . |
| [16] | JI X J, HUANG H, OUYANG P K. Microbial 2,3-butanediol production: a state-of-the-art review[J]. Biotechnology Advances, 2011, 29(3): 351-364. |
| [17] | LIU Z, QIN J Y, GAO C, et al. Production of (2S,3S)-2,3-butanediol and (3S)-acetoin from glucose using resting cells of Klebsiella pneumonia and Bacillus subtilis [J]. Bioresource Technology, 2011, 102(22): 10741-10744. |
| [18] | 生意社(大宗商品网. 异丁醇价格-生意社大宗商品数据[EB/OL]. [2025-02-01]. . |
| Business Society (Bulk Commodity Network). Isobutanol price-business society bulk commodity data[EB/OL]. [2025-02-01]. . | |
| [19] | ChemAnalyst. Iso Butanol Price Trend and Forecast[EB/OL]. [2025-02-01]. . |
| [20] | QYResearch. 1,3 -丁二醇行业剖析:2024年全球市场规模约为2.29亿美元[EB/OL]. (2025-03-07)[2025-02-01]. . |
| QYResearch. Analysis of the 1,3-butanediol industry: the global market size is estimated at USD 229 million in 2024[EB/OL]. (2025-03-07)[2025-02-01]. . | |
| [21] | 生意社(大宗商品网. 1,3 -丁二醇价格(生意社大宗商品数据)[EB/OL]. [2025-02-01]. . |
| Business Society (Bulk Commodity Network). 1,3 -Butanediol price (business society bulk commodity data) [EB/OL]. [2025-02-01]. . | |
| [146] | SUN L, YANG F, ZHU T C, et al. Optimization of 1,2,4-butanetriol synthetic pathway in Escherichia coli [J]. Chinese Journal of Biotechnology, 2016, 32(1): 51-63. |
| [147] | WANG X, XU N N, HU S W, et al. D-1,2,4-butanetriol production from renewable biomass with optimization of synthetic pathway in engineered Escherichia coli [J]. Bioresource Technology, 2018, 250: 406-412. |
| [148] | FENG X J, GAO W J, ZHOU Y F, et al. Coupled biosynthesis and esterification of 1,2,4-butanetriol to simplify its separation from fermentation broth[J]. Engineering in Life Sciences, 2019, 19(6): 444-451. |
| [149] | BAMBA T, YUKAWA T, GUIRIMAND G, et al. Production of 1,2,4-butanetriol from xylose by Saccharomyces cerevisiae through Fe metabolic engineering[J]. Metabolic Engineering, 2019, 56: 17-27. |
| [150] | WANG G L, WANG M Y, YANG J C, et al. De novo synthesis of 2-phenylethanol from glucose by metabolically engineered Escherichia coli [J]. Journal of Industrial Microbiology and Biotechnology, 2022, 49(6): kuac026. |
| [151] | 马晓焉, 王雪芹, 马炼杰, 等. 高级醇的微生物绿色制造[J]. 生物工程学报, 2021, 37(5): 1721-1736. |
| MA X Y, WANG X Q, MA L J, et al. Microbial green manufacturing of higher alcohols[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1721-1736. | |
| [152] | CHEN R B, YANG S, ZHANG L, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23(3): 100879. |
| [153] | WANG P C, YANG X W, LIN B X, et al. Cofactor self-sufficient whole-cell biocatalysts for the production of 2-phenylethanol[J]. Metabolic Engineering, 2017, 44: 143-149. |
| [154] | LIU H Y, WU X X, MA H, et al. High-level production of hydroxytyrosol in engineered Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2022, 11(11): 3706-3713. |
| [155] | XIAO F, LI D F, PAN Y J, et al. Establishing Komagataella phaffii as a cell factory for efficient production of cholesterol sulfate[J]. ACS Sustainable Chemistry & Engineering, 2025, 13(1): 174-186. |
| [156] | ASPACIO D, ZHANG Y L, CUI Y T, et al. Shifting redox reaction equilibria on demand using an orthogonal redox cofactor[J]. Nature Chemical Biology, 2024, 20(11): 1535-1546. |
| [22] | CELIŃSKA E, GRAJEK W. Biotechnological production of 2,3-butanediol—current state and prospects[J]. Biotechnology Advances, 2009, 27(6): 715-725. |
| [23] | MAINA S, PRABHU A A, VIVEK N, et al. Prospects on bio-based 2,3-butanediol and acetoin production: recent progress and advances[J]. Biotechnology Advances, 2022, 54: 107783. |
| [24] | JAISWAL C. 2,3 -Butanediol market research report: by application (solvents, plasticizers, biodegradable polymers, pharmaceuticals, food additives), by end use industry (automotive, cosmetics, food beverage, pharmaceuticals, polymer), by product form (liquid, solid, gas), by production method (fermentation, chemical synthesis, biochemical process) and by regional (North America, Europe, South America, Asia Pacific, Middle East and Africa) - forecast to 2034[R/OL]. Market Research Future, 2025[2025-04-18]. . |
| [25] | 习近平在第七十五届联合国大会一般性辩论上发表重要讲话[EB/OL]. (2020-09-22)[2025-02-01]. . |
| Xi Jinping delivered an important speech at the general debate of the 75th session of the United Nations general assembly[EB/OL]. (2020-09-22)[2025-02-01]. . | |
| [26] | LI C Z, ZHAO X C, WANG A Q, et al. Catalytic transformation of lignin for the production of chemicals and fuels[J]. Chemical Reviews, 2015, 115(21): 11559-11624. |
| [27] | ALONSO D M, WETTSTEIN S G, DUMESIC J A. Bimetallic catalysts for upgrading of biomass to fuels and chemicals[J]. Chemical Society Reviews, 2012, 41(24): 8075-8098. |
| [28] | KUMAR R, SINGH S, SINGH O V. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives[J]. Journal of Industrial Microbiology & Biotechnology, 2008, 35(5): 377-391. |
| [29] | DENG W P, FENG Y C, FU J, et al. Catalytic conversion of lignocellulosic biomass into chemicals and fuels[J]. Green Energy & Environment, 2023, 8(1): 10-114. |
| [30] | NDUKWE N A, IDIKA D I, OKIEI W O. Influence of cellulose-cellulase enzyme digestibility in the production of glucose from lignocellulosic biomass waste[J]. IOP Conference Series: Earth and Environmental Science, 2024, 1322(1): 012005. |
| [31] | DEMEKE M M, ECHEMENDIA D, BELO E, et al. Enhancing xylose-fermentation capacity of engineered Saccharomyces cerevisiae by multistep evolutionary engineering in inhibitor-rich lignocellulose hydrolysate[J]. FEMS Yeast Research, 2024, 24: foae013. |
| [32] | KUSUMOTO S, HIGASHI T, MATSUMOTO Y, et al. Hydrogenative degradation of PEG-functionalized lignin[J]. Polymer Journal, 2024, 56(4): 353-357. |
| [157] | WANG Y X, WAN Z J, ZHU Y T, et al. Enhanced 1,3-propanediol production with high yield from glycerol through a novel Klebsiella-Shewanella co-culture[J]. Biotechnology for Biofuels and Bioproducts, 2023, 16(1): 50. |
| [158] | SCHWARZ K M, GROSSE-HONEBRINK A, DERECKA K, et al. Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum [J]. Metabolic Engineering, 2017, 40: 124-137. |
| [159] | SUN S Q, SHU L, LU X Y, et al. 1,2-Propanediol production from glycerol via an endogenous pathway of Klebsiella pneumoniae [J]. Applied Microbiology and Biotechnology, 2021, 105(23): 9003-9016. |
| [160] | KIM S J, HAHN J S. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing[J]. Metabolic Engineering, 2015, 31: 94-101. |
| [161] | CHEN Z Y, YU S Z, LIU J, et al. Concentration recognition-based auto-dynamic regulation system (CRUISE) enabling efficient production of higher alcohols[J]. Advanced Science, 2024, 11(23): 2310215. |
| [162] | SOH L M J, MAK W S, LIN P P, et al. Engineering a thermostable keto acid decarboxylase using directed evolution and computationally directed protein design[J]. ACS Synthetic Biology, 2017, 6(4): 610-618. |
| [163] | SUTIONO S, CARSTEN J, SIEBER V. Structure-guided engineering of α-keto acid decarboxylase for the production of higher alcohols at elevated temperature[J]. ChemSusChem, 2018, 11(18): 3335-3344. |
| [164] | JANG Y S, YANG J, KIM J K, et al. Adaptive laboratory evolution and transcriptomics-guided engineering of Escherichia coli for increased isobutanol tolerance[J]. Biotechnology Journal, 2024, 19(1): 2300270. |
| [165] | KNOSHAUG E P, ZHANG M. Butanol tolerance in a selection of microorganisms[J]. Applied Biochemistry and Biotechnology, 2009, 153(1): 13-20. |
| [166] | RÜHL J, SCHMID A, BLANK L M. Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations[J]. Applied and Environmental Microbiology, 2009, 75(13): 4653-4656. |
| [167] | ZAKI A M, WIMALASENA T T, GREETHAM D. Phenotypic characterisation of Saccharomyces spp. for tolerance to 1-butanol[J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(11): 1627-1636. |
| [168] | KATAOKA N, TAJIMA T, KATO J, et al. Development of butanol-tolerant Bacillus subtilis strain GRSW2-B1 as a potential bioproduction host[J]. AMB Express, 2011, 1: 10. |
| [33] | MA C Y, LUO X T, XU L H, et al. Structural elucidation and targeted valorization of untractable lignin from pre-hydrolysis liquor of xylose production via a simple and robust separation approach[J]. International Journal of Biological Macromolecules, 2023, 253: 127029. |
| [34] | ZHANG S Z, DUAN Y, TENG C C, et al. Fast and selective degradation of biomass for xylose, glucose and lignin under mild conditions[J]. Molecules, 2023, 28(8): 3306. |
| [35] | 国家统计局. 2023年国民经济回升向好 高质量发展扎实推进[EB/OL].(2024-01-17)[2025-02-01]. . |
| National Bureau of Statistics. The national economy improved in 2023 with solid progress in high-quality development[EB/OL].(2024-01-17)[2025-02-01]. . | |
| [36] | ABDELGHANY A M, ZHANG S R, AZAM M, et al. Natural variation in fatty acid composition of diverse world soybean germplasms grown in China[J]. Agronomy, 2020, 10(1): 24. |
| [37] | ZHANG H L, GAO J R, ZHAO Z D, et al. Esterification of fatty acids from waste cooking oil to biodiesel over a sulfonated resin/PVA composite[J]. Catalysis Science & Technology, 2016, 6(14): 5590-5598. |
| [38] | GANESH I, RAVIKUMAR S, HONG S H. Metabolically engineered Escherichia coli as a tool for the production of bioenergy and biochemicals from glycerol[J]. Biotechnology and Bioprocess Engineering, 2012, 17(4): 671-678. |
| [39] | KRISHNAN A, MCNEIL B A, STUART D T. Biosynthesis of fatty alcohols in engineered microbial cell factories: advances and limitations[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 610936. |
| [40] | CERÓN FERRUSCA M, ROMERO R, MARTÍNEZ S L, et al. Biodiesel production from waste cooking oil: a perspective on catalytic processes[J]. Processes, 2023, 11(7): 1952. |
| [41] | JATOI A S, SHAH A ALI, AHMED J, et al. Hydrothermal liquefaction of lignocellulosic and protein-containing biomass: a comprehensive review[J]. Catalysts, 2022, 12(12): 1621. |
| [42] | ÄYRÄPÄÄ T. Formation of higher alcohols from amino acids derived from yeast proteins[J]. Journal of the Institute of Brewing, 1967, 73(1): 30-33. |
| [43] | EL-DALATONY M M, SAHA S, GOVINDWAR S P, et al. Biological conversion of amino acids to higher alcohols[J]. Trends in Biotechnology, 2019, 37(8): 855-869. |
| [169] | KURTH E G, DOUGHTY D M, BOTTOMLEY P J, et al. Involvement of BmoR and BmoG in n-alkane metabolism in ‘Pseudomonas butanovora’[J]. Microbiology, 2008, 154(Pt 1): 139-147. |
| [170] | KIM N M, SINNOTT R W, ROTHSCHILD L N, et al. Elucidation of sequence-function relationships for an improved biobutanol in vivo biosensor in E. coli [J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 821152. |
| [171] | SATHESH-PRABU C, TIWARI R, KIM D, et al. Inducible and tunable gene expression systems for Pseudomonas putida KT2440[J]. Scientific Reports, 2021, 11: 18079. |
| [172] | COBAN I, LAMPING J P, HIRSCH A G, et al. dsRNA formation leads to preferential nuclear export and gene expression[J]. Nature, 2024, 631(8020): 432-438. |
| [173] | GILBERT L A, LARSON M H, MORSUT L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes[J]. Cell, 2013, 154(2): 442-451. |
| [174] | CANVER M C, SMITH E C, SHER F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis[J]. Nature, 2015, 527(7577): 192-197. |
| [175] | KORKMAZ G, LOPES R, UGALDE A P, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9[J]. Nature Biotechnology, 2016, 34(2): 192-198. |
| [176] | SANJANA N E, WRIGHT J, ZHENG K J, et al. High-resolution interrogation of functional elements in the noncoding genome[J]. Science, 2016, 353(6307): 1545-1549. |
| [177] | REILLY S K, GOSAI S J, GUTIERREZ A, et al. Direct characterization of cis-regulatory elements and functional dissection of complex genetic associations using HCR-FlowFISH[J]. Nature Genetics, 2021, 53(8): 1166-1176. |
| [178] | GASPERINI M, HILL A J, MCFALINE-FIGUEROA J L, et al. A genome-wide framework for mapping gene regulation via cellular genetic screens[J]. Cell, 2019, 176(6): 1516. |
| [179] | THAKORE P I, D’IPPOLITO A M, SONG L Y, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements[J]. Nature Methods, 2015, 12(12): 1143-1149. |
| [180] | NASSER J, BERGMAN D T, FULCO C P, et al. Genome-wide enhancer maps link risk variants to disease genes[J]. Nature, 2021, 593(7858): 238-243. |
| [181] | CHEN Z Y, JAVED N, MOORE M, et al. Integrative dissection of gene regulatory elements at base resolution[J]. Cell Genomics, 2023, 3(6): 100318. |
| [182] | MOORE M M, WEKHANDE S, ISSNER R, et al. Multi-locus CRISPRi targeting with a single truncated guide RNA[J]. Nature Communications, 2025, 16: 1357. |
| [183] | YU Y Y, GAWLITT S, DE ANDRADE E SOUSA L B, et al. Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration[J]. Genome Biology, 2024, 25(1): 13. |
| [44] | 何新媛, 刘杨, 曾祥荷, 等. 基于生物信息学的蛋白质功能预测方法研究进展[J]. 生物工程学报, 2024, 40(7): 2087-2099. |
| HE X Y, LIU Y, ZENG X H, et al. Advances in bioinformatics-based protein function prediction[J]. Chinese Journal of Biotechnology, 2024, 40(7): 2087-2099. | |
| [45] | MU D D, LI P L, MA T G, et al. Advances in the understanding of the production, modification and applications of xylanases in the food industry[J]. Enzyme and Microbial Technology, 2024, 179: 110473. |
| [46] | PARK M K, SHIN D M, CHOI Y S. Comparison of volatile compound profiles derived from various livestock protein alternatives including edible-insect, and plant-based proteins[J]. Food Chemistry: X, 2024, 23: 101570. |
| [47] | CO2·Earth. Daily CO2 [EB/OL]. [2025-02-01]. . |
| [48] | IEA. CO2 emissions in 2023[R/OL]. Paris: IEA, 2024[2025-02-01]. . |
| [49] | IEA. CO2 capture and utilisation[EB/OL]. [2025-04-18]. . |
| [50] | Research and markets. The global market for carbon dioxide(CO2)[R]. 2023. |
| [51] | IEA. Putting CO2 to use[R/OL]. Paris: IEA, 2019[2025-02-01]. . |
| [52] | Processing Hydrocarbon. World’s largest CO2-to-methanol plant starts production[EB/OL]. (2022-10-28)[2025-02-01]. . |
| [53] | BERGGREN M. Global methanol outlook 2023: growth and decarbonization [R/OL]. (2023-06-07)[2024-12-01]. . |
| [54] | PANICH J, TOPPARI E, TEJEDOR-SANZ S, et al. Functional plasticity of HCO3 - uptake and CO2 fixation in Cupriavidus necator H16[EB/OL]. bioRxiv, 2024: 2024.2005. 2007.593039. (2024-05-07)[2024-12-01]. . |
| [55] | MACLEAN M R, CLARK D A, LAIGLE L, et al. Carbon dioxide uptake and fixation by ferrous iron-oxidizing acidophilic bacteria[J]. Minerals Engineering, 2024, 206: 108532. |
| [184] | JUNG D, LIU B Y, HE X P, et al. Accessing previously uncultured marine microbial resources by a combination of alternative cultivation methods[J]. Microbial Biotechnology, 2021, 14(3): 1148-1158. |
| [185] | KAWAI R, TOYA Y, MIYOSHI K, et al. Acceleration of target production in co-culture by enhancing intermediate consumption through adaptive laboratory evolution[J]. Biotechnology and Bioengineering, 2022, 119(3): 936-945. |
| [186] | PANDE S, MERKER H, BOHL K, et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria[J]. The ISME Journal, 2014, 8(5): 953-962. |
| [187] | KARIM A, ISLAM M A, KHALID Z BIN, et al. Microbial lipid accumulation through bioremediation of palm oil mill effluent using a yeast-bacteria co-culture[J]. Renewable Energy, 2021, 176: 106-114. |
| [188] | SUN Y, LIU W C, SHI X, et al. Inducing secondary metabolite production of Aspergillus sydowii through microbial co-culture with Bacillus subtilis [J]. Microbial Cell Factories, 2021, 20(1): 42. |
| [189] | LIN Z N, LIU H J, CONG W, et al. Continuous fermentation coupled with online gas stripping for effective biobutanol production[J]. Fermentation, 2023, 9(11): 942. |
| [190] | BAEZ A, CHO K M, LIAO J C. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal[J]. Applied Microbiology and Biotechnology, 2011, 90(5): 1681-1690. |
| [191] | DING T T, LIANG Z Y, YANG Y, et al. Rapidly engineering an osmotic-pressure-tolerant gut bacterium for efficient non-sterile production of bulk chemicals[J]. Chemical Engineering Journal, 2024, 491: 152076. |
| [192] | BASEN M, SCHUT G J, NGUYEN D M, et al. Single gene insertion drives bioalcohol production by a thermophilic archaeon[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(49): 17618-17623. |
| [193] | XU P, LI L Y, ZHANG F M, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. |
| [194] | DAVID F, NIELSEN J, SIEWERS V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2016, 5(3): 224-233. |
| [195] | ANDREOZZI S, CHAKRABARTI A, SOH K C, et al. Identification of metabolic engineering targets for the enhancement of 1,4-butanediol production in recombinant E. coli using large-scale kinetic models[J]. Metabolic Engineering, 2016, 35: 148-159. |
| [196] | BOOB A G, ZHU Z X, INTASIAN P, et al. CRISPR-COPIES: an in silico platform for discovery of neutral integration sites for CRISPR/Cas-facilitated gene integration[J]. Nucleic Acids Research, 2024, 52(6): e30. |
| [197] | ABRAMSON J, ADLER J, DUNGER J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 630(8016): 493-500. |
| [56] | WANNICKE N, STÜEKEN E E, BAUERSACHS T, et al. Exploring the influence of atmospheric CO2 and O2 levels on the utility of nitrogen isotopes as proxy for biological N2 fixation[EB/OL]. bioRxiv, 2024: 2024.2003. 2028.587259. (2024-04-02)[2024-12-01]. . |
| [57] | CHEN H C, WANG D Q, WANG C C, et al. Lower micromolar activity of the antifungal imidazoles on the bacterial-type bifunctional aldehyde/alcohol dehydrogenase (AdhE) in Cryptosporidium parvum and in vitro efficacy against the zoonotic parasite[J]. International Journal for Parasitology Drugs and Drug Resistance, 2024, 25: 100551. |
| [58] | BROSSIER C, JARDOU M, JANASZKIEWICZ A, et al. Gut microbiota biotransformation of drug glucuronides leading to gastrointestinal toxicity: Therapeutic potential of bacterial β-glucuronidase inhibition in mycophenolate-induced enteropathy[J]. Life Sciences, 2024, 351: 122792. |
| [59] | ATSUMI S, CANN A F, CONNOR M R, et al. Metabolic engineering of Escherichia coli for 1-butanol production[J]. Metabolic Engineering, 2008, 10(6): 305-311. |
| [60] | MAINGUET S E, LIAO J C. Bioengineering of microorganisms for C3 to C5 alcohols production[J]. Biotechnology Journal, 2010, 5(12): 1297-1308. |
| [61] | SCHADEWEG V, BOLES E. n-Butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA[J]. Biotechnology for Biofuels, 2016, 9: 44. |
| [62] | KURIAN J V. A new polymer platform for the future—sorona® from corn derived 1,3-propanediol[J]. Journal of Polymers and the Environment, 2005, 13(2): 159-167. |
| [63] | LI Z H, WU Z Y, CEN X C, et al. Efficient production of 1,3-propanediol from diverse carbohydrates via a non-natural pathway using 3-hydroxypropionic acid as an intermediate[J]. ACS Synthetic Biology, 2021, 10(3): 478-486. |
| [64] | LEE Y G, SEO J H. Production of 2,3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2019, 12(1): 204. |
| [65] | KOU M Y, CUI Z Z, FU J, et al. Metabolic engineering of Corynebacterium glutamicum for efficient production of optically pure (2R,3R)-2,3-butanediol[J]. Microbial Cell Factories, 2022, 21(1): 150. |
| [66] | YIM H, HASELBECK R, NIU W, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol[J]. Nature Chemical Biology, 2011, 7(7): 445-452. |
| [67] | CEN X C, LIU Y J, ZHU F H, et al. Metabolic engineering of Escherichia coli for high production of 1,5-pentanediol via a cadaverine-derived pathway[J]. Metabolic Engineering, 2022, 74: 168-177. |
| [68] | ATSUMI S, LIAO J C. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli [J]. Applied and Environmental Microbiology, 2008, 74(24): 7802-7808. |
| [69] | CHOI Y J, PARK J H, KIM T Y, et al. Metabolic engineering of Escherichia coli for the production of 1-propanol[J]. Metabolic Engineering, 2012, 14(5): 477-486. |
| [70] | INOKUMA K, LIAO J C, OKAMOTO M, et al. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping[J]. Journal of Bioscience and Bioengineering, 2010, 110(6): 696-701. |
| [71] | SOMA Y, INOKUMA K, TANAKA T, et al. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system[J]. Journal of Bioscience and Bioengineering, 2012, 114(1): 80-85. |
| [72] | JOJIMA T, INUI M, YUKAWA H. Production of isopropanol by metabolically engineered Escherichia coli [J]. Applied Microbiology and Biotechnology, 2008, 77(6): 1219-1224. |
| [73] | LEE J M, JANG Y S, CHOI S J, et al. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation[J]. Applied and Environmental Microbiology, 2012, 78(5): 1416-1423. |
| [74] | DUSSÉAUX S, CROUX C, SOUCAILLE P, et al. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture[J]. Metabolic Engineering, 2013, 18: 1-8. |
| [75] | LAN E I, LIAO J C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide[J]. Metabolic Engineering, 2011, 13(4): 353-363. |
| [76] | LAN E I, LIAO J C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6018-6023. |
| [77] | JANG Y S, LEE J Y, LEE J, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum [J]. mBio, 2012, 3(5): e00314-12. |
| [78] | YU M R, DU Y M, JIANG W Y, et al. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum [J]. Applied Microbiology and Biotechnology, 2012, 93(2): 881-889. |
| [79] | STEEN E J, CHAN R, PRASAD N, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol[J]. Microbial Cell Factories, 2008, 7: 36. |
| [80] | NIELSEN D R, LEONARD E, YOON S H, et al. Engineering alternative butanol production platforms in heterologous bacteria[J]. Metabolic Engineering, 2009, 11(4-5): 262-273. |
| [81] | BEREZINA O V, ZAKHAROVA N V, BRANDT A, et al. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis [J]. Applied Microbiology and Biotechnology, 2010, 87(2): 635-646. |
| [82] | KÖPKE M, HELD C, HUJER S, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(29): 13087-13092. |
| [83] | SHEN C R, LAN E I, DEKISHIMA Y, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli [J]. Applied and Environmental Microbiology, 2011, 77(9): 2905-2915. |
| [84] | YU H, WANG N, HUO W B, et al. Establishment of BmoR-based biosensor to screen isobutanol overproducer[J]. Microbial Cell Factories, 2019, 18(1): 30. |
| [85] | ATSUMI S, HIGASHIDE W, LIAO J C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde[J]. Nature Biotechnology, 2009, 27(12): 1177-1180. |
| [86] | SU H F, LIN J F, WANG G W. Metabolic engineering of Corynebacterium crenatium for enhancing production of higher alcohols[J]. Scientific Reports, 2016, 6: 39543. |
| [1] | 朱欣悦, 陈恬恬, 邵恒煊, 唐曼玉, 华威, 程艳玲. 益生菌辅助防治恶性肿瘤的研究进展[J]. 合成生物学, 2025, 6(4): 899-919. |
| [2] | 吴晓燕, 宋琪, 许睿, 丁陈君, 陈方, 郭勍, 张波. 合成生物学研发竞争态势对比分析[J]. 合成生物学, 2025, 6(4): 940-955. |
| [3] | 张建康, 王文君, 郭洪菊, 白北辰, 张亚飞, 袁征, 李彦辉, 李航. 基于机器视觉的高通量微生物克隆挑选工作站研制及应用[J]. 合成生物学, 2025, 6(4): 956-971. |
| [4] | 李全飞, 陈乾, 刘浩, 贺坤东, 潘亮, 雷鹏, 谷益安, 孙良, 李莎, 邱溢彬, 王瑞, 徐虹. 高黏性蛋白材料的合成生物学及应用[J]. 合成生物学, 2025, 6(4): 806-828. |
| [5] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [6] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [7] | 李永珠, 陈禹. 酵母基因组规模模型进展及发展趋势[J]. 合成生物学, 2025, 6(3): 585-602. |
| [8] | 章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700. |
| [9] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [10] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [11] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| [12] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [13] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [14] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [15] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||