合成生物学 ›› 2023, Vol. 4 ›› Issue (5): 1020-1035.DOI: 10.12211/2096-8280.2023-025
单细胞拉曼光谱测试分选装备研制及应用进展
刁志钿, 王喜先, 孙晴, 徐健, 马波
- 中国科学院青岛生物能源与过程研究所,单细胞中心,山东 青岛 266101
-
收稿日期:2023-03-21修回日期:2022-05-17出版日期:2023-10-31发布日期:2023-11-15 -
通讯作者:马波 -
作者简介:刁志钿 (1995—),男,在读博士研究生。研究方向为液滴微流控技术、高通量流式拉曼分选技术等。 E-mail:diaozd@qibebt.ac.cn马波 (1976—),男,博士,研究员,博士生导师。研究方向为单细胞关键技术与仪器、微流控技术等,长期从事基于拉曼光谱的单细胞分析/分选及后续的单细胞测序等关键技术和仪器研究。E-mail:mabo@qibebt.ac.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2018YFA090290);天津市合成生物技术创新能力提升行动项目(TSBICIP-PTJS-003-05)
Advances and applications of single-cell Raman spectroscopy testing and sorting equipment
DIAO Zhidian, WANG Xixian, SUN Qing, XU Jian, MA Bo
- Single-cell Center,Qingdao Institute of Bioenergy and Bioprocess Technology,Chinese Academy of Sciences,Qingdao 266101,Shandong,China
-
Received:2023-03-21Revised:2022-05-17Online:2023-10-31Published:2023-11-15 -
Contact:MA Bo
摘要:
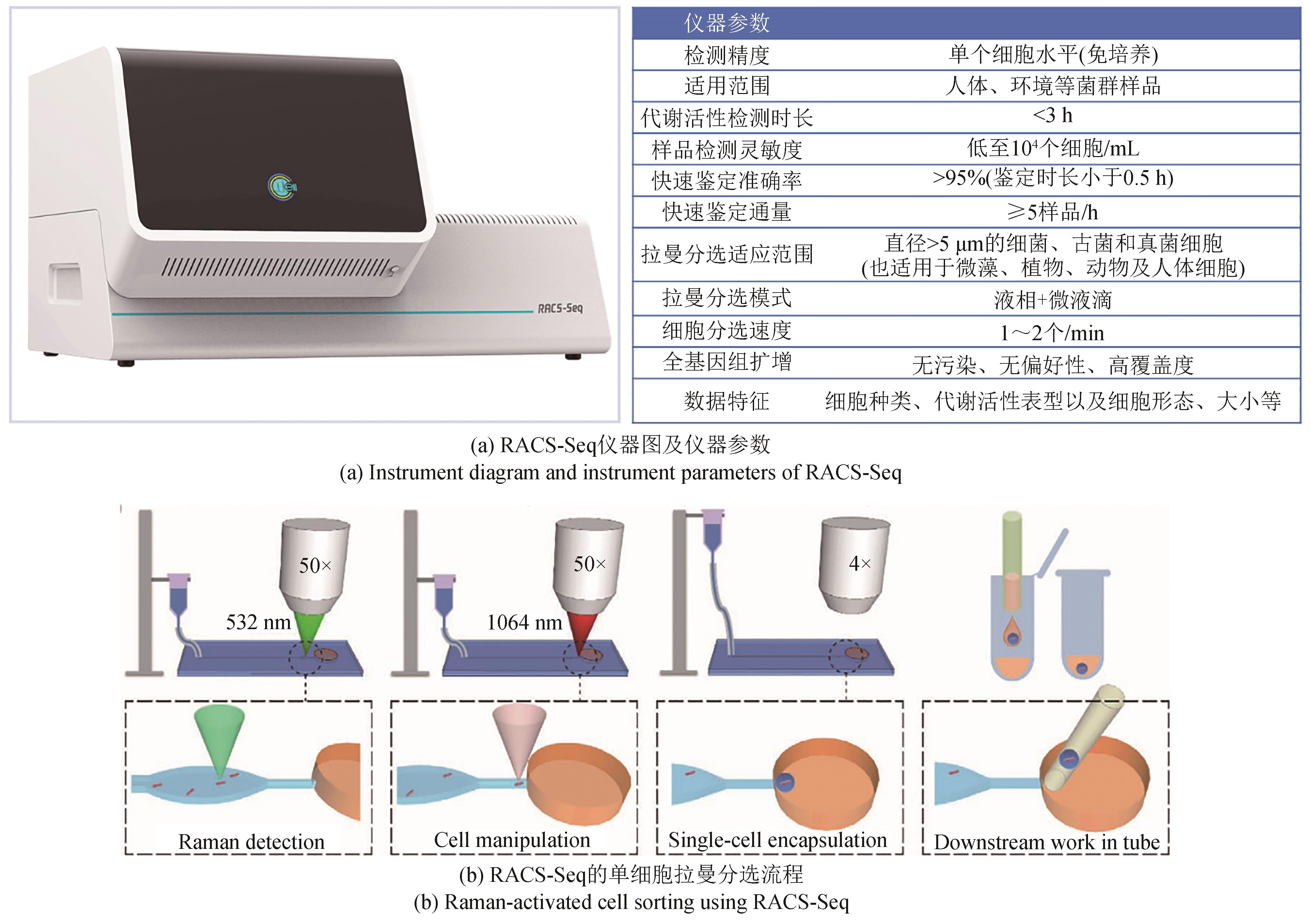
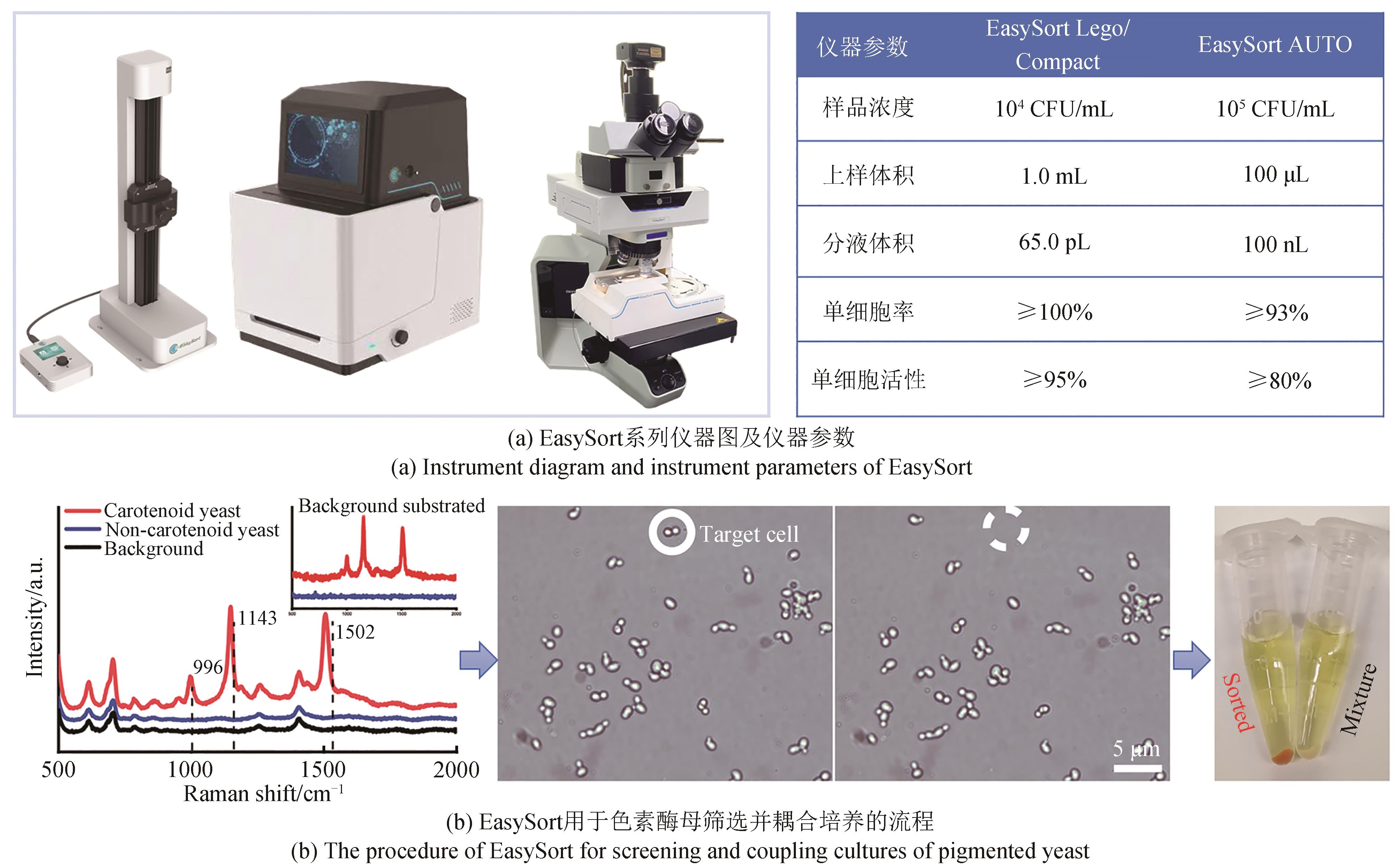
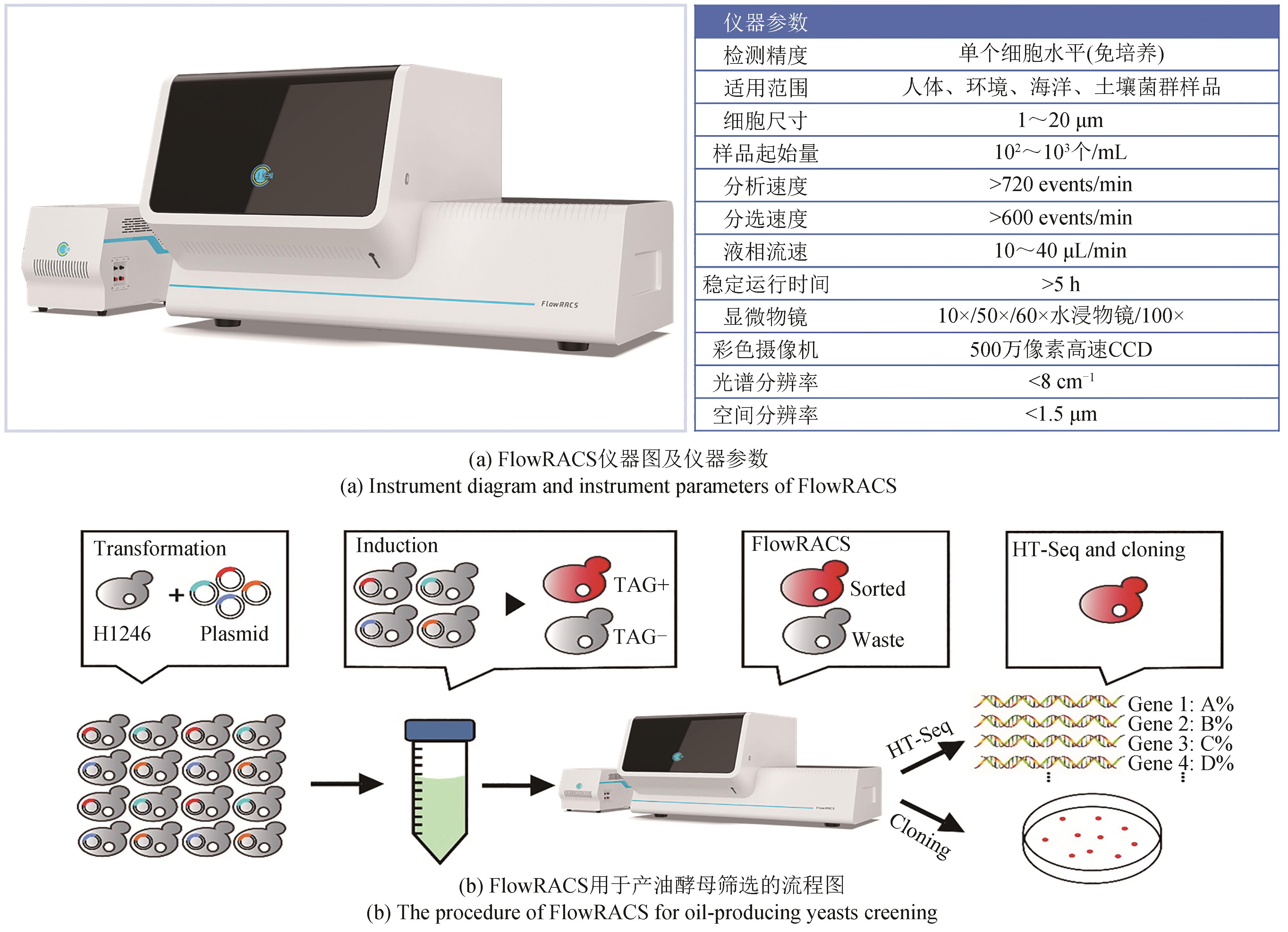
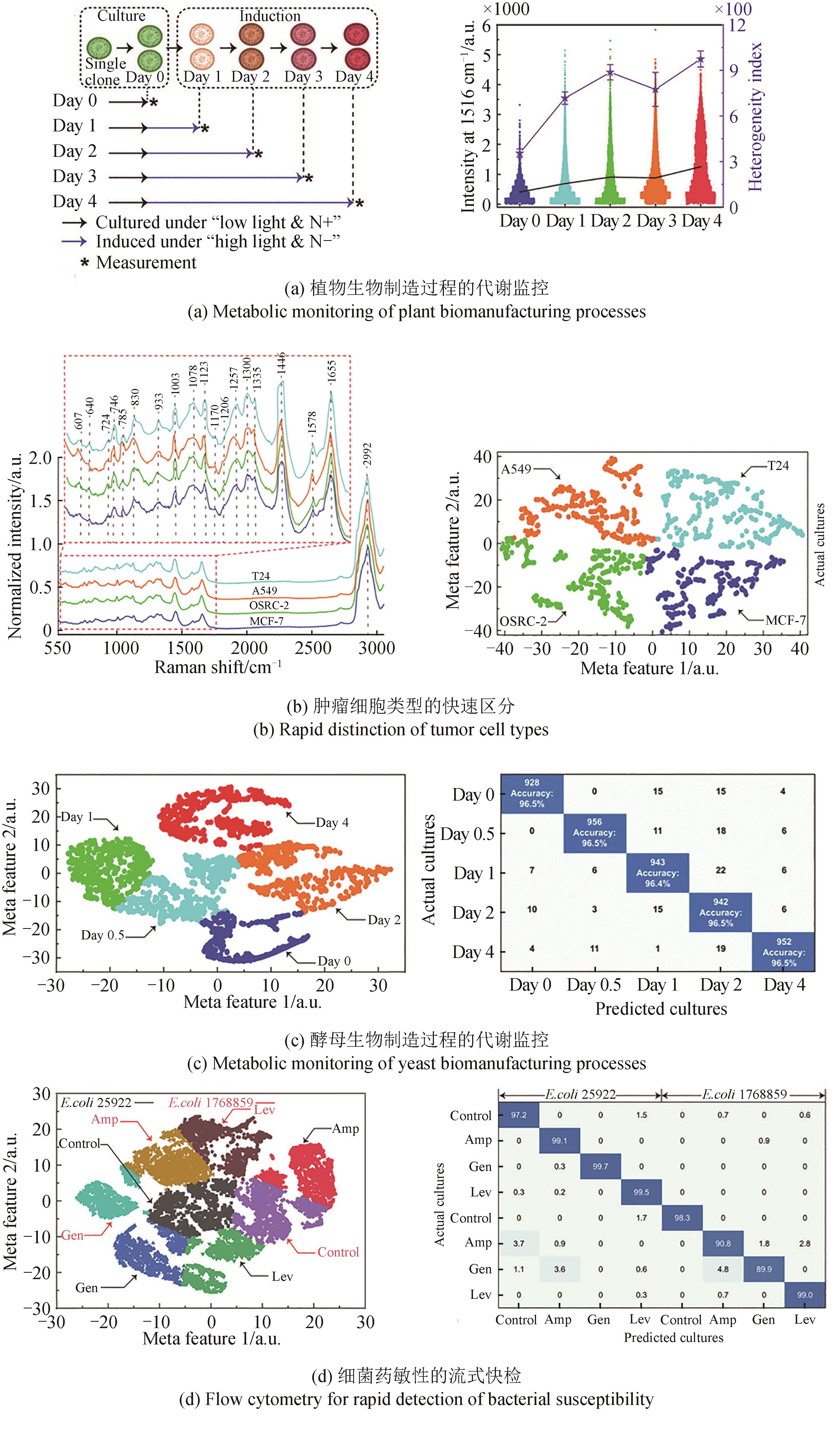
合成生物学的跨越式发展,取决于“设计-构建-测试-学习”(design-build-test-learn)这四大环节的突破。随着基因组测序、编辑、合成以及人工智能技术的日新月异,业界设计和构建突变体甚至人工细胞工厂的能力已经突飞猛进。然而,合成生物学至今仍面临的困境之一便是“大体系的复杂性难以处理”,一旦体系变大,细胞表型测试与分选的工作量就非常艰巨,甚至不可完成。单细胞拉曼光谱(SCRS)技术能够在活体单细胞水平、非标记状态下识别全景信息从而分辨复杂功能表型,且具有快速、低成本、能够与下游细胞组学研究耦联等优势,被视为全新的单细胞表型识别技术。目前,基于SCRS技术强大的表型识别能力已发展了系列合成表型的测试与分选装备,并进行了广泛的应用示范,展示了其助力合成生物学表型测试与分选的巨大潜力。本文选取自主研制的单细胞拉曼光镊分选仪(RACS-Seq)、单细胞微液滴分选系统(EasySort)和高通量流式拉曼分选仪(FlowRACS)为典型仪器装备,分别概述其技术原理和技术迭代以及特色应用案例等。本文最后对当前基于SCRS技术的合成表型测试分选装备所存在的问题及潜在解决策略进行了探讨和展望。
中图分类号:
引用本文
刁志钿, 王喜先, 孙晴, 徐健, 马波. 单细胞拉曼光谱测试分选装备研制及应用进展[J]. 合成生物学, 2023, 4(5): 1020-1035.
DIAO Zhidian, WANG Xixian, SUN Qing, XU Jian, MA Bo. Advances and applications of single-cell Raman spectroscopy testing and sorting equipment[J]. Synthetic Biology Journal, 2023, 4(5): 1020-1035.
| 1 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 2 | BENNER S A, SISMOUR A M. Synthetic biology[J]. Nature Reviews Genetics, 2005, 6(7): 533-543. |
| 3 | SCHOBER L, BÜTTNER E, LASKE C, et al. Cell dispensing in low-volume range with the immediate drop-on-demand technology (I-DOT)[J]. Journal of Laboratory Automation, 2015, 20(2): 154-163. |
| 4 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 5 | SANDER J D, JOUNG J K. CRISPR-Cas systems for editing, regulating and targeting genomes[J]. Nature Biotechnology, 2014, 32(4): 347-355. |
| 6 | SMITH H O, HUTCHISON C A, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| 7 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 8 | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 9 | WANG X X, REN L H, DIAO Z D, et al. Robust spontaneous Raman flow cytometry for single-cell metabolic phenome profiling via pDEP-DLD-RFC[J]. Advanced Science, 2023: 2207497. |
| 10 | ZINCHENKO A, DEVENISH S R A, KINTSES B, et al. One in a million: flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution[J]. Analytical Chemistry, 2014, 86(5): 2526-2533. |
| 11 | BREHM-STECHER B F, JOHNSON E A. Single-cell microbiology: tools, technologies, and applications[J]. Microbiology and Molecular Biology Reviews, 2004, 68(3): 538-559. |
| 12 | 杨建花, 苏晓岚, 朱蕾蕾. 高通量筛选系统在定向改造中的新进展[J]. 生物工程学报, 2021, 37(7): 2197-2210. |
| YANG J H, SU X L, ZHU L L. Advances of high-throughput screening system in reengineering of biological entities[J]. Chinese Journal of Biotechnology, 2021, 37(7): 2197-2210. | |
| 13 | ALI A, ABOULEILA Y, SHIMIZU Y,et al. Single-cell metabolomics by mass spectrometry: advances, challenges, and future applications[J]. TrAC Trends in Analytical Chemistry, 2019, 120: 115436. |
| 14 | SPITZER M H, NOLAN G P. Mass cytometry: single cells, many features[J]. Cell, 2016, 165(4): 780-791. |
| 15 | AMANTONICO A, URBAN P L, ZENOBI R. Analytical techniques for single-cell metabolomics: state of the art and trends[J]. Analytical and Bioanalytical Chemistry, 2010, 398(6): 2493-2504. |
| 16 | RAMAN C V, KRISHNAN K S. A new type of secondary radiation[J]. Nature, 1928, 121(3048): 501-502. |
| 17 | XU J, MA B, SU X Q, et al. Emerging trends for microbiome analysis: From single-cell functional imaging to microbiome big data[J]. Engineering, 2017, 3(1): 66-70. |
| 18 | HE Y H, WANG X X, MA B, et al. Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution[J]. Biotechnology Advances, 2019, 37(6): 107388. |
| 19 | LEE K S, LANDRY Z, PEREIRA F C, et al. Raman microspectroscopy for microbiology[J]. Nature Reviews Methods Primers, 2021, 1(80): 1-25. |
| 20 | YAN S S, QIU J X, GUO L, et al. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level[J].Applied Microbiology and Biotechnology, 2021, 105(4): 1315-1331. |
| 21 | WANG Y, JI Y T, WHARFE E S, et al. Raman activated cell ejection for isolation of single cells[J]. Analytical Chemistry, 2013, 85(22): 10697-10701. |
| 22 | HUANG W E, WARD A D, WHITELEY A S. Raman tweezers sorting of single microbial cells[J]. Environmental Microbiology Reports, 2009, 1(1): 44-49. |
| 23 | XIE C G, CHEN D, LI Y Q. Raman sorting and identification of single living micro-organisms with optical tweezers[J]. Optics Letters, 2005, 30(14): 1800-1802. |
| 24 | XU T, GONG Y H, SU X L, et al. Phenome-genome profiling of single bacterial cell by Raman-activated gravity-driven encapsulation and sequencing [J]. Small, 2020, 16(30): 2001172. |
| 25 | LEE K S, PALATINSZKY M, PEREIRA F C, et al. An automated Raman-based platform for the sorting of live cells by functional properties[J]. Nature Microbiology, 2019, 4(6): 1035-1048. |
| 26 | ZHANG P R, REN L H, ZHANG X, et al. Raman-activated cell sorting based on dielectrophoretic single-cell trap and release[J]. Analytical Chemistry, 2015, 87(4): 2282-2289. |
| 27 | WANG X X, REN L H, SU Y T, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 28 | WANG X X, XIN Y, REN L H, et al. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo [J]. Science Advances, 2020, 6(32): eabb3521. |
| 29 | NITTA N, IINO T, ISOZAKI A, et al. Raman image-activated cell sorting[J]. Nature Communications, 2020, 11(1): 3452. |
| 30 | LINDLEY M, DE PABLO J G, PETERSON W, et al. High-throughput Raman-activated cell sorting in the fingerprint region[J]. Advanced Materials Technologies, 2022, 7(10): 2101567. |
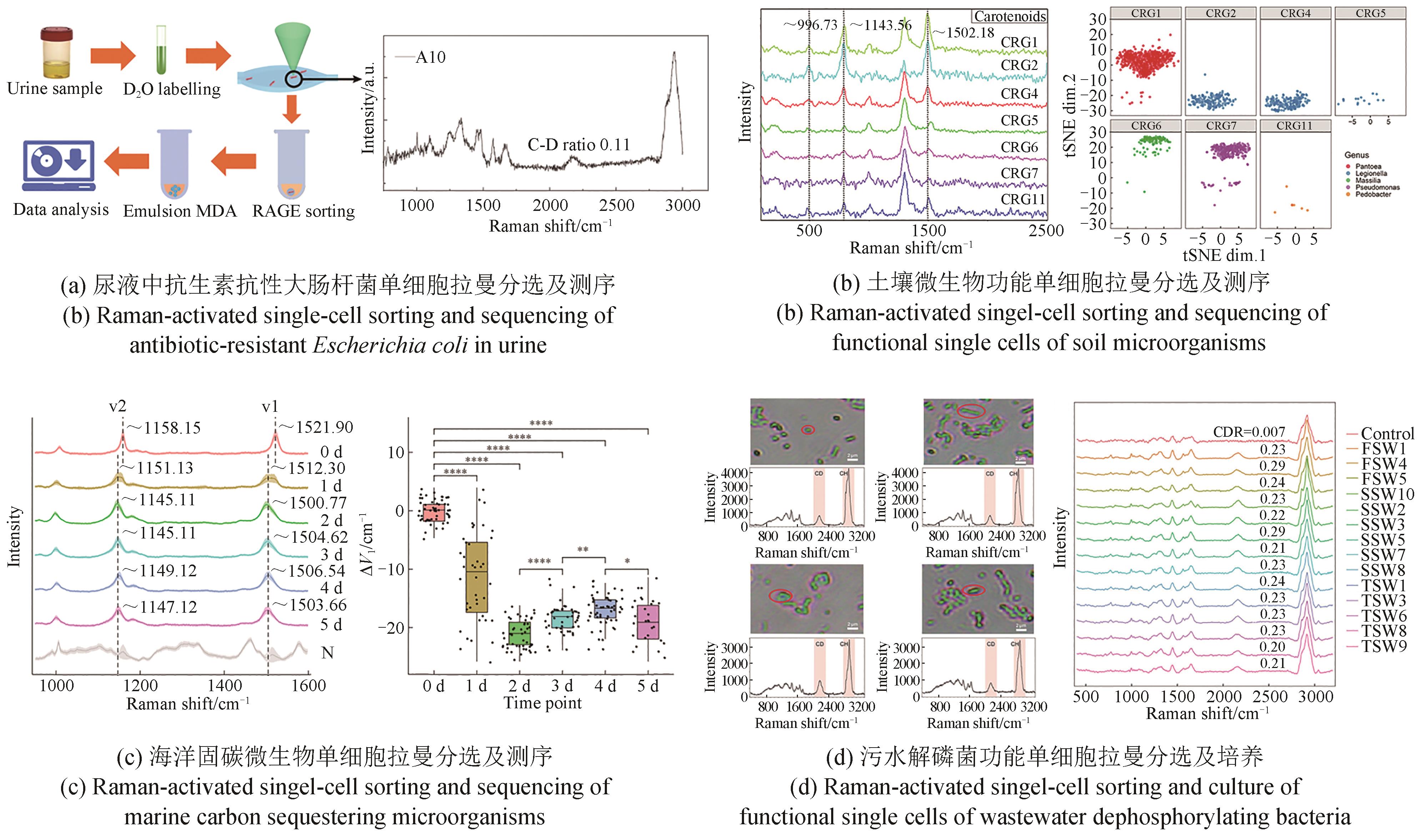
| 31 | JING X Y, GONG Y H, XU T, et al. One-cell metabolic phenotyping and sequencing of soil microbiome by Raman-activated gravity-driven encapsulation (RAGE)[J]. mSystems, 2021, 6(3): e00181-21. |
| 32 | JING X Y, GONG Y H, XU T, et al. Revealing CO2-fixing SAR11 bacteria in the ocean by Raman-based single-cell metabolic profiling and genomics[J]. BioDesign Research, 2022, 2022: 9782712. |
| 33 | JING X Y, GONG Y H, PAN H H, et al. Single-cell Raman-activated sorting and cultivation (scRACS-Culture) for assessing and mining in situ phosphate-solubilizing microbes from nature[J]. ISME Communications, 2022, 2: 106. |
| 34 | XU T, LI Y D, HAN X, et al. Versatile, facile and low-cost single-cell isolation, culture and sequencing by optical tweezer-assisted pool-screening[J]. Lab on a Chip, 2023, 23(1): 125-135. |
| 35 | DIAO Z D, KAN L Y, ZHAO Y L, et al. Artificial intelligence-assisted automatic and index-based microbial single-cell sorting system for One-Cell-One-Tube[J]. mLife, 2022, 1(4): 448-459. |
| 36 | XIN Y, SHEN C, SHE Y T, et al. Biosynthesis of triacylglycerol molecules with a tailored PUFA profile in industrial microalgae[J]. Molecular Plant, 2019, 12(4): 474-488. |
| 37 | XIN Y, LU Y D, LEE Y Y, et al. Producing designer oils in industrial microalgae by rational modulation of Co-evolving type-2 diacylglycerol acyltransferases[J]. Molecular Plant, 2017, 10(12): 1523-1539. |
| 38 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 39 | BERRY D, MADER E, LEE T K, et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(2): E194-203. |
| 40 | JING X Y, GOU H L, GONG Y H, et al. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean[J]. Environmental Microbiology, 2018, 20(6): 2241-2255. |
| 41 | SONG Y Z, KASTER A K, VOLLMERS J, et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea[J]. Microbial Biotechnology, 2017, 10(1): 125-137. |
| 42 | WANG T T, JI Y T, WANG Y, et al. Quantitative dynamics of triacylglycerol accumulation in microalgae populations at single-cell resolution revealed by Raman microspectroscopy[J].Biotechnology for Biofuels, 2014, 7: 58. |
| 43 | JI Y T, HE Y H, CUI Y B, et al. Raman spectroscopy provides a rapid, non-invasive method for quantitation of starch in live, unicellular microalgae[J]. Biotechnology Journal, 2014, 9(12): 1512-1518. |
| 44 | HE Y H, ZHANG P, HUANG S, et al. Label-free, simultaneous quantification of starch, protein and triacylglycerol in single microalgal cells[J].Biotechnology for Biofuels, 2017, 10(1): 275. |
| 45 | TAO Y F, WANG Y, HUANG S, et al. Metabolic-activity-based assessment of antimicrobial effects by D2O-labeled single-cell Raman microspectroscopy[J]. Analytical Chemistry, 2017, 89(7): 4108-4115. |
| 46 | TENG L, WANG X, WANG X J, et al. Label-free, rapid and quantitative phenotyping of stress response in E. coli via ramanome[J]. Scientific Reports, 2016, 6: 34359. |
| 47 | HEKMATARA M, HEIDARI BALADEHI M, JI Y T, et al. D2O-probed Raman microspectroscopy distinguishes the metabolic dynamics of macromolecules in organellar anticancer drug response[J]. Analytical Chemistry, 2021, 93(4): 2125-2134. |
| 48 | WANG Y, SONG Y Z, TAO Y F, et al. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level[J]. Analytical Chemistry, 2016, 88(19): 9443-9450. |
| 49 | HE Y H, HUANG S, ZHANG P, et al. Intra-ramanome correlation analysis unveils metabolite conversion network from an isogenic population of cells[J]. mBio, 2021, 12(4): e0147021. |
| 50 | HEIDARI BALADEHI M, HEKMATARA M, HE Y H, et al. Culture-free identification and metabolic profiling of microalgal single cells via ensemble learning of ramanomes[J]. Analytical Chemistry, 2021, 93(25): 8872-8880. |
| [1] | 应汉杰, 柳东, 王振宇, 沈涛, 庄伟, 朱晨杰. 工业生物制造与“碳中和”目标探讨[J]. 合成生物学, 2025, 6(1): 1-7. |
| [2] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||