合成生物学 ›› 2023, Vol. 4 ›› Issue (6): 1300-1320.DOI: 10.12211/2096-8280.2022-052
• 研究论文 • 上一篇
希瓦氏菌Shewanella sp. MR-4在不同末端电子受体条件下的代谢通量分析
郑庆祥
- 漳州人才发展集团有限公司,福建 漳州 363000
-
收稿日期:2022-09-23修回日期:2022-11-16出版日期:2023-12-31发布日期:2024-01-19 -
通讯作者:郑庆祥 -
作者简介:郑庆祥 (1983—),男,博士,工程师,漳州人才发展集团有限公司副总经理。研究方向为微生物代谢工程。E-mail:179949046@qq.com -
基金资助:国家自然科学基金(30970035)
Metabolic flux analysis of Shewanella sp. MR-4 with different terminal electron receptors
ZHENG Qingxiang
- Zhangzhou Talent Development Group,Zhangzhou 363000,Fujian,China
-
Received:2022-09-23Revised:2022-11-16Online:2023-12-31Published:2024-01-19 -
Contact:ZHENG Qingxiang
摘要:
本研究解析了Shewanella sp. MR-4在厌氧条件下以Fe3+、延胡索酸、硝酸和二甲基亚砜(DMSO)为末端电子受体时代谢通量分布,发现在各种不同末端电子受体条件下细胞内代谢通量分布发生显著改变。与延胡索酸相比,在Fe3+和硝酸为电子受体条件下,乙酸生成速率明显降低,而三羧酸循环代谢通量增加。同时,三羧酸循环回补反应存在显著差异:在延胡索酸为电子受体条件下,磷酸烯醇式丙酮酸羧化酶反应是唯一回补反应,而在Fe3+和硝酸条件下苹果酸酶催化成为唯一回补反应。本研究在代谢通量分析基础上定量解析了还原力NADH合成和消耗,分析了电子流向变化,发现在不同末端电子受体条件下,NADH产生途径和反应发生明显变化。研究结果为希瓦氏菌代谢工程改造提供了一定理论依据。
中图分类号:
引用本文
郑庆祥. 希瓦氏菌Shewanella sp. MR-4在不同末端电子受体条件下的代谢通量分析[J]. 合成生物学, 2023, 4(6): 1300-1320.
ZHENG Qingxiang. Metabolic flux analysis of Shewanella sp. MR-4 with different terminal electron receptors[J]. Synthetic Biology Journal, 2023, 4(6): 1300-1320.
碱性溶液 Basic solution | ||
|---|---|---|
成分 Component | 浓度/(mmol/L) Concentration/(mmol/L) | |
| PIPES | 30 | |
| NH4Cl | 28.04 | |
| KCl | 1.34 | |
| NaH2PO4·2H2O | 4.35 | |
| NaCl | 30 | |
| Na2SO4 | 0.7 | |
矿物溶液 Mineral solution | ||
成分 Component | 浓度/(µmol/L) Concentration/ (µmol/L) | |
| C6H9NO6 | 78.49 | |
| MgSO4·7H2O | 121.71 | |
| MnSO4·H2O | 29.58 | |
| FeSO4·7H2O | 3.6 | |
| CaCl2·2H2O | 6.8 | |
| CoCl2·6H2O | 4.2 | |
| ZnCl2 | 9.54 | |
| CuSO4·5H2O | 0.4 | |
| AlK(SO4)2·12H2O | 0.21 | |
| H3BO3 | 1.62 | |
| Na2MoO4·2H2O | 1.03 | |
| NiCl2·6H2O | 1.01 | |
| Na2WO4·2H2O | 0.76 | |
维生素溶液 Vitamin solution | ||
成分 Component | 浓度/(µmol/L) Concentration/(µmol/L) | |
| D-生物素 | D-biotin | 81.87 |
| 叶酸 | folic acid | 45.34 |
| 盐酸吡哆醇 | pyridoxine HCl | 486.38 |
| 核黄素 | riboflavin | 132.84 |
| 盐酸硫胺素 | thiamine HCl | 140.73 |
| 烟碱酸 | nicotinic acid | 406.17 |
| D-泛酸,半钙盐 | D-pantothenic acid, hemi calcium salt | 209.82 |
| 维生素B12 | VB12 | 7.4 |
| 对氨基苯甲酸 | p-amino benzoic acid | 364.62 |
| 硫辛酸 | thioctic acid | 242.37 |
| 氯化胆碱 | choline chloride | 14.3246 |
表1 修改型M1基本培养基
Table 1 Modified M1 minimal medium
碱性溶液 Basic solution | ||
|---|---|---|
成分 Component | 浓度/(mmol/L) Concentration/(mmol/L) | |
| PIPES | 30 | |
| NH4Cl | 28.04 | |
| KCl | 1.34 | |
| NaH2PO4·2H2O | 4.35 | |
| NaCl | 30 | |
| Na2SO4 | 0.7 | |
矿物溶液 Mineral solution | ||
成分 Component | 浓度/(µmol/L) Concentration/ (µmol/L) | |
| C6H9NO6 | 78.49 | |
| MgSO4·7H2O | 121.71 | |
| MnSO4·H2O | 29.58 | |
| FeSO4·7H2O | 3.6 | |
| CaCl2·2H2O | 6.8 | |
| CoCl2·6H2O | 4.2 | |
| ZnCl2 | 9.54 | |
| CuSO4·5H2O | 0.4 | |
| AlK(SO4)2·12H2O | 0.21 | |
| H3BO3 | 1.62 | |
| Na2MoO4·2H2O | 1.03 | |
| NiCl2·6H2O | 1.01 | |
| Na2WO4·2H2O | 0.76 | |
维生素溶液 Vitamin solution | ||
成分 Component | 浓度/(µmol/L) Concentration/(µmol/L) | |
| D-生物素 | D-biotin | 81.87 |
| 叶酸 | folic acid | 45.34 |
| 盐酸吡哆醇 | pyridoxine HCl | 486.38 |
| 核黄素 | riboflavin | 132.84 |
| 盐酸硫胺素 | thiamine HCl | 140.73 |
| 烟碱酸 | nicotinic acid | 406.17 |
| D-泛酸,半钙盐 | D-pantothenic acid, hemi calcium salt | 209.82 |
| 维生素B12 | VB12 | 7.4 |
| 对氨基苯甲酸 | p-amino benzoic acid | 364.62 |
| 硫辛酸 | thioctic acid | 242.37 |
| 氯化胆碱 | choline chloride | 14.3246 |
| 菌种 | 乳酸盐(lactate) 氧气(oxygen) 有氧(aerobic) | 乳酸盐(lactate) 延胡索酸(fumarate) 厌氧(anaerobic) | 葡萄糖(glucose) 延胡索酸(fumarate) 厌氧(anaerobic) | 葡萄糖(glucose) 次氮基三乙酸铁(Fe-NTA) 厌氧(anaerobic) |
|---|---|---|---|---|
| Shewanella putrefaciens CN-32 | + | + | + | - |
| Shewanella loihica PV-4 | + | + | + | + |
| Shewanella sp. MR-4 | + | + | + | + |
| Shewanella amazonensis SB2B | + | + | + | + |
| Shewanella sp. W3-18-1 | + | + | + | - |
| Shewanella baltica OS155 | + | + | + | + |
| Shewanella denitrificans OS217 | + | + | - | - |
| Shewanella oneidensis MR-1 | + | + | - | - |
| Shewanella sp. MR-7 | + | + | - | + |
| Shewanella sp. ANA-3 | + | + | + | + |
| Shewanella pealeana ANG-SQ1 | + | - | - | - |
| Shewanella frigidimarina NCIMB400 | + | + | - | - |
表2 希瓦氏菌12株菌株生长表型
Table 2 Growth phenotype of 12 Shewanella sp. strains
| 菌种 | 乳酸盐(lactate) 氧气(oxygen) 有氧(aerobic) | 乳酸盐(lactate) 延胡索酸(fumarate) 厌氧(anaerobic) | 葡萄糖(glucose) 延胡索酸(fumarate) 厌氧(anaerobic) | 葡萄糖(glucose) 次氮基三乙酸铁(Fe-NTA) 厌氧(anaerobic) |
|---|---|---|---|---|
| Shewanella putrefaciens CN-32 | + | + | + | - |
| Shewanella loihica PV-4 | + | + | + | + |
| Shewanella sp. MR-4 | + | + | + | + |
| Shewanella amazonensis SB2B | + | + | + | + |
| Shewanella sp. W3-18-1 | + | + | + | - |
| Shewanella baltica OS155 | + | + | + | + |
| Shewanella denitrificans OS217 | + | + | - | - |
| Shewanella oneidensis MR-1 | + | + | - | - |
| Shewanella sp. MR-7 | + | + | - | + |
| Shewanella sp. ANA-3 | + | + | + | + |
| Shewanella pealeana ANG-SQ1 | + | - | - | - |
| Shewanella frigidimarina NCIMB400 | + | + | - | - |
电子受体 (Electron acceptor) | 用于生物质合成的 葡萄糖的比例 (Percentage of glucose for biomass synthesis) | 比率/[mmol/(gCDW·h)] Specific rate/[mmol/(gCDW·h)] | 碳平衡 (Carbon balance) | |||
|---|---|---|---|---|---|---|
葡萄糖摄取 (Glucose uptake) | 醋酸盐分泌 (Acetate secretion) | EPS形成 (EPS formation) | CO2净生成 (Net CO2 formation) | |||
| 硝酸盐(Nitrate) | 12.2% ± 0.5% | 8.27 ± 0.24 | 4.19 ± 0.21 | 2.49 ± 0.11 | 17.47 ± 0.94 | 94.5% |
| Fe | 5.1% ± 0.2% | 4.86 ± 0.20 | 3.74 ± 0.19 | 1.28 ± 0.04 | 10.72 ± 0.35 | 93.9% |
| 延胡索酸(Fumarate) | 11.7%± 0.4% | 5.04 ± 0.17 | 8.33 ± 0.54 | 0.31 ± 0.01 | 7.23 ± 0.38 | 96.8% |
| DMSO | 5.9% ± 0.2% | 5.38 ± 0.15 | 9.48 ± 0.47 | 0.47 ± 0.02 | 6.92 ± 0.21 | 94.9% |
表3 Shewanella sp. MR-4在各种末端电子受体上厌氧生长参数
Table 3 Anaerobic growth parameters of Shewanella sp. MR-4 exposed to various terminal electron acceptors
电子受体 (Electron acceptor) | 用于生物质合成的 葡萄糖的比例 (Percentage of glucose for biomass synthesis) | 比率/[mmol/(gCDW·h)] Specific rate/[mmol/(gCDW·h)] | 碳平衡 (Carbon balance) | |||
|---|---|---|---|---|---|---|
葡萄糖摄取 (Glucose uptake) | 醋酸盐分泌 (Acetate secretion) | EPS形成 (EPS formation) | CO2净生成 (Net CO2 formation) | |||
| 硝酸盐(Nitrate) | 12.2% ± 0.5% | 8.27 ± 0.24 | 4.19 ± 0.21 | 2.49 ± 0.11 | 17.47 ± 0.94 | 94.5% |
| Fe | 5.1% ± 0.2% | 4.86 ± 0.20 | 3.74 ± 0.19 | 1.28 ± 0.04 | 10.72 ± 0.35 | 93.9% |
| 延胡索酸(Fumarate) | 11.7%± 0.4% | 5.04 ± 0.17 | 8.33 ± 0.54 | 0.31 ± 0.01 | 7.23 ± 0.38 | 96.8% |
| DMSO | 5.9% ± 0.2% | 5.38 ± 0.15 | 9.48 ± 0.47 | 0.47 ± 0.02 | 6.92 ± 0.21 | 94.9% |
| 反应编号 (Reaction No.) | 酶 (Enzyme) | 基因 (Gene) | 反应化学计量学 (Stoichiometric reaction) | 碳原子转换 (Carbon atom transition) |
|---|---|---|---|---|
| v1 | glucokinase | Shewmr4_1763 | Glucose => G6P | #ABCDEF => #ABCDEF |
| v2 | glucose-6-phosphate 1-dehydrogenase | Shewmr4_2046 | G6P => 6PG | #ABCDEF => #ABCDEF |
| 6-phosphogluconolactonase | Shewmr4_2045 | |||
| v3 | phosphogluconate dehydratase | Shewmr4_2044 | 6PG => KDPG | #ABCDEF => #ABCDEF |
| 2-keto-3-deoxy-phosphogluconate aldolase | Shewmr4_2043 | KDPG => GAP + PYR | #ABCDEF => #DEF + #ABC | |
| v4 | glucose-6-phosphate isomerase | Shewmr4_2968 | G6P <=> F6P | #ABCDEF <=> #ABCDEF |
| v5 | 6-phosphogluconate dehydrogenase | Shewmr4_1609 | 6PG => P5P + CO2 | #ABCDEF => #BCDEF + #A |
| v6 | transketolase | Shewmr4_0773 | S7P + GAP <=> P5P + P5P | #abcdefg + #ABC <=> #cdefg + #abABC |
| v7 | transaldolase | Shewmr4_2967 | S7P + GAP <=> F6P + E4P | #abcdefg + #ABC <=> #abcABC + #defg |
| v8 | transketolase | Shewmr4_0773 | F6P + GAP <=> P5P + E4P | #abcdef + #ABC <=> #abABC+ #cdef |
| v9 | glyceraldehy-3-phosphate dehydrogenase | Shewmr4_0536 | GAP <=> 1,3-BPG | #ABC <=> #ABC |
| phosphoglycerate kinase | Shewmr4_0775 | 1,3-BPG <=> 3PG | #ABC <=> #ABC | |
| phosphoglyceromutase | Shewmr4_0045 | 3PG <=> 2PG | #ABC <=> #ABC | |
| enolase | Shewmr4_1115 | 2PG <=> PEP | #ABC <=> #ABC | |
| v10 | pyruvate kinase | Shewmr4_2048 | PEP => PYR | #ABC => #ABC |
| v11 | phosphoenolpyruvate carboxylase | Shewmr4_3710 | PEP + CO2 => OAA | #ABC + #D => #ABCD |
| v12 | phosphoenolpyruvate carboxykinase | Shewmr4_0152 | OAA => PEP + CO2 | #ABCD => #ABC + #D |
| v13 | NAD-dependent malic enzyme | Shewmr4_3188 | MAL <=> PYR + CO2 | #ABCD <=> #ABC + #D |
| NADP-dependent malic enzyme | Shewmr4_3492 | |||
| v14 | pyruvate dehydrogenase subunit E1 | Shewmr4_0428 | PYR =>ACoA + CO2 | #ABC=>#BC + #A |
| pyruvate dehydrogenase complex dihydrolipoamide acetyltransferase | Shewmr4_0429 | |||
| dihydrolipoamide dehydrogenase | Shewmr4_0430 | |||
| v15 | type Ⅱ citrate synthase | Shewmr4_1630 | ACoA + OAA => ICT | #abcd+#AB => #dcbaBA |
| bifunctional aconitate hydratase | Shewmr4_0437 | |||
| v16 | isocitrate dehydrogenase, NADP-dependent | Shewmr4_1606 | ICT => AKG + CO2 | #abcdef => #abcef + #d |
| isocitrate dehydrogenase [NAD] | Shewmr4_2720 | |||
| v17 | 2-oxoglutarate dehydrogenase E1 component | Shewmr4_1635 | AKG => FUM + CO2 | #ABCDE => #BCDE+#A |
| dihydrolipoamide dehydrogenase | Shewmr4_0430 | |||
| 2-oxoglutarate dehydrogenase E2 component | Shewmr4_1636 | |||
| succinyl-CoA synthetase subunit beta | Shewmr4_1637 | |||
| fumarate reductase cytochrome b-556 subunit | Shewmr4_0402 | |||
| fumarase | Shewmr4_2124 | FUM <=> MAL | #ABCD <=> #ABCD #ABCD => #DCBA | |
| v18 | malate dehydrogenase | Shewmr4_3339 | MAL <=> OAA | #ABCD <=> #ABCD |
| v19 | biomass synthesis | |||
| v20 | acetate formation | ACoA => Ace | ||
| v21 | EPS synthesis | G6P => EPS | ||
表4 Shewanella sp. MR-4中心代谢网络反应化学计量学和相应碳原子转换
Table 4 Central metabolic network in Shewanella sp. MR-4 with the stoichiometric reactions and the corresponding carbon atom transitions
| 反应编号 (Reaction No.) | 酶 (Enzyme) | 基因 (Gene) | 反应化学计量学 (Stoichiometric reaction) | 碳原子转换 (Carbon atom transition) |
|---|---|---|---|---|
| v1 | glucokinase | Shewmr4_1763 | Glucose => G6P | #ABCDEF => #ABCDEF |
| v2 | glucose-6-phosphate 1-dehydrogenase | Shewmr4_2046 | G6P => 6PG | #ABCDEF => #ABCDEF |
| 6-phosphogluconolactonase | Shewmr4_2045 | |||
| v3 | phosphogluconate dehydratase | Shewmr4_2044 | 6PG => KDPG | #ABCDEF => #ABCDEF |
| 2-keto-3-deoxy-phosphogluconate aldolase | Shewmr4_2043 | KDPG => GAP + PYR | #ABCDEF => #DEF + #ABC | |
| v4 | glucose-6-phosphate isomerase | Shewmr4_2968 | G6P <=> F6P | #ABCDEF <=> #ABCDEF |
| v5 | 6-phosphogluconate dehydrogenase | Shewmr4_1609 | 6PG => P5P + CO2 | #ABCDEF => #BCDEF + #A |
| v6 | transketolase | Shewmr4_0773 | S7P + GAP <=> P5P + P5P | #abcdefg + #ABC <=> #cdefg + #abABC |
| v7 | transaldolase | Shewmr4_2967 | S7P + GAP <=> F6P + E4P | #abcdefg + #ABC <=> #abcABC + #defg |
| v8 | transketolase | Shewmr4_0773 | F6P + GAP <=> P5P + E4P | #abcdef + #ABC <=> #abABC+ #cdef |
| v9 | glyceraldehy-3-phosphate dehydrogenase | Shewmr4_0536 | GAP <=> 1,3-BPG | #ABC <=> #ABC |
| phosphoglycerate kinase | Shewmr4_0775 | 1,3-BPG <=> 3PG | #ABC <=> #ABC | |
| phosphoglyceromutase | Shewmr4_0045 | 3PG <=> 2PG | #ABC <=> #ABC | |
| enolase | Shewmr4_1115 | 2PG <=> PEP | #ABC <=> #ABC | |
| v10 | pyruvate kinase | Shewmr4_2048 | PEP => PYR | #ABC => #ABC |
| v11 | phosphoenolpyruvate carboxylase | Shewmr4_3710 | PEP + CO2 => OAA | #ABC + #D => #ABCD |
| v12 | phosphoenolpyruvate carboxykinase | Shewmr4_0152 | OAA => PEP + CO2 | #ABCD => #ABC + #D |
| v13 | NAD-dependent malic enzyme | Shewmr4_3188 | MAL <=> PYR + CO2 | #ABCD <=> #ABC + #D |
| NADP-dependent malic enzyme | Shewmr4_3492 | |||
| v14 | pyruvate dehydrogenase subunit E1 | Shewmr4_0428 | PYR =>ACoA + CO2 | #ABC=>#BC + #A |
| pyruvate dehydrogenase complex dihydrolipoamide acetyltransferase | Shewmr4_0429 | |||
| dihydrolipoamide dehydrogenase | Shewmr4_0430 | |||
| v15 | type Ⅱ citrate synthase | Shewmr4_1630 | ACoA + OAA => ICT | #abcd+#AB => #dcbaBA |
| bifunctional aconitate hydratase | Shewmr4_0437 | |||
| v16 | isocitrate dehydrogenase, NADP-dependent | Shewmr4_1606 | ICT => AKG + CO2 | #abcdef => #abcef + #d |
| isocitrate dehydrogenase [NAD] | Shewmr4_2720 | |||
| v17 | 2-oxoglutarate dehydrogenase E1 component | Shewmr4_1635 | AKG => FUM + CO2 | #ABCDE => #BCDE+#A |
| dihydrolipoamide dehydrogenase | Shewmr4_0430 | |||
| 2-oxoglutarate dehydrogenase E2 component | Shewmr4_1636 | |||
| succinyl-CoA synthetase subunit beta | Shewmr4_1637 | |||
| fumarate reductase cytochrome b-556 subunit | Shewmr4_0402 | |||
| fumarase | Shewmr4_2124 | FUM <=> MAL | #ABCD <=> #ABCD #ABCD => #DCBA | |
| v18 | malate dehydrogenase | Shewmr4_3339 | MAL <=> OAA | #ABCD <=> #ABCD |
| v19 | biomass synthesis | |||
| v20 | acetate formation | ACoA => Ace | ||
| v21 | EPS synthesis | G6P => EPS | ||
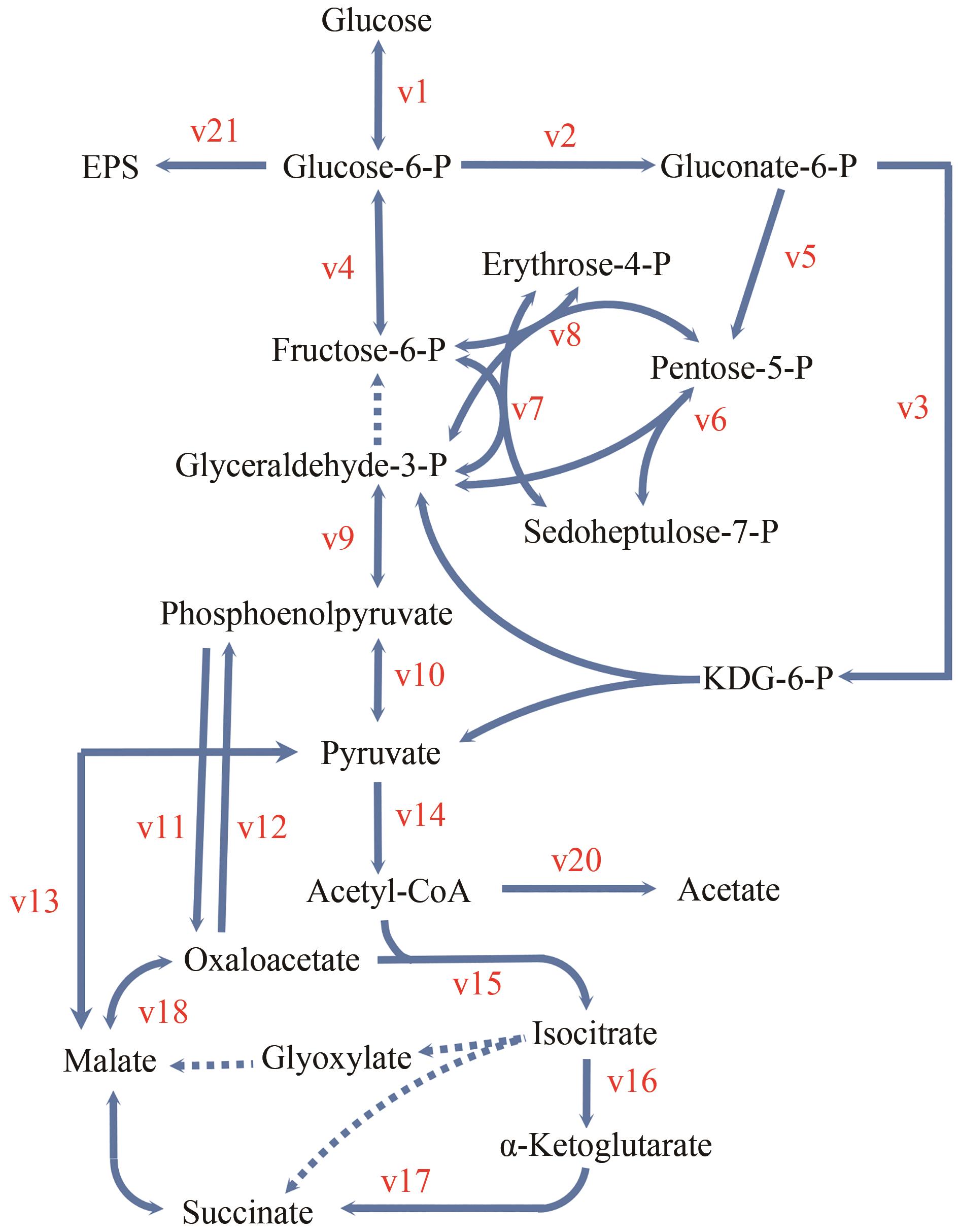
图1 Shewanella sp. MR-4中心碳代谢生物反应网络(虚线箭头表示本研究中鉴定为无活性反应或途径。双向箭头表示推测为可逆反应)
Fig. 1 Bioreaction network of central carbon metabolism of Shewanella sp. MR-4(Dashed arrows indicate reactions or pathways identified to be inactive in this study. Double-headed arrows indicate reactions assumed to be reversible)
代谢物 (Metabolite) | Shewanella sp. MR-4在总池中的比例 (Percentage of total pool in Shewanella sp. MR-4) | |||
|---|---|---|---|---|
| 硝酸盐(nitrate) | Fe | DMSO | 延胡索酸(fumarate) | |
| PEP from PP pathway (ub) | 2.2 % | 3.2 % | 6.7 % | 1.9 % |
| PEP from OAA (ub) | 2.7 % | 2.7 % | 6.7 % | 0.0 % |
| PYR from OAA (ub) | 7.2 % | 11.8 % | 29.8 % | 0.5 % |
| PYR from OAA (lb) | 3.4 % | 9.7 % | 11.7 % | 0.5 % |
| OAA from PEP | 0.0 % | 0.0 % | 80.7 % | 93.1 % |
| MAL from PYR | 33.5 % | 84.3 % | 0.0 % | 0.0 % |
| OAA from glyoxylate shunt | 0.0 % | 0.0 % | 0.0 % | 0.0 % |
| Ser from Gly | 0.0 % | 28.6 % | 18.6 % | 23.0 % |
| Gly from Ser | 84.2 % | 77.0 % | 100.0 % | 86.1 % |
表5 通过流比例分析确定中间代谢物来源
Table 5 Origins of metabolic intermediates determined by flux ratio analysis
代谢物 (Metabolite) | Shewanella sp. MR-4在总池中的比例 (Percentage of total pool in Shewanella sp. MR-4) | |||
|---|---|---|---|---|
| 硝酸盐(nitrate) | Fe | DMSO | 延胡索酸(fumarate) | |
| PEP from PP pathway (ub) | 2.2 % | 3.2 % | 6.7 % | 1.9 % |
| PEP from OAA (ub) | 2.7 % | 2.7 % | 6.7 % | 0.0 % |
| PYR from OAA (ub) | 7.2 % | 11.8 % | 29.8 % | 0.5 % |
| PYR from OAA (lb) | 3.4 % | 9.7 % | 11.7 % | 0.5 % |
| OAA from PEP | 0.0 % | 0.0 % | 80.7 % | 93.1 % |
| MAL from PYR | 33.5 % | 84.3 % | 0.0 % | 0.0 % |
| OAA from glyoxylate shunt | 0.0 % | 0.0 % | 0.0 % | 0.0 % |
| Ser from Gly | 0.0 % | 28.6 % | 18.6 % | 23.0 % |
| Gly from Ser | 84.2 % | 77.0 % | 100.0 % | 86.1 % |
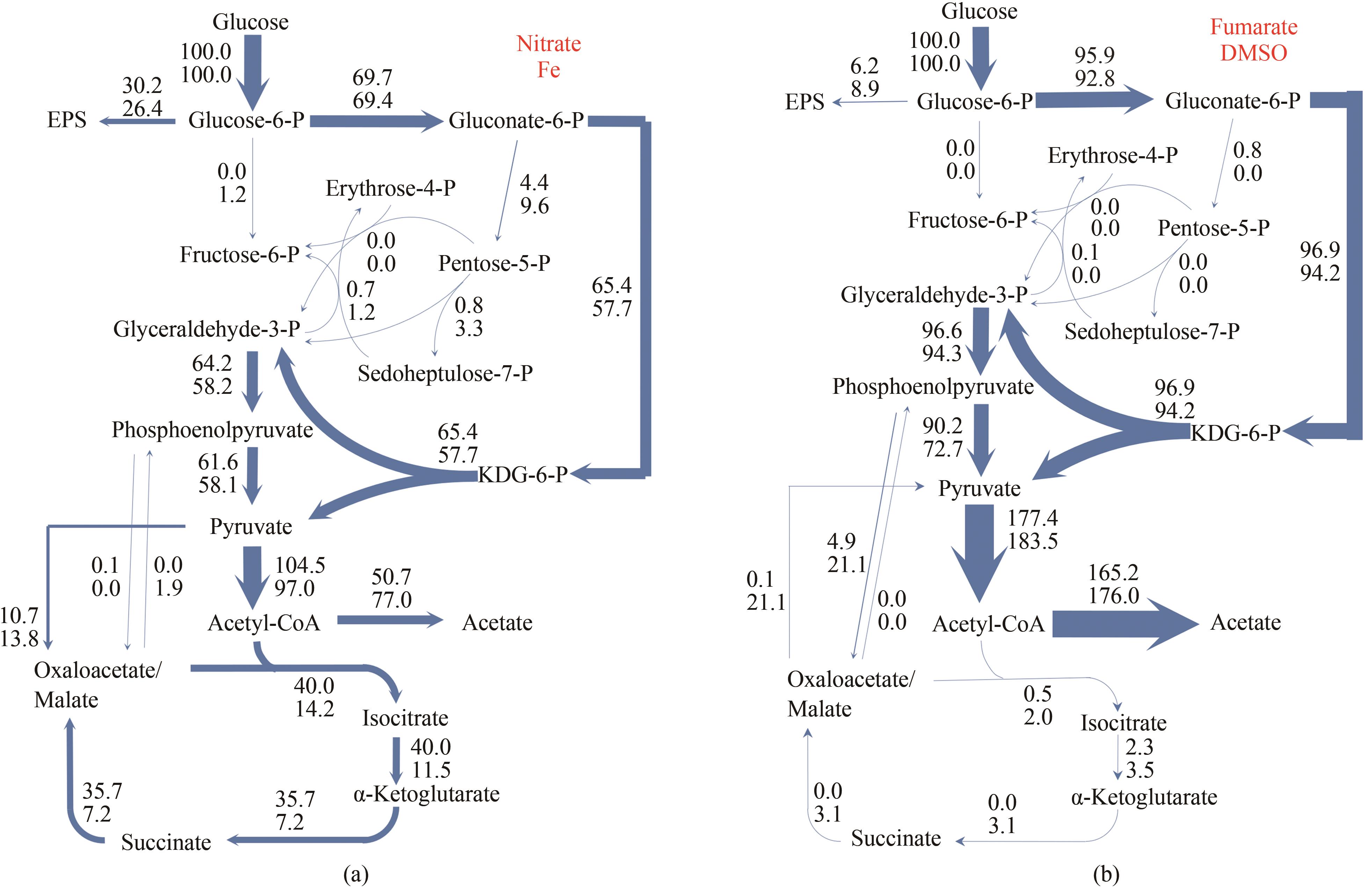
图2 Shewanella sp. MR-4在以硝酸、Fe3+(a)和延胡索酸、二甲基亚砜(b)作为末端电子受体时胞内代谢通量分布[代谢反应旁边数字为该反应净通量(net flux),是以底物葡萄糖比摄取速率进行归一化后数值](a)上,硝酸;下,Fe3+。(b)上,延胡索酸;下,二甲基亚砜
Fig. 2 Intracellular metabolic flux distributions in Shewanella sp. MR-4 with (a) nitrate, Fe3+, (b) fumarate or DMSO as terminal electron acceptor[The number beside a metabolic reaction denotes its net flux, which was normalized by specific rate of glucose uptake](a) top, nitrate; bottom, Fe3+; (b) Top, fumarate; bottom, DMSO
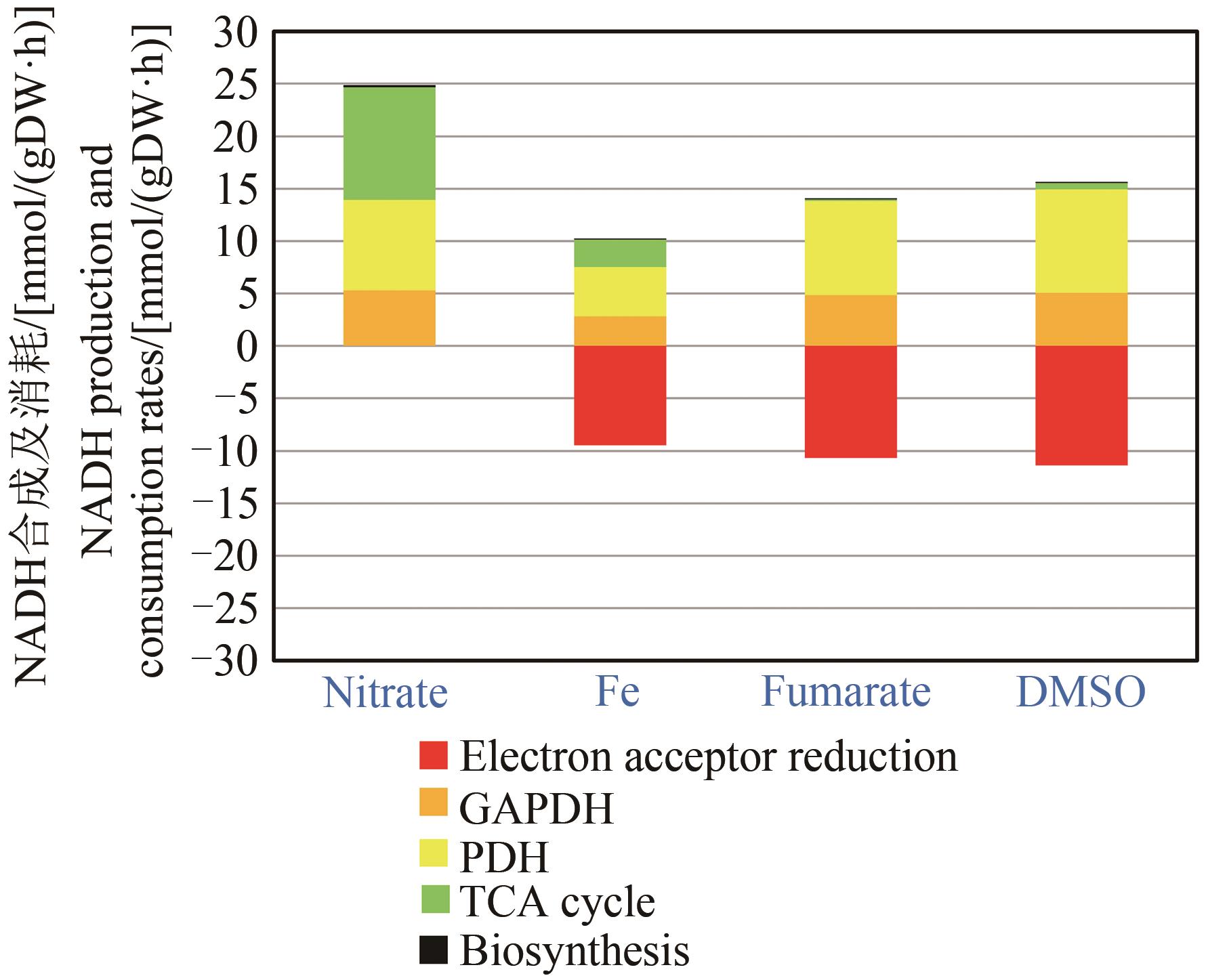
图3 基于代谢通量NADH平衡(NADH生成来自丙酮酸脱氢酶复合体、三羧酸循环和生物质合成。NADH消耗经由电子受体还原。以硝酸作为末端电子受体时NADH消耗不确定)
Fig. 3 NADH balancing based on metabolic flux distributions(NADH is formed by the PDH complex, the TCA cycle and biomass synthesis. NADH was consumed via electron acceptor reduction. NADH consumption rate cannot be determined with nitrate as terminal electron acceptor)
片段 Fragments | 碳原子 C-atom | 前体 precursor | 前体的碳原子 C-atom of precursor | m | m+1 | m+2 | m+3 | m+4 | m+5 | m+6 | m+7 | m+8 | m+9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
硝酸盐 Nitrate (U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7771 | 0.0586 | 0.0285 | 0.1358 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.8027 | 0.0475 | 0.1497 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.6223 | 0.1759 | 0.1167 | 0.0677 | 0.0173 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6698 | 0.1857 | 0.1123 | 0.0321 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.7723 | 0.1108 | 0.1169 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5469 | 0.1780 | 0.1864 | 0.0622 | 0.0207 | 0.0059 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5910 | 0.1856 | 0.1753 | 0.0357 | 0.0124 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.7247 | 0.1880 | 0.0874 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8083 | 0.0612 | 0.1305 | ||||||||
| M_gly_085 | 1 | 1 | 0.8354 | 0.1646 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.5165 | 0.1481 | 0.2009 | 0.0912 | 0.0432 | 0.0000 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5553 | 0.1702 | 0.1861 | 0.0620 | 0.0202 | 0.0063 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.5395 | 0.0884 | 0.2743 | 0.0305 | 0.0650 | 0.0023 | 0.0000 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5507 | 0.1606 | 0.2028 | 0.0551 | 0.0254 | 0.0054 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.5194 | 0.1706 | 0.1497 | 0.0992 | 0.0416 | 0.0145 | 0.0049 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.5333 | 0.1854 | 0.1893 | 0.0636 | 0.0204 | 0.0080 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6538 | 0.2656 | 0.0807 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6959 | 0.2209 | 0.0353 | 0.0297 | 0.0137 | 0.0045 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.7233 | 0.2133 | 0.0389 | 0.0216 | 0.0030 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4987 | 0.1004 | 0.0907 | 0.1524 | 0.0706 | 0.0388 | 0.0302 | 0.0125 | 0.0032 | 0.0026 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.5004 | 0.0965 | 0.1771 | 0.0872 | 0.0709 | 0.0361 | 0.0240 | 0.0046 | 0.0031 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8280 | 0.0179 | 0.1541 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3803 | 0.1470 | 0.1487 | 0.0851 | 0.1094 | 0.0456 | 0.0586 | 0.0252 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5628 | 0.1626 | 0.1632 | 0.0812 | 0.0226 | 0.0076 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.6054 | 0.1808 | 0.1667 | 0.0343 | 0.0127 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7811 | 0.0469 | 0.0218 | 0.1502 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7845 | 0.0653 | 0.1501 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.8107 | 0.0185 | 0.1708 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6021 | 0.1815 | 0.1296 | 0.0636 | 0.0233 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6554 | 0.1878 | 0.1305 | 0.0262 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.6432 | 0.2531 | 0.1037 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6424 | 0.0767 | 0.1348 | 0.1113 | 0.0118 | 0.0229 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6590 | 0.0690 | 0.2307 | 0.0153 | 0.0260 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7932 | 0.0604 | 0.1464 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4878 | 0.1020 | 0.1013 | 0.1531 | 0.0730 | 0.0398 | 0.0269 | 0.0128 | 0.0032 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4889 | 0.1005 | 0.1861 | 0.0923 | 0.0695 | 0.0325 | 0.0252 | 0.0037 | 0.0014 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.8181 | 0.0223 | 0.1596 | |||||||
| Fe (U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.6653 | 0.0687 | 0.0481 | 0.2180 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.7027 | 0.0516 | 0.2457 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.5139 | 0.1980 | 0.0784 | 0.1563 | 0.0534 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5856 | 0.1580 | 0.1399 | 0.1165 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.6452 | 0.1865 | 0.1683 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.4265 | 0.1428 | 0.2328 | 0.1277 | 0.0408 | 0.0294 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.4863 | 0.1174 | 0.3028 | 0.0394 | 0.0541 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6145 | 0.2689 | 0.1166 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7073 | 0.1256 | 0.1672 | ||||||||
| M_gly_085 | 1 | 1 | 0.7732 | 0.2268 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.3812 | 0.1562 | 0.1741 | 0.1614 | 0.0538 | 0.0524 | 0.0208 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.4351 | 0.1340 | 0.2355 | 0.1232 | 0.0412 | 0.0311 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.3875 | 0.0690 | 0.3376 | 0.0590 | 0.1262 | 0.0072 | 0.0134 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.3941 | 0.1712 | 0.2546 | 0.1107 | 0.0524 | 0.0169 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.3937 | 0.1582 | 0.1451 | 0.1789 | 0.0628 | 0.0405 | 0.0209 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.4262 | 0.1373 | 0.2278 | 0.1236 | 0.0429 | 0.0421 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.5390 | 0.3428 | 0.1181 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3944 | 0.2637 | 0.1093 | 0.1351 | 0.0809 | 0.0167 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.4615 | 0.2480 | 0.1384 | 0.1233 | 0.0288 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.3504 | 0.0882 | 0.1024 | 0.1830 | 0.0992 | 0.0689 | 0.0587 | 0.0279 | 0.0108 | 0.0106 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.3535 | 0.0849 | 0.2022 | 0.1126 | 0.1116 | 0.0599 | 0.0527 | 0.0115 | 0.0110 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.7337 | 0.0211 | 0.2451 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.2751 | 0.1470 | 0.1734 | 0.1093 | 0.1352 | 0.0607 | 0.0709 | 0.0284 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5409 | 0.1403 | 0.1547 | 0.1071 | 0.0337 | 0.0233 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5292 | 0.0996 | 0.2819 | 0.0382 | 0.0511 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.6402 | 0.1096 | 0.0697 | 0.1805 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.6570 | 0.1600 | 0.1830 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7130 | 0.0548 | 0.2321 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.5067 | 0.1940 | 0.0814 | 0.1609 | 0.0571 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5781 | 0.1512 | 0.1539 | 0.1168 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.5460 | 0.2992 | 0.1549 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5007 | 0.0748 | 0.1878 | 0.1625 | 0.0208 | 0.0535 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.5210 | 0.0661 | 0.3278 | 0.0241 | 0.0611 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6948 | 0.0725 | 0.2327 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.3363 | 0.0843 | 0.1144 | 0.1806 | 0.1059 | 0.0691 | 0.0615 | 0.0273 | 0.0104 | 0.0102 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.3358 | 0.0837 | 0.2071 | 0.1196 | 0.1076 | 0.0653 | 0.0558 | 0.0121 | 0.0131 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.7298 | 0.0199 | 0.2504 | |||||||
延胡索酸 Fumarate(U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7549 | 0.0769 | 0.0532 | 0.1149 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.8079 | 0.0369 | 0.1553 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.8566 | 0.0824 | 0.0227 | 0.0326 | 0.0057 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8834 | 0.0649 | 0.0371 | 0.0146 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9028 | 0.0649 | 0.0322 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.7139 | 0.0853 | 0.1649 | 0.0262 | 0.0071 | 0.0027 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.7417 | 0.0774 | 0.1629 | 0.0098 | 0.0083 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8524 | 0.1177 | 0.0300 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8221 | 0.0534 | 0.1245 | ||||||||
| M_gly_085 | 1 | 1 | 0.8500 | 0.1500 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.6803 | 0.1019 | 0.1614 | 0.0377 | 0.0149 | 0.0038 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.6985 | 0.0910 | 0.1743 | 0.0255 | 0.0079 | 0.0027 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.5413 | 0.0739 | 0.2895 | 0.0299 | 0.0604 | 0.0006 | 0.0045 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5449 | 0.1569 | 0.2142 | 0.0551 | 0.0240 | 0.0048 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.6674 | 0.1133 | 0.1150 | 0.0846 | 0.0119 | 0.0062 | 0.0016 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3&4,3,2 | 0.6967 | 0.0906 | 0.1707 | 0.0279 | 0.0100 | 0.0040 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.7318 | 0.2435 | 0.0247 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6959 | 0.2209 | 0.0353 | 0.0297 | 0.0137 | 0.0045 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.7233 | 0.2133 | 0.0389 | 0.0216 | 0.0030 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4690 | 0.1018 | 0.1024 | 0.1654 | 0.0744 | 0.0383 | 0.0293 | 0.0131 | 0.0033 | 0.0030 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4716 | 0.1001 | 0.1908 | 0.0929 | 0.0788 | 0.0329 | 0.0263 | 0.0036 | 0.0029 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8166 | 0.0181 | 0.1653 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3993 | 0.1696 | 0.1489 | 0.0949 | 0.0984 | 0.0384 | 0.0504 | |||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6515 | 0.0727 | 0.1395 | 0.1168 | 0.0162 | 0.0033 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.7504 | 0.0787 | 0.1576 | 0.0060 | 0.0073 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7535 | 0.0701 | 0.0430 | 0.1334 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7630 | 0.1045 | 0.1325 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7998 | 0.0287 | 0.1715 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.8495 | 0.0858 | 0.0229 | 0.0346 | 0.0072 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8791 | 0.0609 | 0.0440 | 0.0160 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.7799 | 0.1869 | 0.0332 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6138 | 0.0911 | 0.1594 | 0.1045 | 0.0120 | 0.0193 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6486 | 0.0641 | 0.2490 | 0.0122 | 0.0261 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7650 | 0.0993 | 0.1357 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4588 | 0.1002 | 0.1068 | 0.1695 | 0.0761 | 0.0369 | 0.0308 | 0.0138 | 0.0029 | 0.0043 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4680 | 0.0996 | 0.1933 | 0.0916 | 0.0816 | 0.0338 | 0.0243 | 0.0042 | 0.0037 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.8064 | 0.0214 | 0.1722 | |||||||
| DMSO(U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7596 | 0.0505 | 0.0361 | 0.1538 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.7778 | 0.0509 | 0.1713 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.6192 | 0.1729 | 0.0738 | 0.1092 | 0.0249 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6709 | 0.1582 | 0.1114 | 0.0596 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.7382 | 0.1339 | 0.1279 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5272 | 0.1559 | 0.2052 | 0.0792 | 0.0224 | 0.0102 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5823 | 0.1361 | 0.2267 | 0.0291 | 0.0258 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6949 | 0.2244 | 0.0807 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7738 | 0.0761 | 0.1501 | ||||||||
| M_gly_085 | 1 | 1 | 0.8112 | 0.1888 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.5000 | 0.1680 | 0.1581 | 0.1163 | 0.0465 | 0.0112 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5293 | 0.1528 | 0.2062 | 0.0789 | 0.0209 | 0.0119 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.4933 | 0.1014 | 0.3028 | 0.0354 | 0.0606 | 0.0064 | 0.0000 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5086 | 0.1741 | 0.2230 | 0.0623 | 0.0258 | 0.0061 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.5194 | 0.1706 | 0.1497 | 0.0992 | 0.0416 | 0.0145 | 0.0049 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.5333 | 0.1854 | 0.1893 | 0.0636 | 0.0204 | 0.0080 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6538 | 0.2656 | 0.0807 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.5209 | 0.2351 | 0.0858 | 0.1153 | 0.0357 | 0.0073 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.5631 | 0.2369 | 0.1186 | 0.0545 | 0.0269 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.4415 | 0.1047 | 0.1088 | 0.1722 | 0.0743 | 0.0451 | 0.0320 | 0.0133 | 0.0049 | 0.0032 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4505 | 0.1030 | 0.1953 | 0.1010 | 0.0781 | 0.0390 | 0.0256 | 0.0044 | 0.0032 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8022 | 0.0251 | 0.1727 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3191 | 0.1447 | 0.1799 | 0.0898 | 0.1191 | 0.0467 | 0.0694 | 0.0313 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6670 | 0.1257 | 0.0716 | 0.0936 | 0.0306 | 0.0115 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.6131 | 0.1116 | 0.2324 | 0.0130 | 0.0299 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7470 | 0.0797 | 0.0484 | 0.1249 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7718 | 0.1342 | 0.0940 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7695 | 0.0896 | 0.1410 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6153 | 0.1799 | 0.0688 | 0.1098 | 0.0262 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6649 | 0.1601 | 0.1175 | 0.0575 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.6324 | 0.2705 | 0.0970 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6111 | 0.0749 | 0.1489 | 0.1245 | 0.0132 | 0.0274 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6224 | 0.0756 | 0.2601 | 0.0144 | 0.0274 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7795 | 0.0448 | 0.1757 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.4346 | 0.1068 | 0.1137 | 0.1947 | 0.0657 | 0.0496 | 0.0234 | 0.0107 | 0.0000 | 0.0008 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4182 | 0.0981 | 0.1878 | 0.1259 | 0.0942 | 0.0459 | 0.0287 | 0.0000 | 0.0013 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.7929 | 0.0251 | 0.1821 | |||||||
硝酸盐 Nitrate(1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5398 | 0.4523 | 0.0079 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9793 | 0.0207 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.7515 | 0.1998 | 0.0485 | 0.0000 | 0.0002 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8524 | 0.1461 | 0.0015 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.8451 | 0.1546 | 0.0003 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.8287 | 0.1641 | 0.0068 | 0.0000 | 0.0003 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9262 | 0.0711 | 0.0020 | 0.0004 | 0.0004 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8445 | 0.1554 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8995 | 0.0979 | 0.0026 | ||||||||
| M_gly_085 | 1 | 1 | 0.9867 | 0.0133 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.7233 | 0.2029 | 0.0583 | 0.0103 | 0.0041 | 0.0011 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.8226 | 0.1686 | 0.0083 | 0.0002 | 0.0002 | 0.0002 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9312 | 0.0618 | 0.0010 | 0.0054 | 0.0005 | 0.0000 | 0.0001 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9350 | 0.0645 | 0.0003 | 0.0002 | 0.0001 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.4759 | 0.3053 | 0.1717 | 0.0000 | 0.0246 | 0.0083 | 0.0142 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.2194 | 0.0498 | 0.1072 | 0.1872 | 0.3795 | 0.0569 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6946 | 0.2732 | 0.0322 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.7313 | 0.2028 | 0.0503 | 0.0086 | 0.0027 | 0.0042 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.8263 | 0.1619 | 0.0072 | 0.0032 | 0.0014 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8884 | 0.1044 | 0.0041 | 0.0009 | 0.0001 | 0.0007 | 0.0005 | 0.0005 | 0.0003 | 0.0002 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8961 | 0.0939 | 0.0041 | 0.0018 | 0.0006 | 0.0008 | 0.0007 | 0.0015 | 0.0004 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9771 | 0.0229 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7523 | 0.0947 | 0.0332 | 0.0085 | 0.0215 | 0.0207 | 0.0405 | 0.0286 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.8748 | 0.1149 | 0.0000 | 0.0042 | 0.0045 | 0.0016 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9366 | 0.0612 | 0.0011 | 0.0000 | 0.0010 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9605 | 0.0391 | 0.0000 | 0.0004 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9810 | 0.0190 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9667 | 0.0304 | 0.0029 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.7489 | 0.1979 | 0.0517 | 0.0007 | 0.0008 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8435 | 0.1479 | 0.0086 | 0.0000 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.7463 | 0.2252 | 0.0285 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5218 | 0.4572 | 0.0200 | 0.0009 | 0.0000 | 0.0000 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9458 | 0.0511 | 0.0027 | 0.0003 | 0.0002 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.5866 | 0.4116 | 0.0018 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8800 | 0.1030 | 0.0044 | 0.0000 | 0.0011 | 0.0004 | 0.0006 | 0.0006 | 0.0031 | 0.0068 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8890 | 0.0969 | 0.0014 | 0.0011 | 0.0011 | 0.0014 | 0.0002 | 0.0047 | 0.0042 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9779 | 0.0220 | 0.0001 | |||||||
| Fe (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5273 | 0.4640 | 0.0088 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9764 | 0.0230 | 0.0006 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.3476 | 0.4571 | 0.1910 | 0.0043 | 0.0000 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5600 | 0.4314 | 0.0086 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.5890 | 0.4087 | 0.0023 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5616 | 0.4186 | 0.0195 | 0.0002 | 0.0000 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.8990 | 0.0986 | 0.0020 | 0.0000 | 0.0004 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6891 | 0.3048 | 0.0062 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7664 | 0.2334 | 0.0002 | ||||||||
| M_gly_085 | 1 | 1 | 0.9934 | 0.0066 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.3508 | 0.4485 | 0.1854 | 0.0134 | 0.0019 | 0.0000 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5499 | 0.4265 | 0.0229 | 0.0000 | 0.0007 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9468 | 0.0484 | 0.0017 | 0.0029 | 0.0000 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9339 | 0.0657 | 0.0001 | 0.0001 | 0.0002 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/ 1,2,3&4,3,2 | 0.3053 | 0.4567 | 0.2213 | 0.0123 | 0.0005 | 0.0025 | 0.0014 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.4979 | 0.4162 | 0.0247 | 0.0080 | 0.0057 | 0.0474 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8733 | 0.1267 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3424 | 0.4536 | 0.1957 | 0.0079 | 0.0000 | 0.0004 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.5530 | 0.4297 | 0.0165 | 0.0000 | 0.0008 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.8883 | 0.1064 | 0.0045 | 0.0000 | 0.0002 | 0.0001 | 0.0001 | 0.0002 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8976 | 0.0996 | 0.0021 | 0.0002 | 0.0000 | 0.0003 | 0.0001 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9859 | 0.0141 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7505 | 0.0973 | 0.0400 | 0.0097 | 0.0295 | 0.0126 | 0.0372 | 0.0232 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6058 | 0.3772 | 0.0109 | 0.0043 | 0.0011 | 0.0008 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.8992 | 0.0964 | 0.0039 | 0.0002 | 0.0004 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9242 | 0.0736 | 0.0021 | 0.0000 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9848 | 0.0152 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9275 | 0.0696 | 0.0029 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3373 | 0.4592 | 0.2014 | 0.0021 | 0.0000 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5589 | 0.4272 | 0.0135 | 0.0004 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.5019 | 0.4845 | 0.0136 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5165 | 0.4582 | 0.0251 | 0.0000 | 0.0000 | 0.0002 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9413 | 0.0551 | 0.0033 | 0.0000 | 0.0003 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.5793 | 0.4195 | 0.0012 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8856 | 0.1102 | 0.0031 | 0.0000 | 0.0005 | 0.0005 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8974 | 0.0979 | 0.0021 | 0.0018 | 0.0000 | 0.0005 | 0.0002 | 0.0000 | 0.0000 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9857 | 0.0143 | 0.0000 | |||||||
延胡索酸 Fumarate (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5836 | 0.4087 | 0.0077 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9776 | 0.0224 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.9250 | 0.0653 | 0.0090 | 0.0000 | 0.0008 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9468 | 0.0532 | 0.0000 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9549 | 0.0441 | 0.0009 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.9232 | 0.0758 | 0.0000 | 0.0009 | 0.0000 | 0.0001 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9509 | 0.0490 | 0.0000 | 0.0000 | 0.0000 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.9143 | 0.0857 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8501 | 0.1485 | 0.0014 | ||||||||
| M_gly_085 | 1 | 1 | 0.9928 | 0.0072 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.8896 | 0.0983 | 0.0109 | 0.0000 | 0.0009 | 0.0000 | 0.0003 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.9197 | 0.0794 | 0.0003 | 0.0000 | 0.0005 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9294 | 0.0674 | 0.0030 | 0.0000 | 0.0000 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9340 | 0.0657 | 0.0000 | 0.0000 | 0.0002 | 0.0001 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.7096 | 0.2448 | 0.0374 | 0.0065 | 0.0000 | 0.0000 | 0.0017 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.8280 | 0.1000 | 0.0134 | 0.0026 | 0.0091 | 0.0469 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8531 | 0.1469 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9177 | 0.0766 | 0.0000 | 0.0030 | 0.0014 | 0.0013 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.9272 | 0.0717 | 0.0000 | 0.0000 | 0.0011 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8943 | 0.0981 | 0.0068 | 0.0000 | 0.0000 | 0.0004 | 0.0001 | 0.0003 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8958 | 0.0995 | 0.0035 | 0.0002 | 0.0001 | 0.0005 | 0.0003 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9865 | 0.0135 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7512 | 0.0956 | 0.0420 | 0.0092 | 0.0288 | 0.0134 | 0.0346 | 0.0251 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.7781 | 0.0692 | 0.0009 | 0.1459 | 0.0060 | 0.0000 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9442 | 0.0556 | 0.0000 | 0.0000 | 0.0001 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9120 | 0.0857 | 0.0023 | 0.0000 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9798 | 0.0202 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9211 | 0.0789 | 0.0000 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9300 | 0.0609 | 0.0088 | 0.0000 | 0.0003 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9517 | 0.0464 | 0.0019 | 0.0000 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.8347 | 0.1521 | 0.0131 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5676 | 0.4133 | 0.0184 | 0.0000 | 0.0006 | 0.0001 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9485 | 0.0511 | 0.0000 | 0.0001 | 0.0003 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6328 | 0.3672 | 0.0000 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8977 | 0.1009 | 0.0000 | 0.0007 | 0.0006 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.9080 | 0.0882 | 0.0002 | 0.0018 | 0.0016 | 0.0000 | 0.0000 | 0.0000 | 0.0001 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9834 | 0.0166 | 0.0000 | |||||||
| DMSO (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5872 | 0.4050 | 0.0078 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9792 | 0.0208 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.9336 | 0.0586 | 0.0075 | 0.0002 | 0.0000 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9558 | 0.0440 | 0.0000 | 0.0002 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9664 | 0.0336 | 0.0000 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.9268 | 0.0723 | 0.0000 | 0.0009 | 0.0000 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9477 | 0.0500 | 0.0000 | 0.0023 | 0.0000 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8997 | 0.1003 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8728 | 0.1263 | 0.0010 | ||||||||
| M_gly_085 | 1 | 1 | 0.9910 | 0.0090 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.8904 | 0.0931 | 0.0158 | 0.0000 | 0.0000 | 0.0003 | 0.0003 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.9199 | 0.0786 | 0.0011 | 0.0002 | 0.0002 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9369 | 0.0608 | 0.0000 | 0.0016 | 0.0005 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9308 | 0.0688 | 0.0000 | 0.0000 | 0.0003 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.7104 | 0.2644 | 0.0183 | 0.0053 | 0.0013 | 0.0002 | 0.0000 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3&4,3,2 | 0.8848 | 0.0838 | 0.0017 | 0.0073 | 0.0019 | 0.0205 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8678 | 0.1322 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9140 | 0.0749 | 0.0080 | 0.0024 | 0.0000 | 0.0007 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.9399 | 0.0595 | 0.0005 | 0.0000 | 0.0000 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8961 | 0.0999 | 0.0018 | 0.0016 | 0.0000 | 0.0001 | 0.0003 | 0.0001 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8960 | 0.0987 | 0.0045 | 0.0000 | 0.0003 | 0.0002 | 0.0002 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9835 | 0.0165 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7579 | 0.1032 | 0.0376 | 0.0077 | 0.0256 | 0.0115 | 0.0368 | 0.0197 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6492 | 0.0545 | 0.0009 | 0.2896 | 0.0058 | 0.0000 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9422 | 0.0553 | 0.0000 | 0.0024 | 0.0001 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9195 | 0.0803 | 0.0000 | 0.0002 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9858 | 0.0142 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9235 | 0.0728 | 0.0036 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9335 | 0.0616 | 0.0039 | 0.0004 | 0.0006 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9571 | 0.0400 | 0.0000 | 0.0028 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.8412 | 0.1503 | 0.0085 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5633 | 0.4172 | 0.0193 | 0.0000 | 0.0000 | 0.0002 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9400 | 0.0552 | 0.0048 | 0.0000 | 0.0000 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6248 | 0.3687 | 0.0065 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8903 | 0.1039 | 0.0056 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8916 | 0.1056 | 0.0000 | 0.0005 | 0.0016 | 0.0007 | 0.0000 | 0.0000 | 0.0001 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9859 | 0.0141 | 0.0000 | |||||||
(The symbols of fragments denote the cracking patterns of TBDMS-derivatized amino acids:(M-57)+,(M-85)+,(M-159)+ and f302 fragments)
片段 Fragments | 碳原子 C-atom | 前体 precursor | 前体的碳原子 C-atom of precursor | m | m+1 | m+2 | m+3 | m+4 | m+5 | m+6 | m+7 | m+8 | m+9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
硝酸盐 Nitrate (U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7771 | 0.0586 | 0.0285 | 0.1358 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.8027 | 0.0475 | 0.1497 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.6223 | 0.1759 | 0.1167 | 0.0677 | 0.0173 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6698 | 0.1857 | 0.1123 | 0.0321 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.7723 | 0.1108 | 0.1169 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5469 | 0.1780 | 0.1864 | 0.0622 | 0.0207 | 0.0059 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5910 | 0.1856 | 0.1753 | 0.0357 | 0.0124 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.7247 | 0.1880 | 0.0874 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8083 | 0.0612 | 0.1305 | ||||||||
| M_gly_085 | 1 | 1 | 0.8354 | 0.1646 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.5165 | 0.1481 | 0.2009 | 0.0912 | 0.0432 | 0.0000 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5553 | 0.1702 | 0.1861 | 0.0620 | 0.0202 | 0.0063 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.5395 | 0.0884 | 0.2743 | 0.0305 | 0.0650 | 0.0023 | 0.0000 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5507 | 0.1606 | 0.2028 | 0.0551 | 0.0254 | 0.0054 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.5194 | 0.1706 | 0.1497 | 0.0992 | 0.0416 | 0.0145 | 0.0049 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.5333 | 0.1854 | 0.1893 | 0.0636 | 0.0204 | 0.0080 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6538 | 0.2656 | 0.0807 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6959 | 0.2209 | 0.0353 | 0.0297 | 0.0137 | 0.0045 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.7233 | 0.2133 | 0.0389 | 0.0216 | 0.0030 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4987 | 0.1004 | 0.0907 | 0.1524 | 0.0706 | 0.0388 | 0.0302 | 0.0125 | 0.0032 | 0.0026 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.5004 | 0.0965 | 0.1771 | 0.0872 | 0.0709 | 0.0361 | 0.0240 | 0.0046 | 0.0031 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8280 | 0.0179 | 0.1541 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3803 | 0.1470 | 0.1487 | 0.0851 | 0.1094 | 0.0456 | 0.0586 | 0.0252 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5628 | 0.1626 | 0.1632 | 0.0812 | 0.0226 | 0.0076 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.6054 | 0.1808 | 0.1667 | 0.0343 | 0.0127 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7811 | 0.0469 | 0.0218 | 0.1502 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7845 | 0.0653 | 0.1501 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.8107 | 0.0185 | 0.1708 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6021 | 0.1815 | 0.1296 | 0.0636 | 0.0233 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6554 | 0.1878 | 0.1305 | 0.0262 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.6432 | 0.2531 | 0.1037 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6424 | 0.0767 | 0.1348 | 0.1113 | 0.0118 | 0.0229 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6590 | 0.0690 | 0.2307 | 0.0153 | 0.0260 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7932 | 0.0604 | 0.1464 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4878 | 0.1020 | 0.1013 | 0.1531 | 0.0730 | 0.0398 | 0.0269 | 0.0128 | 0.0032 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4889 | 0.1005 | 0.1861 | 0.0923 | 0.0695 | 0.0325 | 0.0252 | 0.0037 | 0.0014 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.8181 | 0.0223 | 0.1596 | |||||||
| Fe (U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.6653 | 0.0687 | 0.0481 | 0.2180 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.7027 | 0.0516 | 0.2457 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.5139 | 0.1980 | 0.0784 | 0.1563 | 0.0534 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5856 | 0.1580 | 0.1399 | 0.1165 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.6452 | 0.1865 | 0.1683 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.4265 | 0.1428 | 0.2328 | 0.1277 | 0.0408 | 0.0294 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.4863 | 0.1174 | 0.3028 | 0.0394 | 0.0541 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6145 | 0.2689 | 0.1166 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7073 | 0.1256 | 0.1672 | ||||||||
| M_gly_085 | 1 | 1 | 0.7732 | 0.2268 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.3812 | 0.1562 | 0.1741 | 0.1614 | 0.0538 | 0.0524 | 0.0208 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.4351 | 0.1340 | 0.2355 | 0.1232 | 0.0412 | 0.0311 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.3875 | 0.0690 | 0.3376 | 0.0590 | 0.1262 | 0.0072 | 0.0134 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.3941 | 0.1712 | 0.2546 | 0.1107 | 0.0524 | 0.0169 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.3937 | 0.1582 | 0.1451 | 0.1789 | 0.0628 | 0.0405 | 0.0209 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.4262 | 0.1373 | 0.2278 | 0.1236 | 0.0429 | 0.0421 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.5390 | 0.3428 | 0.1181 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3944 | 0.2637 | 0.1093 | 0.1351 | 0.0809 | 0.0167 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.4615 | 0.2480 | 0.1384 | 0.1233 | 0.0288 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.3504 | 0.0882 | 0.1024 | 0.1830 | 0.0992 | 0.0689 | 0.0587 | 0.0279 | 0.0108 | 0.0106 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.3535 | 0.0849 | 0.2022 | 0.1126 | 0.1116 | 0.0599 | 0.0527 | 0.0115 | 0.0110 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.7337 | 0.0211 | 0.2451 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.2751 | 0.1470 | 0.1734 | 0.1093 | 0.1352 | 0.0607 | 0.0709 | 0.0284 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5409 | 0.1403 | 0.1547 | 0.1071 | 0.0337 | 0.0233 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5292 | 0.0996 | 0.2819 | 0.0382 | 0.0511 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.6402 | 0.1096 | 0.0697 | 0.1805 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.6570 | 0.1600 | 0.1830 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7130 | 0.0548 | 0.2321 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.5067 | 0.1940 | 0.0814 | 0.1609 | 0.0571 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5781 | 0.1512 | 0.1539 | 0.1168 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.5460 | 0.2992 | 0.1549 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5007 | 0.0748 | 0.1878 | 0.1625 | 0.0208 | 0.0535 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.5210 | 0.0661 | 0.3278 | 0.0241 | 0.0611 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6948 | 0.0725 | 0.2327 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.3363 | 0.0843 | 0.1144 | 0.1806 | 0.1059 | 0.0691 | 0.0615 | 0.0273 | 0.0104 | 0.0102 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.3358 | 0.0837 | 0.2071 | 0.1196 | 0.1076 | 0.0653 | 0.0558 | 0.0121 | 0.0131 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.7298 | 0.0199 | 0.2504 | |||||||
延胡索酸 Fumarate(U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7549 | 0.0769 | 0.0532 | 0.1149 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.8079 | 0.0369 | 0.1553 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.8566 | 0.0824 | 0.0227 | 0.0326 | 0.0057 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8834 | 0.0649 | 0.0371 | 0.0146 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9028 | 0.0649 | 0.0322 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.7139 | 0.0853 | 0.1649 | 0.0262 | 0.0071 | 0.0027 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.7417 | 0.0774 | 0.1629 | 0.0098 | 0.0083 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8524 | 0.1177 | 0.0300 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8221 | 0.0534 | 0.1245 | ||||||||
| M_gly_085 | 1 | 1 | 0.8500 | 0.1500 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.6803 | 0.1019 | 0.1614 | 0.0377 | 0.0149 | 0.0038 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.6985 | 0.0910 | 0.1743 | 0.0255 | 0.0079 | 0.0027 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.5413 | 0.0739 | 0.2895 | 0.0299 | 0.0604 | 0.0006 | 0.0045 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5449 | 0.1569 | 0.2142 | 0.0551 | 0.0240 | 0.0048 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.6674 | 0.1133 | 0.1150 | 0.0846 | 0.0119 | 0.0062 | 0.0016 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3&4,3,2 | 0.6967 | 0.0906 | 0.1707 | 0.0279 | 0.0100 | 0.0040 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.7318 | 0.2435 | 0.0247 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6959 | 0.2209 | 0.0353 | 0.0297 | 0.0137 | 0.0045 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.7233 | 0.2133 | 0.0389 | 0.0216 | 0.0030 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4690 | 0.1018 | 0.1024 | 0.1654 | 0.0744 | 0.0383 | 0.0293 | 0.0131 | 0.0033 | 0.0030 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4716 | 0.1001 | 0.1908 | 0.0929 | 0.0788 | 0.0329 | 0.0263 | 0.0036 | 0.0029 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8166 | 0.0181 | 0.1653 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3993 | 0.1696 | 0.1489 | 0.0949 | 0.0984 | 0.0384 | 0.0504 | |||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6515 | 0.0727 | 0.1395 | 0.1168 | 0.0162 | 0.0033 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.7504 | 0.0787 | 0.1576 | 0.0060 | 0.0073 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7535 | 0.0701 | 0.0430 | 0.1334 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7630 | 0.1045 | 0.1325 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7998 | 0.0287 | 0.1715 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.8495 | 0.0858 | 0.0229 | 0.0346 | 0.0072 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8791 | 0.0609 | 0.0440 | 0.0160 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.7799 | 0.1869 | 0.0332 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6138 | 0.0911 | 0.1594 | 0.1045 | 0.0120 | 0.0193 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6486 | 0.0641 | 0.2490 | 0.0122 | 0.0261 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7650 | 0.0993 | 0.1357 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.4588 | 0.1002 | 0.1068 | 0.1695 | 0.0761 | 0.0369 | 0.0308 | 0.0138 | 0.0029 | 0.0043 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4680 | 0.0996 | 0.1933 | 0.0916 | 0.0816 | 0.0338 | 0.0243 | 0.0042 | 0.0037 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.8064 | 0.0214 | 0.1722 | |||||||
| DMSO(U-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.7596 | 0.0505 | 0.0361 | 0.1538 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.7778 | 0.0509 | 0.1713 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.6192 | 0.1729 | 0.0738 | 0.1092 | 0.0249 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6709 | 0.1582 | 0.1114 | 0.0596 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.7382 | 0.1339 | 0.1279 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5272 | 0.1559 | 0.2052 | 0.0792 | 0.0224 | 0.0102 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.5823 | 0.1361 | 0.2267 | 0.0291 | 0.0258 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6949 | 0.2244 | 0.0807 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7738 | 0.0761 | 0.1501 | ||||||||
| M_gly_085 | 1 | 1 | 0.8112 | 0.1888 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.5227 | 0.1609 | 0.1080 | 0.1325 | 0.0357 | 0.0329 | 0.0073 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.5169 | 0.1612 | 0.2063 | 0.0689 | 0.0355 | 0.0112 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.7984 | 0.0361 | 0.1654 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.5000 | 0.1680 | 0.1581 | 0.1163 | 0.0465 | 0.0112 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5293 | 0.1528 | 0.2062 | 0.0789 | 0.0209 | 0.0119 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.4933 | 0.1014 | 0.3028 | 0.0354 | 0.0606 | 0.0064 | 0.0000 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.5086 | 0.1741 | 0.2230 | 0.0623 | 0.0258 | 0.0061 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.5194 | 0.1706 | 0.1497 | 0.0992 | 0.0416 | 0.0145 | 0.0049 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.5333 | 0.1854 | 0.1893 | 0.0636 | 0.0204 | 0.0080 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6538 | 0.2656 | 0.0807 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.5209 | 0.2351 | 0.0858 | 0.1153 | 0.0357 | 0.0073 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.5631 | 0.2369 | 0.1186 | 0.0545 | 0.0269 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.4415 | 0.1047 | 0.1088 | 0.1722 | 0.0743 | 0.0451 | 0.0320 | 0.0133 | 0.0049 | 0.0032 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4505 | 0.1030 | 0.1953 | 0.1010 | 0.0781 | 0.0390 | 0.0256 | 0.0044 | 0.0032 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.8022 | 0.0251 | 0.1727 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.3191 | 0.1447 | 0.1799 | 0.0898 | 0.1191 | 0.0467 | 0.0694 | 0.0313 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6670 | 0.1257 | 0.0716 | 0.0936 | 0.0306 | 0.0115 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.6131 | 0.1116 | 0.2324 | 0.0130 | 0.0299 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.7470 | 0.0797 | 0.0484 | 0.1249 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.7718 | 0.1342 | 0.0940 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.7695 | 0.0896 | 0.1410 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.6153 | 0.1799 | 0.0688 | 0.1098 | 0.0262 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.6649 | 0.1601 | 0.1175 | 0.0575 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.6324 | 0.2705 | 0.0970 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.6111 | 0.0749 | 0.1489 | 0.1245 | 0.0132 | 0.0274 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.6224 | 0.0756 | 0.2601 | 0.0144 | 0.0274 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.7795 | 0.0448 | 0.1757 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.4346 | 0.1068 | 0.1137 | 0.1947 | 0.0657 | 0.0496 | 0.0234 | 0.0107 | 0.0000 | 0.0008 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.4182 | 0.0981 | 0.1878 | 0.1259 | 0.0942 | 0.0459 | 0.0287 | 0.0000 | 0.0013 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.7929 | 0.0251 | 0.1821 | |||||||
硝酸盐 Nitrate(1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5398 | 0.4523 | 0.0079 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9793 | 0.0207 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.7515 | 0.1998 | 0.0485 | 0.0000 | 0.0002 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8524 | 0.1461 | 0.0015 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.8451 | 0.1546 | 0.0003 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.8287 | 0.1641 | 0.0068 | 0.0000 | 0.0003 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9262 | 0.0711 | 0.0020 | 0.0004 | 0.0004 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8445 | 0.1554 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8995 | 0.0979 | 0.0026 | ||||||||
| M_gly_085 | 1 | 1 | 0.9867 | 0.0133 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.7233 | 0.2029 | 0.0583 | 0.0103 | 0.0041 | 0.0011 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.8226 | 0.1686 | 0.0083 | 0.0002 | 0.0002 | 0.0002 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9312 | 0.0618 | 0.0010 | 0.0054 | 0.0005 | 0.0000 | 0.0001 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9350 | 0.0645 | 0.0003 | 0.0002 | 0.0001 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.4759 | 0.3053 | 0.1717 | 0.0000 | 0.0246 | 0.0083 | 0.0142 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.2194 | 0.0498 | 0.1072 | 0.1872 | 0.3795 | 0.0569 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.6946 | 0.2732 | 0.0322 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.7313 | 0.2028 | 0.0503 | 0.0086 | 0.0027 | 0.0042 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.8263 | 0.1619 | 0.0072 | 0.0032 | 0.0014 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8884 | 0.1044 | 0.0041 | 0.0009 | 0.0001 | 0.0007 | 0.0005 | 0.0005 | 0.0003 | 0.0002 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8961 | 0.0939 | 0.0041 | 0.0018 | 0.0006 | 0.0008 | 0.0007 | 0.0015 | 0.0004 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9771 | 0.0229 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7523 | 0.0947 | 0.0332 | 0.0085 | 0.0215 | 0.0207 | 0.0405 | 0.0286 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.8748 | 0.1149 | 0.0000 | 0.0042 | 0.0045 | 0.0016 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9366 | 0.0612 | 0.0011 | 0.0000 | 0.0010 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9605 | 0.0391 | 0.0000 | 0.0004 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9810 | 0.0190 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9667 | 0.0304 | 0.0029 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.7489 | 0.1979 | 0.0517 | 0.0007 | 0.0008 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.8435 | 0.1479 | 0.0086 | 0.0000 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.7463 | 0.2252 | 0.0285 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5218 | 0.4572 | 0.0200 | 0.0009 | 0.0000 | 0.0000 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9458 | 0.0511 | 0.0027 | 0.0003 | 0.0002 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.5866 | 0.4116 | 0.0018 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8800 | 0.1030 | 0.0044 | 0.0000 | 0.0011 | 0.0004 | 0.0006 | 0.0006 | 0.0031 | 0.0068 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8890 | 0.0969 | 0.0014 | 0.0011 | 0.0011 | 0.0014 | 0.0002 | 0.0047 | 0.0042 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9779 | 0.0220 | 0.0001 | |||||||
| Fe (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5273 | 0.4640 | 0.0088 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9764 | 0.0230 | 0.0006 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.3476 | 0.4571 | 0.1910 | 0.0043 | 0.0000 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5600 | 0.4314 | 0.0086 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.5890 | 0.4087 | 0.0023 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.5616 | 0.4186 | 0.0195 | 0.0002 | 0.0000 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.8990 | 0.0986 | 0.0020 | 0.0000 | 0.0004 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.6891 | 0.3048 | 0.0062 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.7664 | 0.2334 | 0.0002 | ||||||||
| M_gly_085 | 1 | 1 | 0.9934 | 0.0066 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.3508 | 0.4485 | 0.1854 | 0.0134 | 0.0019 | 0.0000 | 0.0000 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.5499 | 0.4265 | 0.0229 | 0.0000 | 0.0007 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9468 | 0.0484 | 0.0017 | 0.0029 | 0.0000 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9339 | 0.0657 | 0.0001 | 0.0001 | 0.0002 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/ 1,2,3&4,3,2 | 0.3053 | 0.4567 | 0.2213 | 0.0123 | 0.0005 | 0.0025 | 0.0014 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.4979 | 0.4162 | 0.0247 | 0.0080 | 0.0057 | 0.0474 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8733 | 0.1267 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3424 | 0.4536 | 0.1957 | 0.0079 | 0.0000 | 0.0004 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.5530 | 0.4297 | 0.0165 | 0.0000 | 0.0008 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3& 1, 2, 3, 4 | 0.8883 | 0.1064 | 0.0045 | 0.0000 | 0.0002 | 0.0001 | 0.0001 | 0.0002 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8976 | 0.0996 | 0.0021 | 0.0002 | 0.0000 | 0.0003 | 0.0001 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9859 | 0.0141 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7505 | 0.0973 | 0.0400 | 0.0097 | 0.0295 | 0.0126 | 0.0372 | 0.0232 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6058 | 0.3772 | 0.0109 | 0.0043 | 0.0011 | 0.0008 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.8992 | 0.0964 | 0.0039 | 0.0002 | 0.0004 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9242 | 0.0736 | 0.0021 | 0.0000 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9848 | 0.0152 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9275 | 0.0696 | 0.0029 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.3373 | 0.4592 | 0.2014 | 0.0021 | 0.0000 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.5589 | 0.4272 | 0.0135 | 0.0004 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.5019 | 0.4845 | 0.0136 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5165 | 0.4582 | 0.0251 | 0.0000 | 0.0000 | 0.0002 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9413 | 0.0551 | 0.0033 | 0.0000 | 0.0003 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.5793 | 0.4195 | 0.0012 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8856 | 0.1102 | 0.0031 | 0.0000 | 0.0005 | 0.0005 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8974 | 0.0979 | 0.0021 | 0.0018 | 0.0000 | 0.0005 | 0.0002 | 0.0000 | 0.0000 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9857 | 0.0143 | 0.0000 | |||||||
延胡索酸 Fumarate (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5836 | 0.4087 | 0.0077 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9776 | 0.0224 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.9250 | 0.0653 | 0.0090 | 0.0000 | 0.0008 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9468 | 0.0532 | 0.0000 | 0.0000 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9549 | 0.0441 | 0.0009 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.9232 | 0.0758 | 0.0000 | 0.0009 | 0.0000 | 0.0001 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9509 | 0.0490 | 0.0000 | 0.0000 | 0.0000 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.9143 | 0.0857 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8501 | 0.1485 | 0.0014 | ||||||||
| M_gly_085 | 1 | 1 | 0.9928 | 0.0072 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.8896 | 0.0983 | 0.0109 | 0.0000 | 0.0009 | 0.0000 | 0.0003 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.9197 | 0.0794 | 0.0003 | 0.0000 | 0.0005 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9294 | 0.0674 | 0.0030 | 0.0000 | 0.0000 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9340 | 0.0657 | 0.0000 | 0.0000 | 0.0002 | 0.0001 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.7096 | 0.2448 | 0.0374 | 0.0065 | 0.0000 | 0.0000 | 0.0017 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3& 4,3,2 | 0.8280 | 0.1000 | 0.0134 | 0.0026 | 0.0091 | 0.0469 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8531 | 0.1469 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9177 | 0.0766 | 0.0000 | 0.0030 | 0.0014 | 0.0013 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.9272 | 0.0717 | 0.0000 | 0.0000 | 0.0011 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8943 | 0.0981 | 0.0068 | 0.0000 | 0.0000 | 0.0004 | 0.0001 | 0.0003 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8958 | 0.0995 | 0.0035 | 0.0002 | 0.0001 | 0.0005 | 0.0003 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9865 | 0.0135 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7512 | 0.0956 | 0.0420 | 0.0092 | 0.0288 | 0.0134 | 0.0346 | 0.0251 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.7781 | 0.0692 | 0.0009 | 0.1459 | 0.0060 | 0.0000 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9442 | 0.0556 | 0.0000 | 0.0000 | 0.0001 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9120 | 0.0857 | 0.0023 | 0.0000 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9798 | 0.0202 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9211 | 0.0789 | 0.0000 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9300 | 0.0609 | 0.0088 | 0.0000 | 0.0003 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9517 | 0.0464 | 0.0019 | 0.0000 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.8347 | 0.1521 | 0.0131 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5676 | 0.4133 | 0.0184 | 0.0000 | 0.0006 | 0.0001 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9485 | 0.0511 | 0.0000 | 0.0001 | 0.0003 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6328 | 0.3672 | 0.0000 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8977 | 0.1009 | 0.0000 | 0.0007 | 0.0006 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.9080 | 0.0882 | 0.0002 | 0.0018 | 0.0016 | 0.0000 | 0.0000 | 0.0000 | 0.0001 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9834 | 0.0166 | 0.0000 | |||||||
| DMSO (1-13C) | |||||||||||||
| M_ala_057 | 1, 2, 3 | PYR | 1, 2, 3 | 0.5872 | 0.4050 | 0.0078 | 0.0000 | ||||||
| M_ala_085 | 2, 3 | PYR | 2, 3 | 0.9792 | 0.0208 | 0.0000 | |||||||
| M_asx_057 | 1, 2, 3, 4 | OAA | 1, 2, 3, 4 | 0.9336 | 0.0586 | 0.0075 | 0.0002 | 0.0000 | |||||
| M_asx_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9558 | 0.0440 | 0.0000 | 0.0002 | ||||||
| M_asx_302 | 1, 2 | OAA | 1, 2 | 0.9664 | 0.0336 | 0.0000 | |||||||
| M_glx_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.9268 | 0.0723 | 0.0000 | 0.0009 | 0.0000 | 0.0000 | ||||
| M_glx_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9477 | 0.0500 | 0.0000 | 0.0023 | 0.0000 | |||||
| M_glx_302 | 1, 2 | AKG | 1, 2 | 0.8997 | 0.1003 | 0.0000 | |||||||
| M_gly_057 | 1, 2 | 1, 2 | 0.8728 | 0.1263 | 0.0010 | ||||||||
| M_gly_085 | 1 | 1 | 0.9910 | 0.0090 | |||||||||
| M_his_057 | 1, 2, 3, 4, 5, 6 | R5P | 5, 4, 3, 2, 1 | 0.4425 | 0.4269 | 0.1183 | 0.0116 | 0.0002 | 0.0005 | 0.0000 | |||
| M_his_159 | 2, 3, 4, 5, 6 | R5P | 4, 3, 2, 1 | 0.4720 | 0.4370 | 0.0742 | 0.0107 | 0.0044 | 0.0017 | ||||
| M_his_302 | 1, 2 | R5P | 5, 4 | 0.8912 | 0.1088 | 0.0000 | |||||||
| M_ile_015 | 1, 2, 3, 4, 5, 6 | OAA&PYR | 1,2,3,4&2,3 | 0.8904 | 0.0931 | 0.0158 | 0.0000 | 0.0000 | 0.0003 | 0.0003 | |||
| M_ile_085 | 2, 3, 4, 5, 6 | OAA&PYR | 2,3,4&2,3 | 0.9199 | 0.0786 | 0.0011 | 0.0002 | 0.0002 | 0.0000 | ||||
| M_leu_015 | 1, 2, 3, 4, 5, 6 | AcoA&PYR | 1,2&2,2,3,3 | 0.9369 | 0.0608 | 0.0000 | 0.0016 | 0.0005 | 0.0000 | 0.0002 | |||
| M_leu_085 | 2, 3, 4, 5, 6 | AcoA&PYR | 2&2,2,3,3 | 0.9308 | 0.0688 | 0.0000 | 0.0000 | 0.0003 | 0.0000 | ||||
| M_lys_057 | 1, 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 1,2,3,4&3,2/1,2,3&4,3,2 | 0.7104 | 0.2644 | 0.0183 | 0.0053 | 0.0013 | 0.0002 | 0.0000 | |||
| M_lys_159 | 2, 3, 4, 5, 6 | OAA&PYR/PYR&OAA | 2,3,4&3,2/2,3&4,3,2 | 0.8848 | 0.0838 | 0.0017 | 0.0073 | 0.0019 | 0.0205 | ||||
| M_lys_302 | 1,2 | OAA/PYR | 1,2/1,2 | 0.8678 | 0.1322 | 0.0000 | |||||||
| M_met_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9140 | 0.0749 | 0.0080 | 0.0024 | 0.0000 | 0.0007 | ||||
| M_met_085 | 2,3,4 | OAA | 2,3,4 | 0.9399 | 0.0595 | 0.0005 | 0.0000 | 0.0000 | |||||
| M_phe_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8961 | 0.0999 | 0.0018 | 0.0016 | 0.0000 | 0.0001 | 0.0003 | 0.0001 | 0.0000 | 0.0000 |
| M_phe_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8960 | 0.0987 | 0.0045 | 0.0000 | 0.0003 | 0.0002 | 0.0002 | 0.0001 | 0.0000 | |
| M_phe_302 | 1, 2 | PEP | 1, 2 | 0.9835 | 0.0165 | 0.0000 | |||||||
| M_phe_sc | 3,4,5,6,7,8,9 | PEP&E4P | 3,2,3&1,2,3,4 | 0.7579 | 0.1032 | 0.0376 | 0.0077 | 0.0256 | 0.0115 | 0.0368 | 0.0197 | ||
| M_pro_057 | 1, 2, 3, 4, 5 | AKG | 1, 2, 3, 4, 5 | 0.6492 | 0.0545 | 0.0009 | 0.2896 | 0.0058 | 0.0000 | ||||
| M_pro_085 | 2, 3, 4, 5 | AKG | 2, 3, 4, 5 | 0.9422 | 0.0553 | 0.0000 | 0.0024 | 0.0001 | |||||
| M_ser_057 | 1, 2, 3 | 1, 2, 3 | 0.9195 | 0.0803 | 0.0000 | 0.0002 | |||||||
| M_ser_085 | 2, 3 | 2, 3 | 0.9858 | 0.0142 | 0.0000 | ||||||||
| M_ser_302 | 1, 2 | 1, 2 | 0.9235 | 0.0728 | 0.0036 | ||||||||
| M_thr_057 | 1,2,3,4 | OAA | 1,2,3,4 | 0.9335 | 0.0616 | 0.0039 | 0.0004 | 0.0006 | |||||
| M_thr_085 | 2, 3, 4 | OAA | 2, 3, 4 | 0.9571 | 0.0400 | 0.0000 | 0.0028 | ||||||
| M_thr_sc | 3,4 | OAA | 3,4 | 0.8412 | 0.1503 | 0.0085 | |||||||
| M_val_057 | 1, 2, 3, 4, 5 | PYR | 1, 2, 2, 3, 3 | 0.5633 | 0.4172 | 0.0193 | 0.0000 | 0.0000 | 0.0002 | ||||
| M_val_085 | 2, 3, 4, 5 | PYR | 2, 2, 3, 3 | 0.9400 | 0.0552 | 0.0048 | 0.0000 | 0.0000 | |||||
| M_val_302 | 1, 2 | PYR | 1, 2 | 0.6248 | 0.3687 | 0.0065 | |||||||
| M_tyr_057 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 1, 2, 3, 2, 3&1, 2, 3, 4 | 0.8903 | 0.1039 | 0.0056 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| M_tyr_085 | 2, 3, 4, 5, 6, 7, 8, 9 | PEP&E4P | 2, 3, 2, 3&1, 2, 3, 4 | 0.8916 | 0.1056 | 0.0000 | 0.0005 | 0.0016 | 0.0007 | 0.0000 | 0.0000 | 0.0001 | |
| M_tyr_302 | 1, 2 | PEP | 1, 2 | 0.9859 | 0.0141 | 0.0000 | |||||||
| 17 | MYERS C R, MYERS J M. Fumarate reductase is a soluble enzyme in anaerobically grown Shewanella putrefaciens MR-1[J]. FEMS Microbiology Letters, 1992, 98(1/2/3): 13-19. |
| 18 | ZHAO J S, MANNO D, BEAULIEU C, et al. Shewanella sediminis sp nov., a novel Na+-requiring and hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading bacterium from marine sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(Pt 4): 1511-1520. |
| 19 | ZHAO J S, MANNO D, THIBOUTOT S, et al. Shewanella canadensis sp nov. and Shewanella atlantica sp. nov., manganese dioxide- and hexahydro-1,3,5-trinitro-1,3,5-triazine-reducing, psychrophilic marine bacteria[J]. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(9): 2155-2162. |
| 20 | HAU H H, GRALNICK J A. Ecology and biotechnology of the genus Shewanella [J]. Annual Review of Microbiology, 2007, 61: 237-258. |
| 21 | 肖翔, 吴勇民, 徐灿灿, 等. 金属还原菌希瓦氏菌厌氧呼吸能力及其在环境修复中的研究进展[J]. 微生物学通报, 2012, 39(11): 1677-1686. |
| XIAO X, WU Y M, XU C C, et al. Anaerobic respiratory capabilities of a metal-reducing microorganism Shewanella and its application in environmental remediation[J]. Microbiology China, 2012, 39(11): 1677-1686. | |
| 22 | TANG Y J J, HWANG J S, WEMMER D E, et al. Shewanella oneidensis MR-1 fluxome under various oxygen conditions[J]. Applied and Environmental Microbiology, 2007, 73(3): 718-729. |
| 23 | TANG Y J J, MARTIN H G, DEUTSCHBAUER A, et al. Invariability of central metabolic flux distribution in Shewanella oneidensis MR-1 under environmental or genetic perturbations[J]. Biotechnology Progress, 2009, 25(5): 1254-1259. |
| 24 | TANG Y J J, MARTIN H G, DEHAL P S, et al. Metabolic flux analysis of Shewanella spp. reveals evolutionary robustness in central carbon metabolism[J]. Biotechnology and Bioengineering, 2009, 102(4): 1161-1169. |
| 25 | TANG Y J J, MEADOWS A L, KIRBY J, et al. Anaerobic central metabolic pathways in Shewanella oneidensis MR-1 reinterpreted in the light of isotopic metabolite labeling[J]. Journal of Bacteriology, 2007, 189(3): 894-901. |
| 26 | STOOKEY L L. Ferrozine— a new spectrophotometric reagent for iron[J]. Analytical Chemistry, 1970, 42(7): 779-781. |
| 27 | ALEF K, KLEINER D. Rapid and sensitive determination of microbial activity in soils and in soil aggregates by dimethylsulfoxide reduction[J]. Biology and Fertility of Soils, 1989, 8(4): 349-355. |
| 28 | MASUKO T, MINAMI A, IWASAKI N, et al. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format[J]. Analytical Biochemistry, 2005, 339(1): 69-72. |
| 29 | ZAMBONI N, FENDT S M, RÜHL M, et al. 13C-based metabolic flux analysis[J]. Nature Protocols, 2009, 4(6): 878-892. |
| 30 | PINCHUK G E, HILL E A, GEYDEBREKHT O V, et al. Constraint-based model of Shewanella oneidensis MR-1 metabolism: a tool for data analysis and hypothesis generation[J]. PLoS Computational Biology, 2010, 6(6): e1000822. |
| 31 | NANCHEN A, FUHRER T, SAUER U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data[M/OL]//Methods in molecular biology: metabolomics. Totowa, NJ, USA: Humana Press. 2007, 358: 177-197 [2022-09-01]. . |
| 32 | AYYASAMY P M, CHUN S H, LEE S H. Desorption and dissolution of heavy metals from contaminated soil using Shewanella sp. (HN-41) amended with various carbon sources and synthetic soil organic matters[J]. Journal of Hazardous Materials, 2009, 161(2/3): 1095-1102. |
| 33 | ONG W K, VU T T, LOVENDAHL K N, et al. Comparisons of Shewanella strains based on genome annotations, modeling, and experiments[J]. BMC Systems Biology, 2014, 8: 31. |
| 34 | CRUZ-GARCÍA C, MURRAY A E, KLAPPENBACH J A, et al. Respiratory nitrate ammonification by Shewanella oneidensis MR-1[J]. Journal of Bacteriology, 2007, 189(2): 656-662. |
| 35 | INOHANA Y, KATSUYA S, KOGA R, et al. Shewanella algae relatives capable of generating electricity from acetate contribute to coastal-sediment microbial fuel cells treating complex organic matter[J]. Microbes and Environments, 2020, 35(2): ME19161. |
| 36 | REWATKAR P, GOEL S. Shewanella putrefaciens powered microfluidic microbial fuel cell with printed circuit board electrodes and soft-lithographic microchannel[J]. Chemosphere, 2022, 286: 131855. |
| 37 | DAI H N, DUONG NGUYEN T A, LE L P MY, et al. Power generation of Shewanella oneidensis MR-1 microbial fuel cells in bamboo fermentation effluent[J]. International Journal of Hydrogen Energy, 2021, 46(31): 16612-16621. |
| 38 | CHAUDHARY S, BANERJEE D K. Metal phase association of chromium in contaminated soils from an industrial area in Delhi[J]. Chemical Speciation & Bioavailability, 2004, 16(4): 145-151. |
| 39 | FREDRICKSON J K, ROMINE M F, BELIAEV A S, et al. Towards environmental systems biology of Shewanella [J]. Nature Reviews Microbiology, 2008, 6(8): 592-603. |
| 40 | LEMAIRE O N, MÉJEAN V, IOBBI-NIVOL C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems[J]. FEMS Microbiology Reviews, 2020, 44(2): 155-170. |
| 41 | FAN G, DUNDAS C M, GRAHAM A J, et al. Shewanella oneidensis as a living electrode for controlled radical polymerization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(18): 4559-4564. |
| 1 | MYERS C R, NEALSON K H. Bacterial Manganese reduction and growth with manganese oxide as the sole electron acceptor[J]. Science, 1988, 240(4857): 1319-1321. |
| 2 | LARSEN I, LITTLE B, NEALSON K H, et al. Manganite reduction by Shewanella putrefaciens MR-4[J]. American Mineralogist, 1998, 83(11/12): 1564-1572. |
| 3 | URRUTIA M M, RODEN E E, FREDRICKSON J K, et al. Microbial and surface chemistry controls on reduction of synthetic Fe(Ⅲ) oxide minerals by the dissimilatory iron-reducing bacterium Shewanella alga [J]. Geomicrobiology Journal, 1998, 15(4): 269-291. |
| 4 | CACCAVO F, BLAKEMORE R P, LOVLEY D R. A hydrogen-oxidizing, Fe(Ⅲ)-reducing microorganism from the Great Bay Estuary, New Hampshire[J]. Applied and Environmental Microbiology, 1992, 58(10): 3211-3216. |
| 5 | WIELINGA B, MIZUBA M M, HANSEL C M, et al. Iron promoted reduction of chromate by dissimilatory iron-reducing bacteria[J]. Environmental Science & Technology, 2001, 35(3): 522-527. |
| 6 | FARRENKOPF A M, DOLLHOPF M E, NI CHADHAIN S, et al. Reduction of iodate in seawater during Arabian Sea shipboard incubations and in laboratory cultures of the marine bacterium Shewanella putrefaciens strain MR-4[J]. Marine Chemistry, 1997, 57(3/4): 347-354. |
| 7 | WILDUNG R E, GORBY Y A, KRUPKA K M, et al. Effect of electron donor and solution chemistry on products of dissimilatory reduction of technetium by Shewanella putrefaciens [J]. Applied and Environmental Microbiology, 2000, 66(6): 2451-2460. |
| 8 | LLOYD J R, YONG P, MACASKIE L E. Biological reduction and removal of Np(Ⅴ) by two microorganisms[J]. Environmental Science & Technology, 2000, 34(7): 1297-1301. |
| 9 | SALTIKOV C W, WILDMAN R A JR, NEWMAN D K. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3[J]. Journal of Bacteriology, 2005, 187(21): 7390-7396. |
| 10 | BOUKHALFA H, ICOPINI G A, REILLY S D, et al. Plutonium(Ⅳ) reduction by the metal-reducing bacteria Geobacter metallireducens GS15 and Shewanella oneidensis MR1[J]. Applied and Environmental Microbiology, 2007, 73(18): 5897-5903. |
| 11 | KLONOWSKA A, HEULIN T, VERMEGLIO A. Selenite and tellurite reduction by Shewanella oneidensis [J]. Applied and Environmental Microbiology, 2005, 71(9): 5607-5609. |
| 12 | CARPENTIER W, SANDRA K, DE SMET I, et al. Microbial reduction and precipitation of vanadium by Shewanella oneidensis [J]. Applied and Environmental Microbiology, 2003, 69(6): 3636-3639. |
| 13 | PERRY K A, KOSTKA J E, LUTHER G W 3 RD,et al. Mediation of sulfur speciation by a Black Sea facultative anaerobe[J]. Science, 1993, 259(5096): 801-803. |
| 14 | KRAUSE B, NEALSON K H. Physiology and enzymology involved in denitrification by Shewanella putrefaciens [J]. Applied and Environmental Microbiology, 1997, 63(7): 2613-2618. |
| 15 | RINGØ E, STENBERG E, STRØM A R. Amino acid and lactate catabolism in trimethylamine oxide respiration of Alteromonas putrefaciens NCMB 1735[J]. Applied and Environmental Microbiology, 1984, 47(5): 1084-1089. |
| 16 | GRALNICK J A, VALI H, LIES D P, et al. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(12): 4669-4674. |
| [1] | 张成辛. 基于文本数据挖掘的蛋白功能预测的机遇与挑战[J]. 合成生物学, 2025, (): 1-14. |
| [2] | 李倩, James E. Ferrell, 陈于平. 细胞质浓度:细胞生物学的老问题、新参数[J]. 合成生物学, 2025, (): 1-18. |
| [3] | 姜百翼, 钱珑. 活细胞记录器在细胞谱系追踪中的应用和前景[J]. 合成生物学, 2025, (): 1-17. |
| [4] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [5] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [6] | 张日新, 田晓军. 合成基因回路面临的细胞‘经济学窘境’[J]. 合成生物学, 2025, (): 1-14. |
| [7] | 谭骁天, 李睿涵, 杨慧. 生物分子传感中的抗体探针:由碳基计算走向硅基计算[J]. 合成生物学, 2025, (): 1-9. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 刘建明, 张炽坚, 张冰, 曾安平. 巴氏梭菌作为工业底盘细胞高效生产1,3-丙二醇——从代谢工程和菌种进化到过程工程和产品分离[J]. 合成生物学, 2024, 5(6): 1386-1403. |
| [10] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [11] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [12] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2024, (): 1-21. |
| [13] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2024, (): 1-36. |
| [14] | 郑梦梦, 刘犇犇, 林芝, 瞿旭东. 重要甾体化合物的化学酶法合成研究进展[J]. 合成生物学, 2024, 5(5): 941-959. |
| [15] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
