• 特约评述 •
基于CRISPR系统的高通量基因组编辑研究进展
滕佳尧1,2,3, 任传宏1,2,3, 朱芮莹1,2,3, 鲍泽华1,2,3,4
- 1.浙江大学化学工程与生物工程学院,生物质化工教育部重点实验室,浙江 杭州 310058
2.浙江大学杭州国际科创中心,全省功能化学品智造重点实验室,浙江 杭州 311215
3.浙江大学化学工程与生物工程学院,生物工程研究所,浙江 杭州 310058
4.浙江大学化学工程与生物工程学院,浙江省智能生物材料重点实验室,浙江 杭州 310058
-
收稿日期:2025-07-15修回日期:2025-10-30出版日期:2025-11-03 -
通讯作者:鲍泽华 -
作者简介:滕佳尧 (2002—),男,硕士研究生。研究方向为合成生物学。 E-mail:22428049@zju.edu.cn任传宏 (1993—),男,博士,助理研究员。研究方向为植物天然产物合成调控。E-mail:chuanhongren@163.com鲍泽华 (1988—),男,博士,研究员,博士生导师。研究方向为合成生物学、基因编辑、人工转录因子和定向进化等。 E-mail:zbao@zju.edu.cn -
基金资助:国家重点研发计划(2023YFF1204500);国家自然科学基金(22308316);中央高校基本科研业务费专项资金(226-2025-00043)
Recent advances in CRISPR-based high-throughput genome editing
TENG Jiayao1,2,3, REN Chuanhong1,2,3, ZHU Ruiying1,2,3, BAO Zehua1,2,3,4
- 1.Key Laboratory of Biomass Chemical Engineering of Ministry of Education,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310058,Zhejiang,China
2.Zhejiang Key Laboratory of Intelligent Manufacturing for Functional Chemicals,ZJU-Hangzhou Global Scientific and Technological Innovation Center,Zhejiang University,Hangzhou 311215,Zhejiang,China
3.Institute of Bioengineering,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310058,Zhejiang,China
4.Zhejiang Key Laboratory of Smart Biomaterials,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310058,Zhejiang,China
-
Received:2025-07-15Revised:2025-10-30Online:2025-11-03 -
Contact:BAO Zehua
摘要:
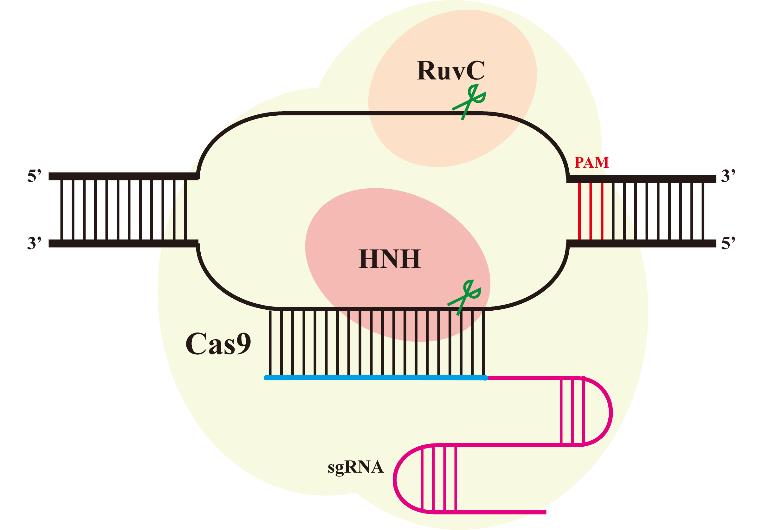
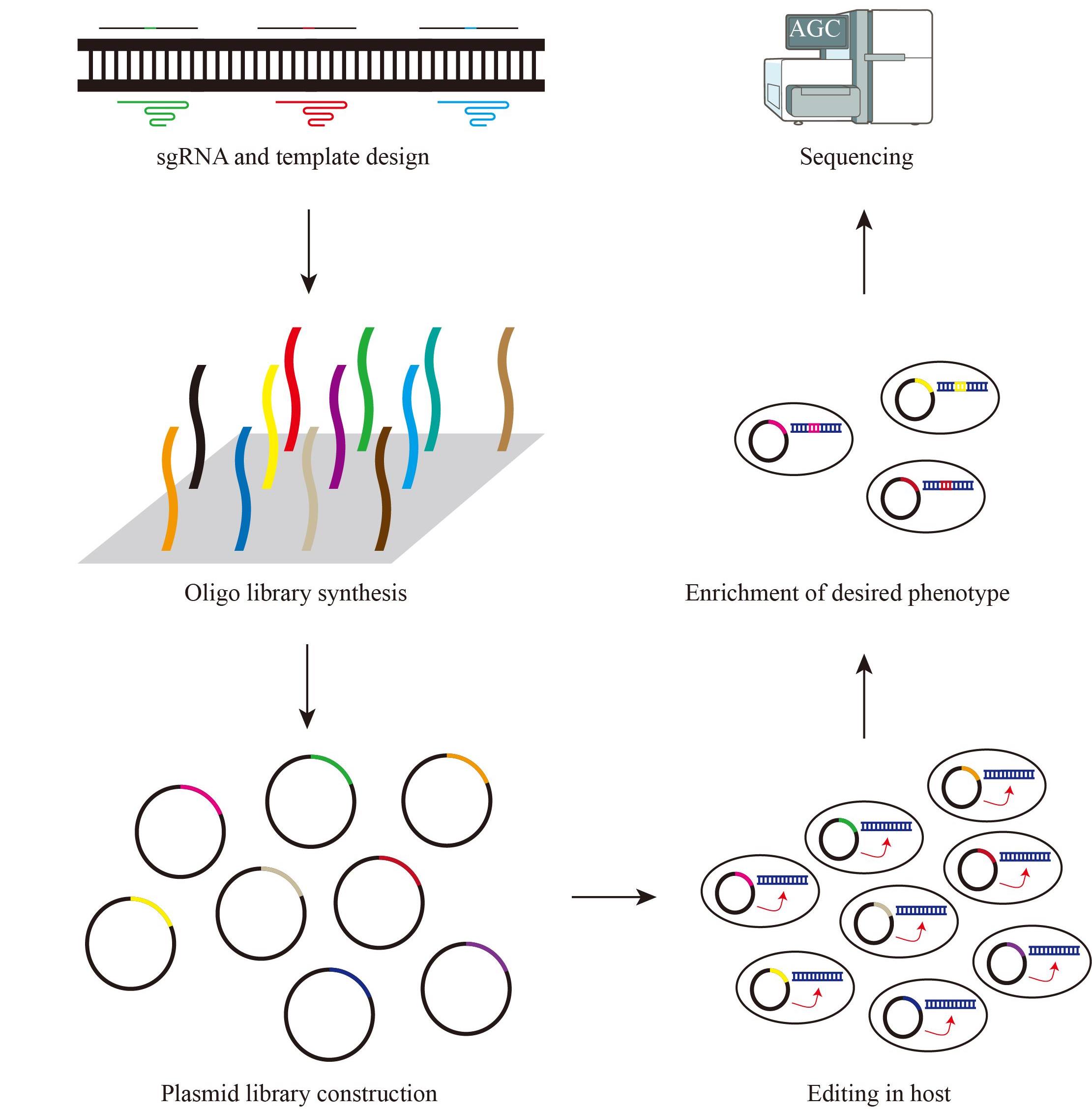
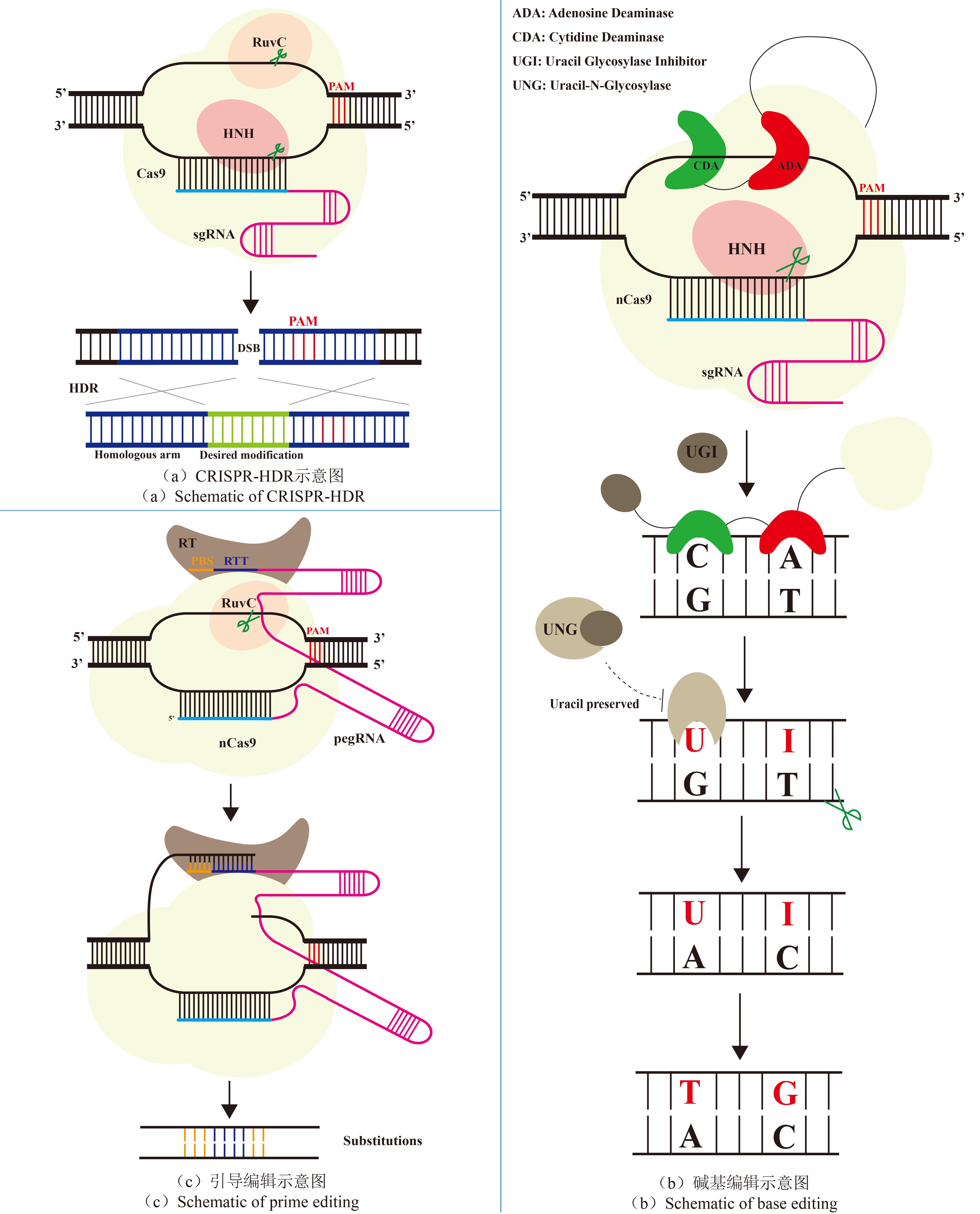
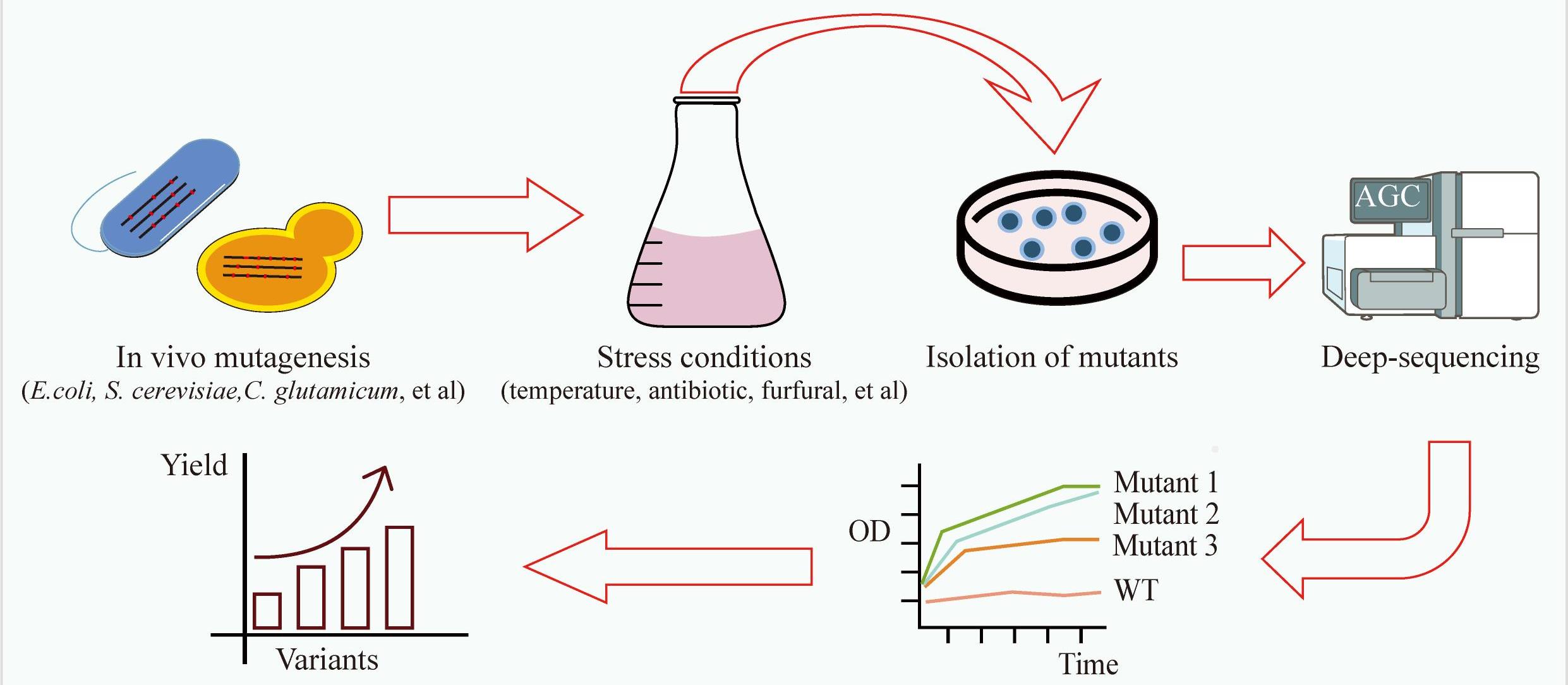
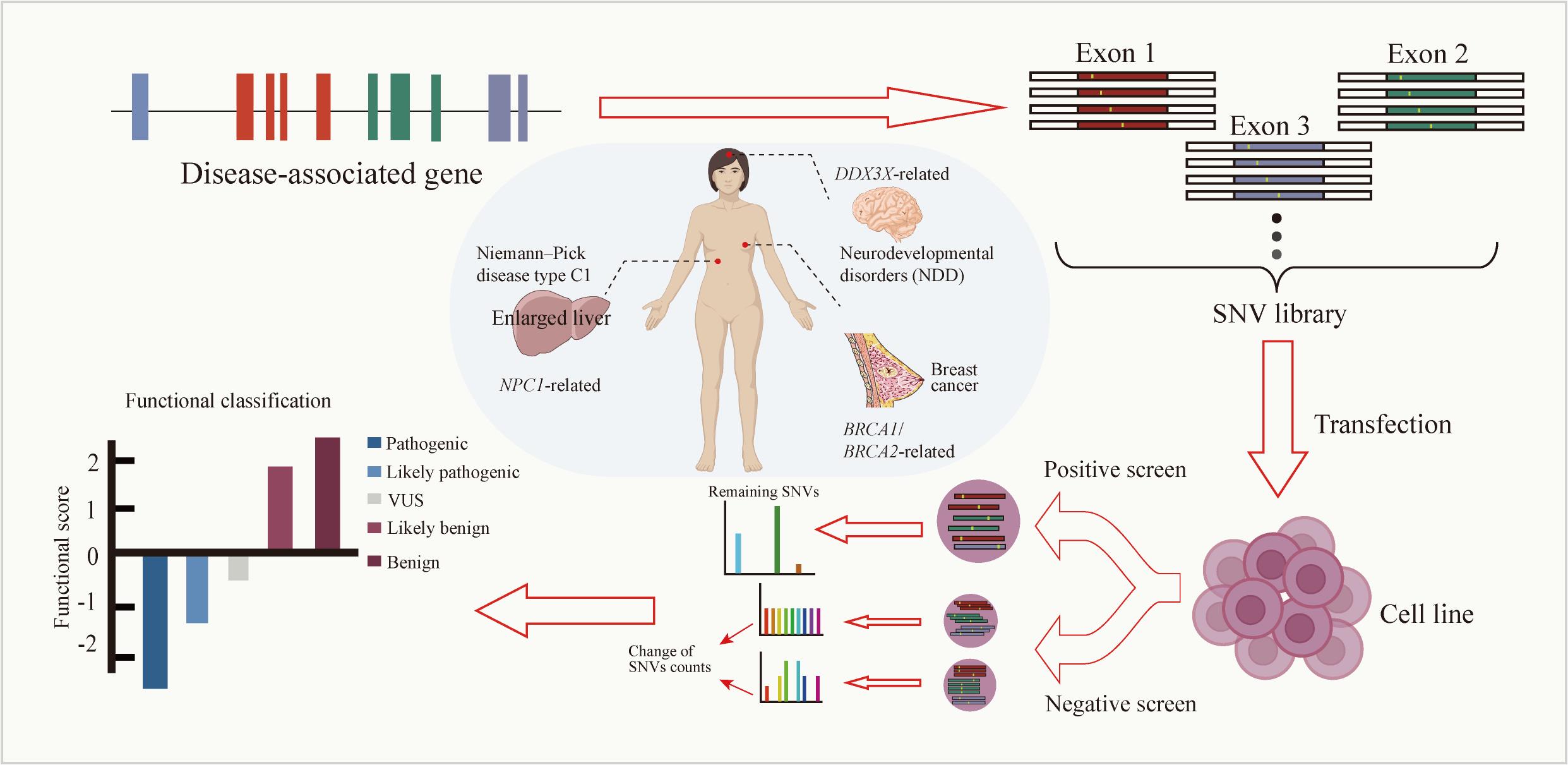
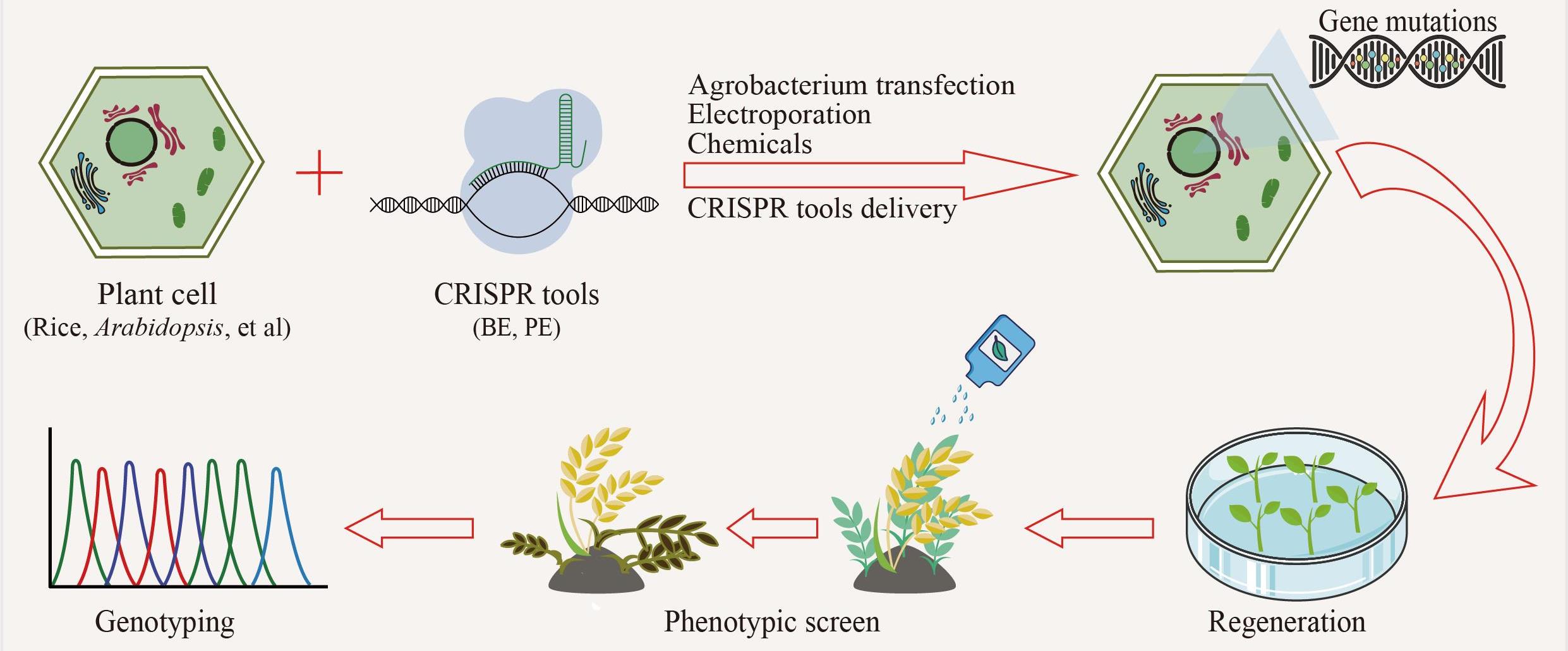
高通量基因组编辑是快速分析大量基因突变功能和进行遗传育种的有效方法。相比于传统随机诱变,基于规律间隔成簇短回文重复序列(CRISPR)系统的基因组编辑具有效率高、可靶向的优点。通过设计靶向目标基因的向导RNA文库可以实现高通量基因组编辑和筛选。近年来,多种CRISPR系统以及CRISPR衍生基因编辑技术的开发进一步丰富了高通量基因组编辑工具箱。本文主要介绍基于CRISPR系统的高通量基因组编辑方法,包括CRISPR辅助的同源定向修复、碱基编辑系统、引导编辑系统等,并介绍了这些方法在不同领域的应用,如工业微生物育种、人类功能基因组学和作物改良。最后,对相关方法存在的物种适用性有限、突变多样性低、编辑范围窄、多基因编辑困难等问题以及潜在的解决方法进行讨论和展望。
中图分类号:
引用本文
滕佳尧, 任传宏, 朱芮莹, 鲍泽华. 基于CRISPR系统的高通量基因组编辑研究进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-073.
TENG Jiayao, REN Chuanhong, ZHU Ruiying, BAO Zehua. Recent advances in CRISPR-based high-throughput genome editing[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-073.
| 编辑方法 | 方法名称 | 物种 | Cas蛋白 | PAM | 效应蛋白 | 参考文献 |
|---|---|---|---|---|---|---|
| CRISPR-HDR | Saturation editing | 哺乳动物细胞 | SpCas9 | NGG | / | [ |
| CasPER | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CREATE | 大肠杆菌 | SpCas9 | NGG | / | [ | |
| CHAnGE | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| MAGESTIC | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CRISPEY | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CHASE | 酿酒酵母 | SpiG(SpCas9突变体) | NGN | / | [ | |
| CRAIDE | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| Base editing | TAM | 哺乳动物细胞 | dSpCas9 | NGG | 胞苷脱氨酶AID-P182X(AIDx) | [ |
| CRISPR-X | 哺乳动物细胞 | dSpCas9 | NGG | 胞苷脱氨酶AID/AID*∆ | [ | |
| BARBEKO | 哺乳动物细胞 | nSpCas9 | NGG | 胞苷脱氨酶Anc689 APOBEC | [ | |
| HACE | 哺乳动物细胞 | nSpCas9 | NGG | 胞苷脱氨酶AID*∆ 解旋酶PcrA M6 | [ | |
| MACBETH | 谷氨酸棒状杆菌 | nSpCas9 | NGG | 胞苷脱氨酶AID-P47S(AIDTS) | [ | |
| CoMuTER | 酿酒酵母 | Cas3 | AAG | 胞苷脱氨酶rAPOBEC1 | [ | |
| ScBE3 | 大肠杆菌 | nScCas9 | NNG | 胞苷脱氨酶rAPOBEC1 | [ | |
STEME STEME-NG | 水稻 | nSpCas9 nCas9-NG | NGG NG | 胞苷脱氨酶APOBEC3 腺苷脱氨酶ecTadA-ecTadA7.10 | [ | |
| MoBE | 水稻 | nSpCas9 | NGG | 胞苷脱氨酶CDA1 腺苷脱氨酶TadA9 | [ | |
| Prime editing | PLSM | 水稻 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于pPE2) | [ |
| SPE | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE2) | [ | |
| PRIME | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE3) | [ | |
| PEER-seq | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ | |
| MOSAIC | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE2) | [ | |
| 高通量基因敲除筛选的PE平台 | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ | |
| PRESENT | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ |
表1 CRISPR辅助的高通量基因组编辑方法
Table 1 CRISPR-assisted high-throughput genome editing methods
| 编辑方法 | 方法名称 | 物种 | Cas蛋白 | PAM | 效应蛋白 | 参考文献 |
|---|---|---|---|---|---|---|
| CRISPR-HDR | Saturation editing | 哺乳动物细胞 | SpCas9 | NGG | / | [ |
| CasPER | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CREATE | 大肠杆菌 | SpCas9 | NGG | / | [ | |
| CHAnGE | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| MAGESTIC | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CRISPEY | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| CHASE | 酿酒酵母 | SpiG(SpCas9突变体) | NGN | / | [ | |
| CRAIDE | 酿酒酵母 | SpCas9 | NGG | / | [ | |
| Base editing | TAM | 哺乳动物细胞 | dSpCas9 | NGG | 胞苷脱氨酶AID-P182X(AIDx) | [ |
| CRISPR-X | 哺乳动物细胞 | dSpCas9 | NGG | 胞苷脱氨酶AID/AID*∆ | [ | |
| BARBEKO | 哺乳动物细胞 | nSpCas9 | NGG | 胞苷脱氨酶Anc689 APOBEC | [ | |
| HACE | 哺乳动物细胞 | nSpCas9 | NGG | 胞苷脱氨酶AID*∆ 解旋酶PcrA M6 | [ | |
| MACBETH | 谷氨酸棒状杆菌 | nSpCas9 | NGG | 胞苷脱氨酶AID-P47S(AIDTS) | [ | |
| CoMuTER | 酿酒酵母 | Cas3 | AAG | 胞苷脱氨酶rAPOBEC1 | [ | |
| ScBE3 | 大肠杆菌 | nScCas9 | NNG | 胞苷脱氨酶rAPOBEC1 | [ | |
STEME STEME-NG | 水稻 | nSpCas9 nCas9-NG | NGG NG | 胞苷脱氨酶APOBEC3 腺苷脱氨酶ecTadA-ecTadA7.10 | [ | |
| MoBE | 水稻 | nSpCas9 | NGG | 胞苷脱氨酶CDA1 腺苷脱氨酶TadA9 | [ | |
| Prime editing | PLSM | 水稻 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于pPE2) | [ |
| SPE | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE2) | [ | |
| PRIME | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE3) | [ | |
| PEER-seq | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ | |
| MOSAIC | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PE2) | [ | |
| 高通量基因敲除筛选的PE平台 | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ | |
| PRESENT | 哺乳动物细胞 | nSpCas9 | NGG | 逆转录酶MMLV-RT(基于PEmax) | [ |
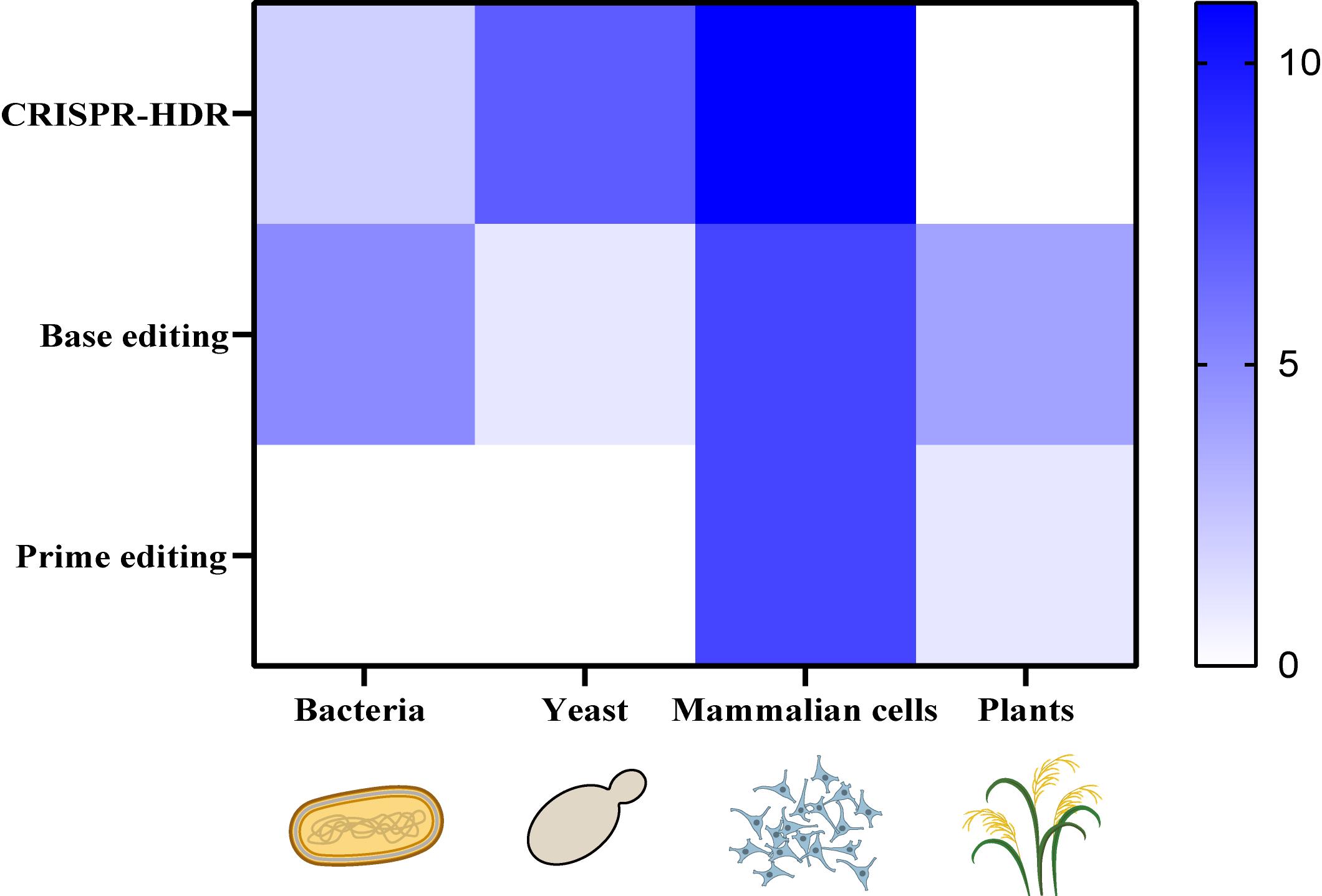
图7 CRISPR辅助的高通量基因组编辑方法在不同物种中的适用性 (热图程度代表本文所引相关文献数)
Fig. 7 Applicability of CRISPR-assisted high-throughput genome editing methods in different species (The heat map indicates the number of relevant publications cited in this review)
| [1] | 曹中正, 张心怡, 徐艺源, 等. 基因组编辑技术及其在合成生物学中的应用[J]. 合成生物学, 2020, 1(4): 413-426. |
| CAO Z, ZHANG X, XU Y, et al. Genome editing technology and its applications in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(4): 413-426. | |
| [2] | DRUMMOND D A, IVERSON B L, GEORGIOU G, et al. Why high-error-rate random mutagenesis libraries are enriched in functional and improved proteins[J]. Journal of Molecular Biology, 2005, 350(4): 806-816. |
| [3] | ZHENG L, BAUMANN U, J-L REYMOND. An efficient one-step site-directed and site-saturation mutagenesis protocol[J]. Nucleic Acids Research, 2004, 32(14): e115. |
| [4] | STEMMER W P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(22): 10747-10751. |
| [5] | ZHAO H, GIVER L, SHAO Z, et al. Molecular evolution by staggered extension process (StEP) in vitro recombination[J]. Nature Biotechnology, 1998, 16(3): 258-261. |
| [6] | FRYER T, WOLFF D S, OVERATH M D, et al. Post-assembly plasmid amplification for increased transformation yields in E. coli and S. cerevisiae [J]. Chem & Bio Engineering, 2025, 2(2): 87-96. |
| [7] | MOLINA R S, RIX G, MENGISTE A A, et al. In vivo hypermutation and continuous evolution[J]. Nature Reviews Methods Primers, 2022, 2(1): 1-22. |
| [8] | CHEN X, ZHANG J. The genomic landscape of position effects on protein expression level and noise in yeast[J]. Cell Systems, 2016, 2(5): 347-354. |
| [9] | ERDOGAN M, FABRITIUS A, BASQUIN J, et al. Targeted in situ protein diversification and intra-organelle validation in mammalian cells[J]. Cell Chemical Biology, 2020, 27(5): 610-621.e5. |
| [10] | LYNCH M, ACKERMAN M S, J-F GOUT, et al. Genetic drift, selection and the evolution of the mutation rate[J]. Nature Reviews Genetics, 2016, 17(11): 704-714. |
| [11] | LEWIS J A, MORRAN L T. Advantages of laboratory natural selection in the applied sciences[J]. Journal of Evolutionary Biology, 2022, 35(1): 5-22. |
| [12] | WANNIER T M, CIACCIA P N, ELLINGTON A D, et al. Recombineering and MAGE[J]. Nature Reviews. Methods Primers, 2021, 1: 7. |
| [13] | DICARLO J E, CONLEY A J, PENTTILÄ M, et al. Yeast oligo-mediated genome engineering (YOGE)[J]. ACS synthetic biology, 2013, 2(12): 741-749. |
| [14] | BARBIERI E M, MUIR P, AKHUETIE-ONI B O, et al. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes[J]. Cell, 2017, 171(6): 1453-1467.e13. |
| [15] | CIACCIA P N, LIANG Z, SCHWEITZER A Y, et al. Enhanced eMAGE applied to identify genetic factors of nuclear hormone receptor dysfunction via combinatorial gene editing[J]. Nature Communications, 2024, 15(1): 5218. |
| [16] | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| [17] | WANG H H, KIM H, CONG L, et al. Genome-scale promoter engineering by coselection MAGE[J]. Nature Methods, 2012, 9(6): 591-593. |
| [18] | CARR P A, WANG H H, STERLING B, et al. Enhanced multiplex genome engineering through co-operative oligonucleotide co-selection[J]. Nucleic Acids Research, 2012, 40(17): e132. |
| [19] | NYERGES Á, CSÖRGŐ B, NAGY I, et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(9): 2502-2507. |
| [20] | GHOSH K, VAN DUYNE G D. Cre-loxP biochemistry[J]. Methods, 2002, 28(3): 374-383. |
| [21] | KANG Y, DURFEE T, GLASNER J D, et al. Systematic mutagenesis of the Escherichia coli genome[J]. Journal of Bacteriology, 2004, 186(15): 4921-4930. |
| [22] | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| [23] | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| [24] | PÂQUES F, HABER J E. Multiple pathways of recombination induced by double-strand breaks in saccharomyces cerevisiae [J]. Microbiology and Molecular Biology Reviews, 1999, 63(2): 349-404. |
| [25] | CUI Y, DONG H, MA Y, et al. Strategies for applying nonhomologous end joining-mediated genome editing in prokaryotes[J]. ACS Synthetic Biology, 2019, 8(10): 2194-2202. |
| [26] | SCULLY R, PANDAY A, ELANGO R, et al. DNA double-strand break repair-pathway choice in somatic mammalian cells[J]. Nature Reviews Molecular Cell Biology, 2019, 20(11): 698-714. |
| [27] | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2013, 152(5): 1173-1183. |
| [28] | MALI P, AACH J, STRANGES P B, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering[J]. Nature Biotechnology, 2013, 31(9): 833-838. |
| [29] | XU X, QI L S. A CRISPR-dCas toolbox for genetic engineering and synthetic biology[J]. Journal of Molecular Biology, 2019, 431(1): 34-47. |
| [30] | SANG Y, XU L, BAO Z. Development of artificial transcription factors and their applications in cell reprograming, genetic screen, and disease treatment[J]. Molecular Therapy: The Journal of the American Society of Gene Therapy, 2024, 32(12): 4208-4234. |
| [31] | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| [32] | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| [33] | 梁丽亚, 刘嵘明. 靶向DNA的Ⅱ类CRISPR/Cas系统的蛋白工程化改造[J]. 合成生物学, 2023, 4(1): 86-101. |
| LIANG L, LIU R. Protein engineering of DNA targeting type II CRISPR/Cas systems[J]. Synthetic Biology Journal, 2023, 4(1): 86-101. | |
| [34] | BROWN A D, CLAYBON A B, BISHOP A J R. A conditional mouse model for measuring the frequency of homologous recombination events in vivo in the absence of essential genes[J]. Molecular and Cellular Biology, 2011, 31(17): 3593-3602. |
| [35] | ROUET P, SMIH F, JASIN M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease[J]. Molecular and Cellular Biology, 1994, 14(12): 8096-8106. |
| [36] | EID A, MAHFOUZ M M. Genome editing: the road of CRISPR/Cas9 from bench to clinic[J]. Experimental & Molecular Medicine, 2016, 48(10): e265-e265. |
| [37] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [38] | FINDLAY G M, BOYLE E A, HAUSE R J, et al. Saturation editing of genomic regions by multiplex homology-directed repair[J]. Nature, 2014, 513(7516): 120-123. |
| [39] | FINDLAY G M, DAZA R M, MARTIN B, et al. Accurate classification of BRCA1 variants with saturation genome editing[J]. Nature, 2018, 562(7726): 217-222. |
| [40] | WATERS A J, BRENDLER-SPAETH T, SMITH D, et al. Saturation genome editing of BAP1 functionally classifies somatic and germline variants[J]. Nature Genetics, 2024, 56(7): 1434-1445. |
| [41] | JIA X, BURUGULA B B, CHEN V, et al. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk[J]. The American Journal of Human Genetics, 2021, 108(1): 163-175. |
| [42] | RADFORD E J, TAN H-K, ANDERSSON M H L, et al. Saturation genome editing of DDX3X clarifies pathogenicity of germline and somatic variation[J]. Nature Communications, 2023, 14(1): 7702. |
| [43] | JAKOČIŪNAS T, PEDERSEN L E, LIS A V, et al. CasPER, a method for directed evolution in genomic contexts using mutagenesis and CRISPR/Cas9[J]. Metabolic Engineering, 2018, 48: 288-296. |
| [44] | GARST A D, BASSALO M C, PINES G, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering[J]. Nature Biotechnology, 2017, 35(1): 48-55. |
| [45] | DEWACHTER L, BROOKS A N, NOON K, et al. Deep mutational scanning of essential bacterial proteins can guide antibiotic development[J]. Nature Communications, 2023, 14(1): 241. |
| [46] | BAO Z, HAMEDIRAD M, XUE P, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision[J]. Nature Biotechnology, 2018, 36(6): 505-508. |
| [47] | ROY K R, SMITH J D, VONESCH S C, et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast[J]. Nature Biotechnology, 2018, 36(6): 512-520. |
| [48] | GUO X, CHAVEZ A, TUNG A, et al. High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR-Cas9 in yeast[J]. Nature Biotechnology, 2018, 36(6): 540-546. |
| [49] | SHARON E, CHEN S-A A, KHOSLA N M, et al. Functional genetic variants revealed by massively parallel precise genome editing[J]. Cell, 2018, 175(2): 544-557.e16. |
| [50] | DENG L, ZHOU Y-L, CAI Z, et al. Massively parallel CRISPR-assisted homologous recombination enables saturation editing of full-length endogenous genes in yeast[J]. Science Advances, 2024, 10(20): eadj9382. |
| [51] | STORICI F, BEBENEK K, KUNKEL T A, et al. RNA-templated DNA repair[J]. Nature, 2007, 447(7142): 338-341. |
| [52] | JENSEN E D, LALOUX M, LEHKA B J, et al. A synthetic RNA-mediated evolution system in yeast[J]. Nucleic Acids Research, 2021, 49(15): e88. |
| [53] | KOSICKI M, TOMBERG K, BRADLEY A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements[J]. Nature Biotechnology, 2018, 36(8): 765-771. |
| [54] | CULLOT G, BOUTIN J, TOUTAIN J, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations[J]. Nature Communications, 2019, 10(1): 1136. |
| [55] | ADIKUSUMA F, PILTZ S, CORBETT M A, et al. Large deletions induced by Cas9 cleavage[J]. Nature, 2018, 560(7717): E8-E9. |
| [56] | LEIBOWITZ M L, PAPATHANASIOU S, DOERFLER P A, et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing[J]. Nature Genetics, 2021, 53(6): 895-905. |
| [57] | KARANAM K, KAFRI R, LOEWER A, et al. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase[J]. Molecular Cell, 2012, 47(2): 320-329. |
| [58] | PLOESSL D, ZHAO Y, CAO M, et al. A repackaged CRISPR platform increases homology-directed repair for yeast engineering[J]. Nature Chemical Biology, 2022, 18(1): 38-46. |
| [59] | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| [60] | ZHANG X, ZHU B, CHEN L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells[J]. Nature Biotechnology, 2020, 38(7): 856-860. |
| [61] | TONG H, LIU N, WEI Y, et al. Programmable deaminase-free base editors for G-to-Y conversion by engineered glycosylase[J]. National Science Review, 2023, 10(8): nwad143. |
| [62] | YE L, ZHAO D, LI J, et al. Glycosylase-based base editors for efficient T-to-G and C-to-G editing in mammalian cells[J]. Nature Biotechnology, 2024, 42(10): 1538-1547. |
| [63] | MA Y, ZHANG J, YIN W, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells[J]. Nature Methods, 2016, 13(12): 1029-1035. |
| [64] | HESS G T, FRÉSARD L, HAN K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells[J]. Nature Methods, 2016, 13(12): 1036-1042. |
| [65] | XU P, LIU Z, LIU Y, et al. Genome-wide interrogation of gene functions through base editor screens empowered by barcoded sgRNAs[J]. Nature Biotechnology, 2021, 39(11): 1403-1413. |
| [66] | CHEN X D, CHEN Z, WYTHES G, et al. Helicase-assisted continuous editing for programmable mutagenesis of endogenous genomes[J]. Science, 2024, 386(6718): eadn5876. |
| [67] | HALPERIN S O, TOU C J, WONG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717): 248-252. |
| [68] | WANG Y, LIU Y, LIU J, et al. MACBETH: Multiplex automated Corynebacterium glutamicum base editing method[J]. Metabolic Engineering, 2018, 47: 200-210. |
| [69] | ZIMMERMANN A, PRIETO-VIVAS J E, CAUTEREELS C, et al. A Cas3-base editing tool for targetable in vivo mutagenesis[J]. Nature Communications, 2023, 14(1): 3389. |
| [70] | GAWLITT S, COLLINS S P, YU Y, et al. Expanding the flexibility of base editing for high-throughput genetic screens in bacteria[J]. Nucleic Acids Research, 2024, 52(7): 4079-4097. |
| [71] | LI C, ZHANG R, MENG X, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors[J]. Nature Biotechnology, 2020, 38(7): 875-882. |
| [72] | ZHANG A, SHAN T, SUN Y, et al. Directed evolution rice genes with randomly multiplexed sgRNAs assembly of base editors[J]. Plant Biotechnology Journal, 2023, 21(12): 2597-2610. |
| [73] | XU R, LIU X, LI J, et al. Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice[J]. Nature Plants, 2021, 7(7): 888-892. |
| [74] | ERWOOD S, BILY T M I, LEQUYER J, et al. Saturation variant interpretation using CRISPR prime editing[J]. Nature Biotechnology, 2022, 40(6): 885-895. |
| [75] | REN X, YANG H, NIERENBERG J L, et al. High-throughput PRIME-editing screens identify functional DNA variants in the human genome[J]. Molecular Cell, 2023, 83(24): 4633-4645.e9. |
| [76] | KIM Y, H-C OH, LEE S, et al. Saturation profiling of drug-resistant genetic variants using prime editing[J]. Nature Biotechnology, 2024, doi:10.1038/s41587-024-02465-z . |
| [77] | HSU J Y, LAM K C, SHIH J, et al. MOSAIC enables in situ saturation mutagenesis of genes and CRISPR prime editing guide RNA optimization in human cells[J]. bioRxiv, 2024, 591078. |
| [78] | CHARDON F M, SUITER C C, DAZA R M, et al. A multiplex, prime editing framework for identifying drug resistance variants at scale[J]. bioRxiv, 2023, 550902. |
| [79] | HERGER M, KAJBA C M, BUCKLEY M, et al. High-throughput screening of human genetic variants by pooled prime editing[J]. Cell Genomics, 2025, 5(4): 100814. |
| [80] | CIRINCIONE A, SIMPSON D, YAN W, et al. A benchmarked, high-efficiency prime editing platform for multiplexed dropout screening[J]. Nature Methods, 2025, 22(1): 92-101. |
| [81] | NIU X, TANG W, LIU Y, et al. Prime editor-based high-throughput screening reveals functional synonymous mutations in human cells[J]. Nature Biotechnology, 2025, doi:10.1038/s41587-025-02710-z . |
| [82] | PARK J, YU G, SEO S-Y, et al. SynDesign: web-based prime editing guide RNA design and evaluation tool for saturation genome editing[J]. Nucleic Acids Research, 2024, 52(W1): W121-W125. |
| [83] | MAVROMMATI M, DASKALAKI A, PAPANIKOLAOU S, et al. Adaptive laboratory evolution principles and applications in industrial biotechnology[J]. Biotechnology Advances, 2022, 54: 107795. |
| [84] | BASSALO M C, LIU R, GILL R T. Directed evolution and synthetic biology applications to microbial systems[J]. Current Opinion in Biotechnology, 2016, 39: 126-133. |
| [85] | LAM F H, TURANLI-YILDIZ B, LIU D, et al. Engineered yeast tolerance enables efficient production from toxified lignocellulosic feedstocks[J]. Science Advances, 2021, 7(26): eabf7613. |
| [86] | ALPER H, STEPHANOPOULOS G. Global transcription machinery engineering: a new approach for improving cellular phenotype[J]. Metabolic Engineering, 2007, 9(3): 258-267. |
| [87] | LIU Y, WANG R, LIU J, et al. Base editor enables rational genome-scale functional screening for enhanced industrial phenotypes in Corynebacterium glutamicum [J]. Science Advances, 2022, 8(35): eabq2157. |
| [88] | HAO W, CUI W, CHENG Z, et al. Development of a base editor for protein evolution via in situ mutation in vivo [J]. Nucleic Acids Research, 2021, 49(16): 9594-9605. |
| [89] | HAO W, CUI W, LIU Z, et al. A new-generation base editor with an expanded editing window for microbial cell evolution in vivo based on CRISPR‒Cas12b engineering[J]. Advanced Science (Weinheim, Baden-Wurttemberg, Germany), 2024, 11(22): e2309767. |
| [90] | MARTÍNEZ-JIMÉNEZ F, MUIÑOS F, SENTÍS I, et al. A compendium of mutational cancer driver genes[J]. Nature Reviews Cancer, 2020, 20(10): 555-572. |
| [91] | KRAIS J J, JOHNSON N. BRCA1 mutations in cancer: coordinating deficiencies in homologous recombination with tumorigenesis[J]. Cancer Research, 2020, 80(21): 4601-4609. |
| [92] | SANGREE A K, GRIFFITH A L, SZEGLETES Z M, et al. Benchmarking of SpCas9 variants enables deeper base editor screens of BRCA1 and BCL2[J]. Nature Communications, 2022, 13(1): 1318. |
| [93] | HANNA R E, HEGDE M, FAGRE C R, et al. Massively parallel assessment of human variants with base editor screens[J]. Cell, 2021, 184(4): 1064-1080.e20. |
| [94] | KWEON J, A-H JANG, SHIN H R, et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants[J]. Oncogene, 2020, 39(1): 30-35. |
| [95] | HUANG C, LI G, WU J, et al. Identification of pathogenic variants in cancer genes using base editing screens with editing efficiency correction[J]. Genome Biology, 2021, 22(1): 80. |
| [96] | OLVERA-LEÓN R, ZHANG F, OFFORD V, et al. High-resolution functional mapping of RAD51C by saturation genome editing[J]. Cell, 2024, 187(20): 5719-5734.e19. |
| [97] | BUCKLEY M, TERWAGNE C, GANNER A, et al. Saturation genome editing maps the functional spectrum of pathogenic VHL alleles[J]. Nature Genetics, 2024, 56(7): 1446-1455. |
| [98] | SÁNCHEZ-RIVERA F J, DIAZ B J, KASTENHUBER E R, et al. Base editing sensor libraries for high-throughput engineering and functional analysis of cancer-associated single nucleotide variants[J]. Nature Biotechnology, 2022, 40(6): 862-873. |
| [99] | LUE N Z, GARCIA E M, NGAN K C, et al. Base editor scanning charts the DNMT3A activity landscape[J]. Nature Chemical Biology, 2023, 19(2): 176-186. |
| [100] | LI Y, XU T, MA H, et al. Functional profiling of serine, threonine and tyrosine sites[J]. Nature Chemical Biology, 2025, 21(4): 532-543. |
| [101] | HOLME I B, GREGERSEN P L, BRINCH-PEDERSEN H. Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences[J]. Frontiers in Plant Science, 2019, 10: 1468. |
| [102] | KUANG Y, LI S, REN B, et al. Base-editing-mediated artificial evolution of OsALS1 in Planta to develop novel herbicide-tolerant rice germplasms[J]. Molecular Plant, 2020, 13(4): 565-572. |
| [103] | WANG X, PAN W, SUN C, et al. Creating large-scale genetic diversity in Arabidopsis via base editing-mediated deep artificial evolution[J]. Genome Biology, 2024, 25(1): 215. |
| [104] | ZHANG H, MA J, WU Z, et al. BacPE: a versatile prime-editing platform in bacteria by inhibiting DNA exonucleases[J]. Nature Communications, 2024, 15(1): 825. |
| [105] | TUNCEL A, PAN C, CLEM J S, et al. CRISPR-Cas applications in agriculture and plant research[J]. Nature Reviews Molecular Cell Biology, 2025, 26(6): 419-441. |
| [106] | JIN Y-Y, ZHANG P, LIU L-L, et al. Enhancing homology-directed repair efficiency with HDR-boosting modular ssDNA donor[J]. Nature Communications, 2024, 15(1): 6843. |
| [107] | MITOUSIS L, MUSIOL-KROLL E, WOHLLEBEN W. CRISPR-Cas in actinomycetes: still a lot to be discovered[J]. microLife, 2025, 6: uqaf010. |
| [108] | CHEN W, REN Z-H, TANG N, et al. Targeted genetic screening in bacteria with a Cas12k-guided transposase[J]. Cell Reports, 2021, 36(9): 109635. |
| [109] | TONG H, WANG X, LIU Y, et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase[J]. Nature Biotechnology, 2023, 41(8): 1080-1084. |
| [110] | 潘颖佳, 夏思杨, 董昌, 等. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240. |
| PAN Y, XIA S, DONG C, et al. Mutator-driven continuous genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2023, 4(1): 225-240. | |
| [111] | YAO Y, ZHOU Z, WANG X, et al. SpRY-mediated screens facilitate functional dissection of non-coding sequences at single-base resolution[J]. Cell Genomics, 2024, 4(7): 100583. |
| [112] | CAI Z, XIE W, BAO Z. Broadening the targetable space: engineering and discovery of PAM-flexible Cas proteins[J]. Trends in Microbiology, 2024, 32(8): 728-731. |
| [113] | YANG C, ZHOU Z, SUN X, et al. PAMless SpRY exhibits a preference for the seed region for efficient targeting[J]. Cell Reports, 2024, 43(5): 114225. |
| [114] | KIM H K, LEE S, KIM Y, et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells[J]. Nature Biomedical Engineering, 2020, 4(1): 111-124. |
| [115] | ZHANG W, YIN J, ZHANG-DING Z, et al. In-depth assessment of the PAM compatibility and editing activities of Cas9 variants[J]. Nucleic Acids Research, 2021, 49(15): 8785-8795. |
| [116] | SHI H, AL-SAYYAD N, WASKO K M, et al. Rapid two-step target capture ensures efficient CRISPR-Cas9-guided genome editing[J]. Molecular Cell, 2025, 85(9): 1730-1742.e9. |
| [117] | SILVERSTEIN R A, KIM N, A-S KROELL, et al. Custom CRISPR-Cas9 PAM variants via scalable engineering and machine learning[J]. Nature, 2025, 643(8071): 539-550. |
| [118] | JIANG W, FENG S, HUANG S, et al. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity[J]. Cell Research, 2018, 28(8): 855-861. |
| [119] | WANG Y, ZHOU L, LIU N, et al. BE-PIGS: a base-editing tool with deaminases inlaid into Cas9 PI domain significantly expanded the editing scope[J]. Signal Transduction and Targeted Therapy, 2019, 4(1): 1-3. |
| [120] | VILLIGER L, SCHMIDHEINI L, MATHIS N, et al. Replacing the SpCas9 HNH domain by deaminases generates compact base editors with an alternative targeting scope[J]. Molecular Therapy. Nucleic Acids, 2021, 26: 502-510. |
| [121] | LIANG R, HE Z, ZHAO K T, et al. Prime editing using CRISPR-Cas12a and circular RNAs in human cells[J]. Nature Biotechnology, 2024, 42(12): 1867-1875. |
| [122] | CHEN P, LI X, ZHOU Q, et al. Configuration of adaptable template RNA architectures to unfold the editable space of a nuclease prime editor[J]. Nucleic Acids Research, 2025, 53(11): gkaf522. |
| [123] | CHEN F, LIAN M, MA B, et al. Multiplexed base editing through Cas12a variant-mediated cytosine and adenine base editors[J]. Communications Biology, 2022, 5(1): 1-12. |
| [124] | GEURTS M H, GANDHI S, BORETTO M G, et al. One-step generation of tumor models by base editor multiplexing in adult stem cell-derived organoids[J]. Nature Communications, 2023, 14(1): 4998. |
| [125] | WU Y, LI Y, LIU Y, et al. Multiplexed in-situ mutagenesis driven by a dCas12a-based dual-function base editor[J]. Nucleic Acids Research, 2024, 52(8): 4739-4755. |
| [126] | GUPTA A, LIU B, RAZA S, et al. Modularly assembled multiplex prime editors for simultaneous editing of agronomically important genes in rice[J]. Plant Communications, 2024, 5(2): 100741. |
| [1] | 王宏, 陆孔泳, 郑洋洋, 陈涛, 王智文. 基于转录因子生物传感器的构建与应用进展[J]. 合成生物学, 2025, 6(4): 829-845. |
| [2] | 姜百翼, 钱珑. 活细胞记录器在细胞谱系追踪中的应用和前景[J]. 合成生物学, 2025, 6(3): 651-668. |
| [3] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [4] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [5] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [6] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [7] | 雷茹, 陶慧, 刘天罡. 基因组深度挖掘驱动微生物萜类化合物高效发现[J]. 合成生物学, 2024, 5(3): 507-526. |
| [8] | 陈盈盈, 刘扬, 史俊杰, 马俊英, 鞠建华. CRISPR/Cas基因编辑及其新兴技术在丝状真菌研究中的系统应用[J]. 合成生物学, 2024, 5(3): 672-693. |
| [9] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [10] | 杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| [11] | 许志锰, 谢震. 引导编辑研究进展及其应用[J]. 合成生物学, 2024, 5(1): 1-15. |
| [12] | 郭肖杰, 剪兴金, 王立言, 张翀, 邢新会. 合成生物学表型测试生物反应器及其装备化研究进展[J]. 合成生物学, 2024, 5(1): 16-37. |
| [13] | 陈雅如, 曹英秀, 宋浩. 电活性微生物基因编辑与转录调控技术进展与应用[J]. 合成生物学, 2023, 4(6): 1281-1299. |
| [14] | 赵国淼, 杨鑫, 张媛, 王靖, 谭剑, 魏超, 周娜娜, 李凡, 王小艳. 生物设施平台及其工业应用[J]. 合成生物学, 2023, 4(5): 892-903. |
| [15] | 刁志钿, 王喜先, 孙晴, 徐健, 马波. 单细胞拉曼光谱测试分选装备研制及应用进展[J]. 合成生物学, 2023, 4(5): 1020-1035. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||