合成生物学 ›› 2020, Vol. 1 ›› Issue (5): 583-592.DOI: 10.12211/2096-8280.2020-019
天然产物成药性的合成生物学改良
- 中国药科大学生命科学与技术学院化学生物学研究室,江苏 南京 211198
-
收稿日期:2020-03-08修回日期:2020-09-23出版日期:2020-10-31发布日期:2020-12-03 -
通讯作者:陈依军 -
作者简介:作者简介:王清(1994—),女,硕士研究生。研究方向:化学生物学。E-mail:499836857@qq.com
陈依军(1962—),男,教授,主要从事药物合成生物学研究。E-mail:yjchen@cpu.edu.cn -
基金资助:“重大新药创制”国家科技重大专项(2019ZX09721001-004-001);国家自然科学基金(21778076)
Synthetic biology approaches to improve druggability of natural products
- Laboratory of Chemical Biology,School of Life Science and Technology,China Pharmaceutical University,Nanjing 211198,Jiangsu,China
-
Received:2020-03-08Revised:2020-09-23Online:2020-10-31Published:2020-12-03 -
Contact:CHEN Yijun
摘要:
微生物和植物来源的天然产物结构复杂多样,具有抗感染、抗肿瘤、免疫抑制等多种活性,是现代临床治疗药物的重要来源之一。然而,大部分天然产物存在水溶性差、活性不强、结构类似物多以及可及性受限等问题,难以通过简单的化学修饰和改造解决,极大限制了天然产物的成药性及其后续研发。综合基因工程、代谢工程、基因组学、系统生物学、合成化学和计算生物学等学科的合成生物学,为改善天然产物的成药性提供了新机遇。本文针对限制天然产物成药的主要因素,概述了近年来利用合成生物学方法与策略在提高天然产物成药性方面取得的研究进展。通过理性分析天然产物的构效关系、挖掘合成和调控元件、构建系列反应模块和人工合成体系、筛选并优化底盘生物等策略,实现了多种天然产物来源药物或前体在“细胞工厂”中的定向、高效合成。与此同时,合成生物学技术也衍生了结构多样和性质改良的生物活性分子和潜在新药。随着合成生物学、药学和信息科学等方面的发展,可以预见提高和改善天然产物成药性的研究将会进入一个崭新的时代。
中图分类号:
引用本文
王清, 陈依军. 天然产物成药性的合成生物学改良[J]. 合成生物学, 2020, 1(5): 583-592.
WANG Qing, CHEN Yijun. Synthetic biology approaches to improve druggability of natural products[J]. Synthetic Biology Journal, 2020, 1(5): 583-592.
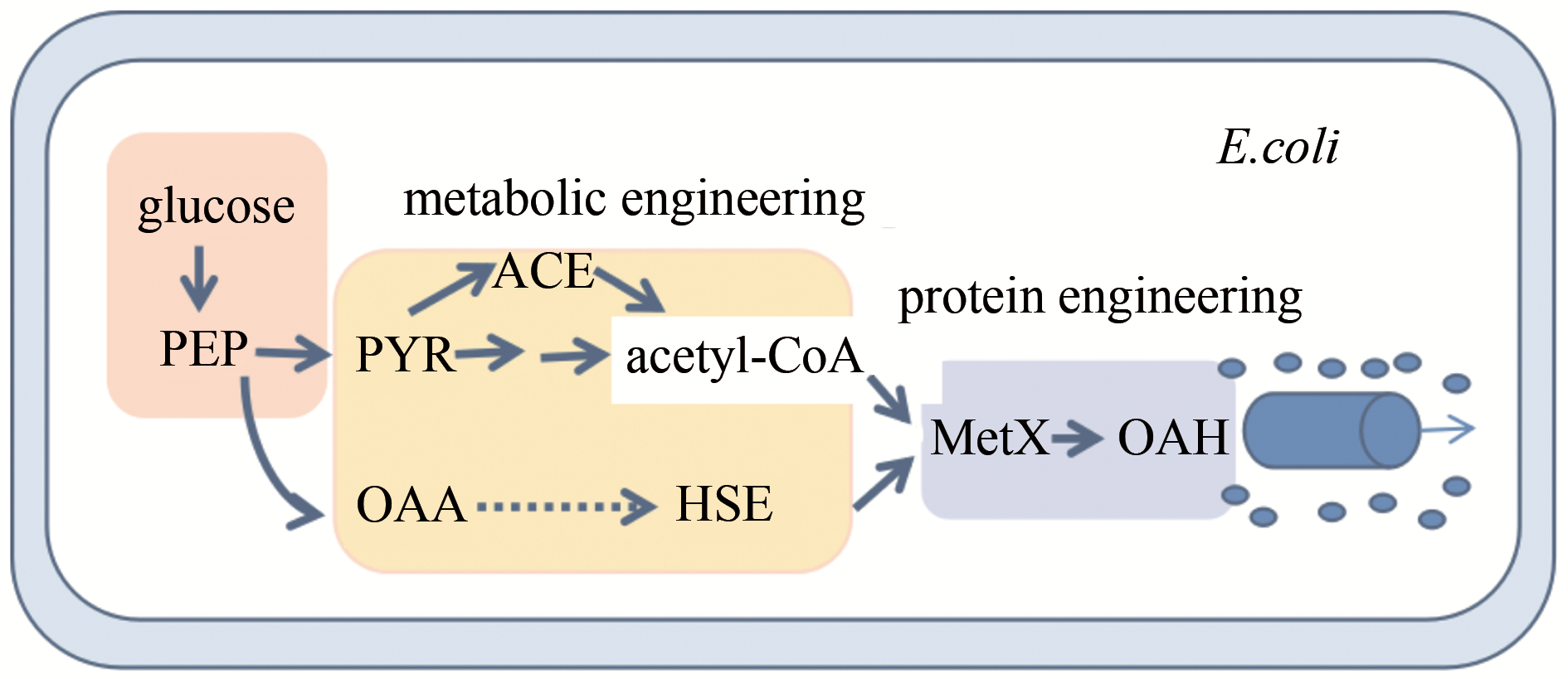
图4 O-乙酰高丝氨酸生物合成途径及改造PEP—磷酸烯醇丙酮酸;PYR—丙酮酸;OAA—草酰乙酸;ACE—醋酸盐;HSE—L-高丝氨酸;MetX—乙酰转移酶
Fig.4 Synthetic biology strategy for high level production of O-acetylhomoserine in Escherichia coli PEP—phosphoenolpyruvate; PYR—pyruvate; OAA—oxaloacetate; ACE—acetate; HSE—L-homoserine; MetX—acetyltransferase
| 1 | DU Lin, ROBLES A J, KING J B, et al. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity [J]. Angewandte Chemie International Edition, 2014, 53(3): 804-809. |
| 2 | BAUER A, BRÖNSTRUP M. Industrial natural product chemistry for drug discovery and development [J]. Natural Product Reports, 2014, 31(1): 35-60. |
| 3 | BROWN D G, LISTER T, MAY-DRACKA T L. New natural products as new leads for antibacterial drug discovery [J]. Bioorganic & Medicinal Chemistry Letters, 2014, 24(2): 413-418. |
| 4 | SCHEEPSTRA M, NIETO L, HIRSCH A K, et al. A natural-product switch for a dynamic protein interface [J]. Angewandte Chemie International Edition, 2014, 53(25): 6443-6448. |
| 5 | HARVEY A L, EDRADA-EBEL R A, QUINN R J. The re-emergence of natural products for drug discovery in the genomics era [J]. Nature Reviews Drug Discovery, 2015, 14(2): 111-129. |
| 6 | ZIMMERMANN T J, ROY S, MARTINEZ N E, et al. Biology-oriented synthesis of a tetrahydroisoquinoline-based compound collection targeting microtubule polymerization [J]. ChemBioChem, 2013, 14(3): 295-300. |
| 7 | LOWE D B. Drug discovery: combichem all over again [J]. Nature Chemistry, 2014, 6(10): 851-852. |
| 8 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the 30 years from 1981 to 2010 [J]. Journal of Natural Products, 2012, 75(3): 311-335. |
| 9 | RODRIGUES T, REKER D, SCHNEIDER P, et al. Counting on natural products for drug design [J]. Nature Chemistry, 2016, 8(6): 531-534. |
| 10 | DEITERS A, CROPP T A, SUMMERER D, et al. Site-specific PEGylation of proteins containing unnatural amino acids [J]. Bioorganic & Medicinal Chemistry Letters, 2004, 14(23): 5743-5745. |
| 11 | NEUMANN H, WANG Kaihang, DAVIS L, et al. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome [J]. Nature, 2010, 464(7287): 441-444. |
| 12 | BALTZ R H. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways [J]. ACS Synthetic Biology, 2014, 3(10): 748-758. |
| 13 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age [J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 14 | BREITLING R, TAKANO E. Synthetic biology advances for pharmaceutical production [J]. Current Opinion in Biotechnology, 2015, 35: 46-51. |
| 15 | TANG Xiaolong, DAI Hong, ZHU Yongxiang, et al. Maytansine-loaded star-shaped folate-core PLA-TPGS nanoparticles enhancing anticancer activity [J]. American Journal of Translational Research, 2014, 6(5): 528-537. |
| 16 | JARAPRAKASH NG, SUROLIA A. Role of glycosylation in nucleating protein folding and stability [J]. Biochemical Journal, 2017, 474(14): 2333-2347. |
| 17 | JI Shuai, LIANG Wenfei, LI Ziwei, et al. Effcient and selective glucosylation of prenylated phenolic compounds by Mucor hiemalis [J]. RSC Advances, 2016, 6(25): 20791-20799. |
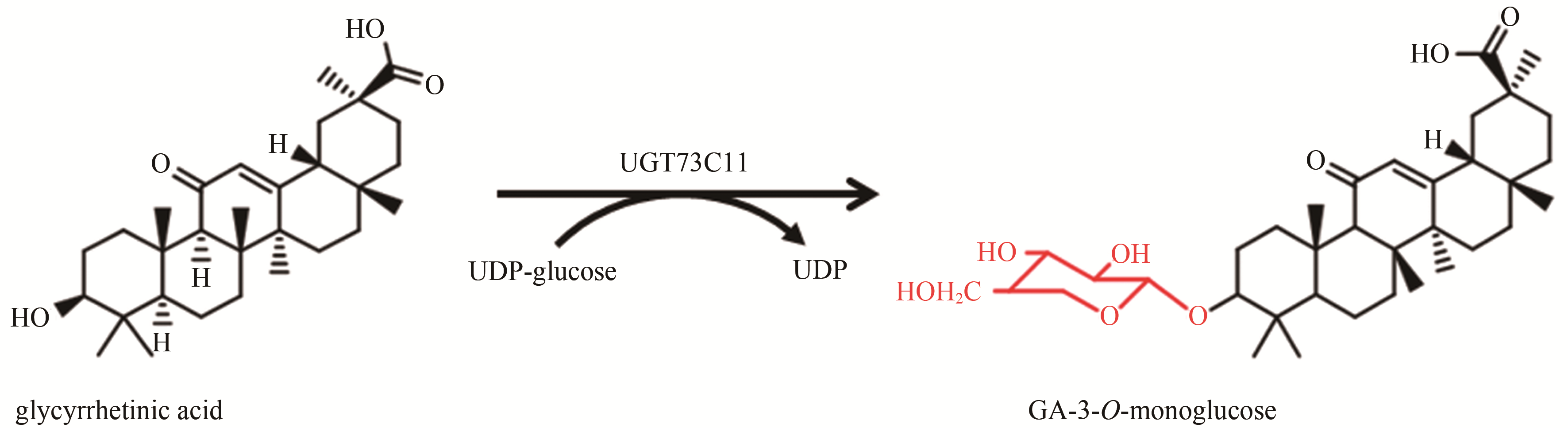
| 18 | LIU Xiaochen, ZHANG Liang. Biosynthesis of glycyrrhetinic acid-3-O-monoglucose using glycosyltransferase UGT73C11 from Barbarea vulgaris [J]. Industrial & Engineering Chemistry Research, 2017, 56(52): 14949-14958. |
| 19 | LIANG Wenfei, LI Ziwei, JI Shuai, et al. Microbial glycosylation of tanshinone IIA by Cunninghamella elegans AS 3.2028 [J]. RSC Advances, 2015, 5(78): 63753-63756. |
| 20 | HARMS J M, WILSON D N, SCHLUENZEN F, et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin [J]. Molecular Cell, 2008, 30(1): 26-38. |
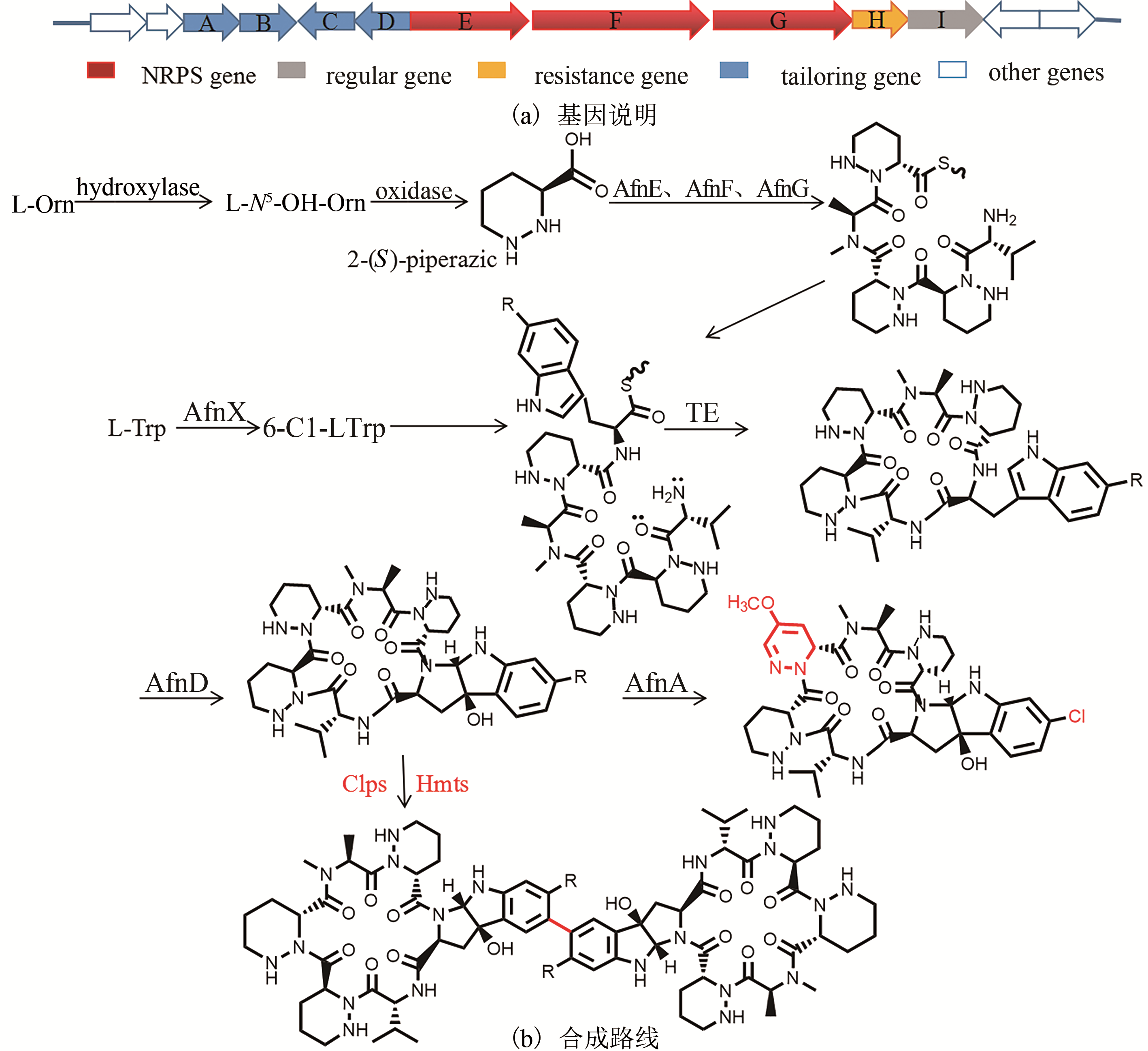
| 21 | ZHENG Qingfei, WANG Shoufeng, LIAO Rijing, et al. Precursor-directed mutational biosynthesis facilitates the functional assignment of two cytochromes P450 in thiostrepton biosynthesis [J]. ACS Chemical Biology, 2016, 11: 2673–2678. |
| 22 | WANG Shoufeng, ZHENG Qingfei, WANG Jianfeng, et al. Target-oriented design and biosynthesis of thiostrepton-derived thiopeptide antibiotics with improved pharmaceutical properties [J]. Organic Chemistry Frontiers, 2015, 2: 106-109. |
| 23 | EVANS B, CHEN Yunqiu, METCALF W, et al. Directed evolution of the nonribosomal peptide synthetase AdmK generates new Andrimid derivatives in vivo [J]. Chemistry & Biology, 2011, 18(5): 601-607. |
| 24 | MATSUDA Y, GOTFREDSEN C H, LARSEN T O. Genetic characterization of neosartorin biosynthesis provides insight into heterodimeric natural product generation [J]. Organic Letters, 2018, 20(22): 7197-7200. |
| 25 | JI Zhiqin, WEI Shaopeng, FAN Lixia, et al. Three novel cyclic hexapeptides from Streptomyces alboflavus 313 and their antibacterial activity [J]. European Journal of Medicinal Chemistry, 2012, 50: 296-303. |
| 26 | FAN Lixia, JI Zhiqin, GUO Zhengyan, et al. NW-G12, a novel nonchlorinated cyclohexapeptide from Streptomyces alboflavus313 [J]. Chemistry of Natural Compounds, 2013, 49(5): 910-913. |
| 27 | GUO Zhengyan, LI Pengwei, CHEN Guozhu, et al. Design and biosynthesis of dimeric alboflavusins with biaryl linkages via regiospecific C—C bond coupling [J]. Journal of the American Chemical Society, 2018, 140(51): 18009-18015. |
| 28 | HINDRA, YANG Dong, TENG Qihui, et al. Genome mining of Streptomyces mobaraensis DSM40847 as a bleomycin producer providing a biotechnology platform to engineer designer bleomycin analogues [J]. Organic Letters, 2017, 19(6): 1386-1389. |
| 29 | BUTLER M S, ROBERTSON A A B, COOPER M A. Natural product and natural product derived drugs in clinical trials [J]. Natural Product Reports, 2014, 31(11): 1612-1661. |
| 30 | CHEN Yun, DENG Wei, WU Jiequn, et al. Genetic modulation of the overexpression of tailoring genes eryK and eryG leading to the improvement of erythromycin A purity and production in Saccharopolyspora erythraea fermentation [J]. Applied and Environmental Microbiology, 2008, 74(6): 1820-1828. |
| 31 | 张万祥, 汪焰胜, 吴杭, 等. 前体代谢工程提高红霉素产量的研究进展[J]. 生物技术通讯, 2019, 30(1): 140-145. |
| ZHANG Wanxiang, WANG Yansheng, WU Hang, et al. Advances in metabolic engineering of precursors for improving Erythromycin production [J]. Letters in Biotechnology, 2019, 30(1): 140-145. | |
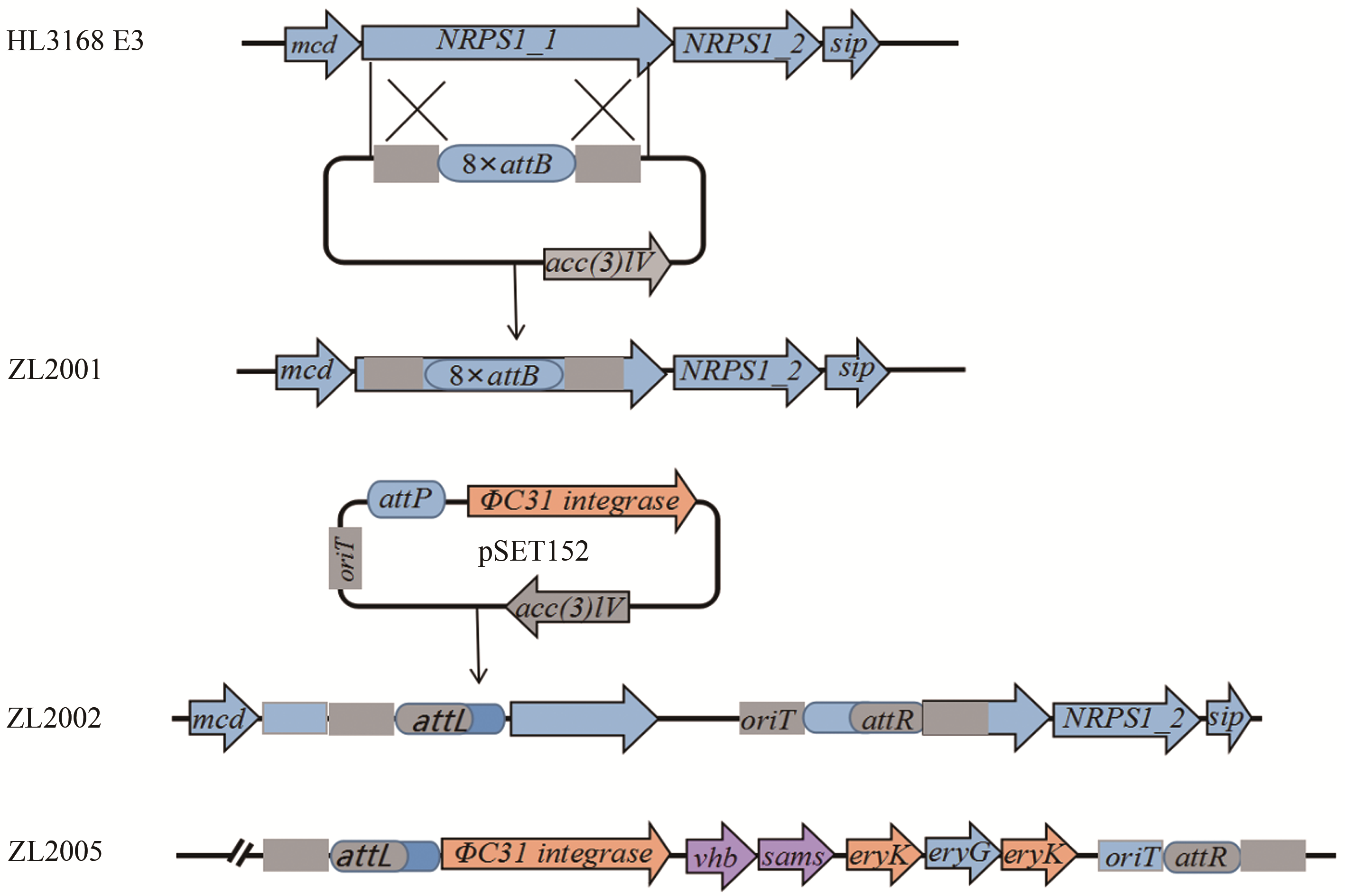
| 32 | WU Jiequn, ZHANG Qinglin, DENG Wei, et al. Toward improvement of erythromycin A production in an industrial Saccharopolyspora erythraea strain via facilitation of genetic manipulation with an artificial attB site for specific recombination [J]. Applied and Environmental Microbiology, 2011, 77(21): 7508-7516. |
| 33 | LIAN Jiazhang, SI Tong, NAIR N U, et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains [J]. Metabolic Engineering, 2014, 24: 139-149. |
| 34 | KRIVORUCHKO A, ZHANG Yiming, SIEWERS V, et al. Microbial acetyl-CoA metabolism and metabolic engineering [J]. Metabolic Engineering, 2015, 28: 28-42. |
| 35 | LIU Yiqi, BAI Chenxiao, LIU Qi, et al. Engineered ethanol-driven biosynthetic system for improving production of acetyl-CoA derived drugs in Crabtree-negative yeast [J]. Metabolic Engineering, 2019, 54: 275-284. |
| 36 | 朱灵英, 郭娟, 张爱丽, 等. 参与植物三萜生物合成的细胞色素P450酶研究进展[J]. 中草药, 2019, 50(22): 5597-5610. |
| ZHU Lingying, GUO Juan, ZHANG Aili, et al. Research progress on CYP450 involved in medicinal plant triterpenoid biosynthesis [J]. Chinese Traditional and Herbal Drugs, 2019, 50(22): 5597-5610. | |
| 37 | 漆丽华, 张媚, 潘海学, 等. 基于生物合成途径改造的一个三欣卡辛类似物的发现[J]. 生命有机化学, 2014, 34(7): 1376-1381. |
| XI Lihua, ZHANG Mei, PAN Haixue, et al. Production of a trioxacarcin analogue by engineering of its biosynthetic pathway [J]. Chinese Journal of Organic Chemistry, 2014, 34(7): 1376-1381. | |
| 38 | MARTIN V J J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids [J]. Nature Biotechnology, 2003, 21(7): 796-802. |
| 39 | 范楚珧, 刘龙英, 沈玥, 等. 吗啡的合成生物学研究和工业化生产[J]. 科学通报, 2016, 61: 1436-1444. |
| FAN Chuyao, LIU Longying, SHEN Yue, et al. Progress of biosynthesis of morphine and its industrial manufacture [J]. Chinese Science Bulletin, 2016, 61: 1436-1444. | |
| 40 | NAKAGAWA A, MINAMI H, KIM Ju-Sung, et al. A bacterial platform for fermentative production of plant alkaloids [J]. Nature Communications, 2011, 2(1): 1-9. |
| 41 | NEUMANN H, NEUMANN-STAUBITZ P. Synthetic biology approaches in drug discovery and pharmaceutical biotechnology [J]. Applied Microbiology and Biotechnology, 2010, 87(1): 75-86. |
| 42 | ENGELS B, DAHM P, JENNEWEIN S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production [J]. Metabolic Engineering, 2008, 10(3/4): 201-206. |
| 43 | KIRBY J, KEASLING J D. Metabolic engineering of microorganisms for isoprenoid production [J]. Natural Product Reports, 2008, 25(4): 656-661. |
| 44 | WALTHER T, CALVAYRAC F, MALBERT Y, et al. Construction of a synthetic metabolic pathway for the production of 2,4-dihydroxybutyric acid from homoserine [J]. Metabolic Engineering, 2018, 45: 237-245. |
| 45 | WEI Liang, WANG Qian, XU Ning, et al. Combining protein and metabolic engineering strategies for high level production of O-acetylhomoserine in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8: 1153-1167. |
| 46 | WILLIAMS T L, YIN Yuhui W, CARTER C W. Selective inhibition of bacterial tryptophanyl-tRNA synthetases by indolmycin is mechanism-based [J]. Journal of Biological Chemistry, 2016, 291(1): 255-265. |
| 47 | DU Yiling, HIGGINS M A, ZHAO Guiyun, et al. Convergent biosynthetic transformations to a bacterial specialized metabolite [J]. Nature Chemical Biology, 2019, 15(11): 1043-1048. |
| 48 | DANGEL V, WESTRICH L, SMITH M C M, et al. Use of an inducible promoter for antibiotic production in a heterologous host [J]. Applied Microbiology and Biotechnology, 2010, 87(1): 261-269. |
| 49 | YAN Fu, BURGARD C, POPOFF A, et al. Synthetic biology approaches and combinatorial biosynthesis towards heterologous lipopeptide production [J]. Chemical Science, 2018, 9(38): 7510-7519. |
| 50 | KOMATSU M, KOMATSU K, KOIWAI H, et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites [J]. ACS Synthetic Biology, 2013, 2(7): 384-396. |
| 51 | LUO Yunzi, HUANG Hua, LIANG Jing, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster [J]. Nature Communications, 2013, 4: 94-105. |
| 52 | TAN Gaoyi, DENG Kunhua, LIU Xinhua, et al. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces [J]. ACS Synthetic Biology, 2017, 6(6): 995-1005. |
| 53 | D'ISCHIA M, WAKAMATSU K, CICOIRA F, et al. Melanins and melanogenesis: from pigment cells to human health and technological applications [J]. Pigment Cell & Melanoma Research, 2015, 28(5): 520-544. |
| 54 | KIM Young Jo, KHETAN A, WU Wei, et al. Evidence of porphyrin-like structures in natural melanin pigments using electrochemical fingerprinting [J]. Advanced Materials, 2016, 28(16): 3173-3180. |
| 55 | WANG Zheng, TSCHIRHART T, SCHULTZHAUS Z, et al. Characterization and application of melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis [J]. Applied and Environmental Microbiology, 2020, 86(5): e02749-19. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 仲泉周, 单依怡, 裴清云, 金艳芸, 王艺涵, 孟璐远, 王歆韵, 张雨鑫, 刘坤媛, 王慧中, 冯尚国. 生物合成法生产α-熊果苷的研究进展[J]. 合成生物学, 2025, 6(1): 118-135. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [11] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [12] | 李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517. |
| [13] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [14] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [15] | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||