合成生物学 ›› 2022, Vol. 3 ›› Issue (4): 658-675.DOI: 10.12211/2096-8280.2021-069
无细胞合成策略在生物材料研究中的应用
吉博涛, 钱志刚, 夏小霞
- 上海交通大学生命科学技术学院,微生物代谢国家重点实验室,教育部代谢与发育科学国际合作联合实验室,上海 200240
-
收稿日期:2021-06-30修回日期:2021-07-22出版日期:2022-08-31发布日期:2022-09-08 -
通讯作者:夏小霞 -
作者简介:吉博涛 (1997—),男,博士研究生。研究方向为无细胞表达系统合成生物材料。E-mail: botao-ji@sjtu.edu.cn夏小霞 (1978—),女,教授,博士生导师。研究方向为蛋白材料合成生物学、人工细胞器、生物大分子自组装等。E-mail: xiaoxiaxia@sjtu.edu.cn -
基金资助:国家重点研发计划(2020YFA0907702);国家自然科学基金(32071414);上海市自然科学基金(21ZR1432100)
Application of cell-free synthesis strategy in biomaterial research
JI Botao, QIAN Zhigang, XIA Xiaoxia
- State Key Laboratory of Microbial Metabolism,Joint International Research Laboratory of Metabolic and Developmental Sciences,School of Life Science & Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2021-06-30Revised:2021-07-22Online:2022-08-31Published:2022-09-08 -
Contact:XIA Xiaoxia
摘要:
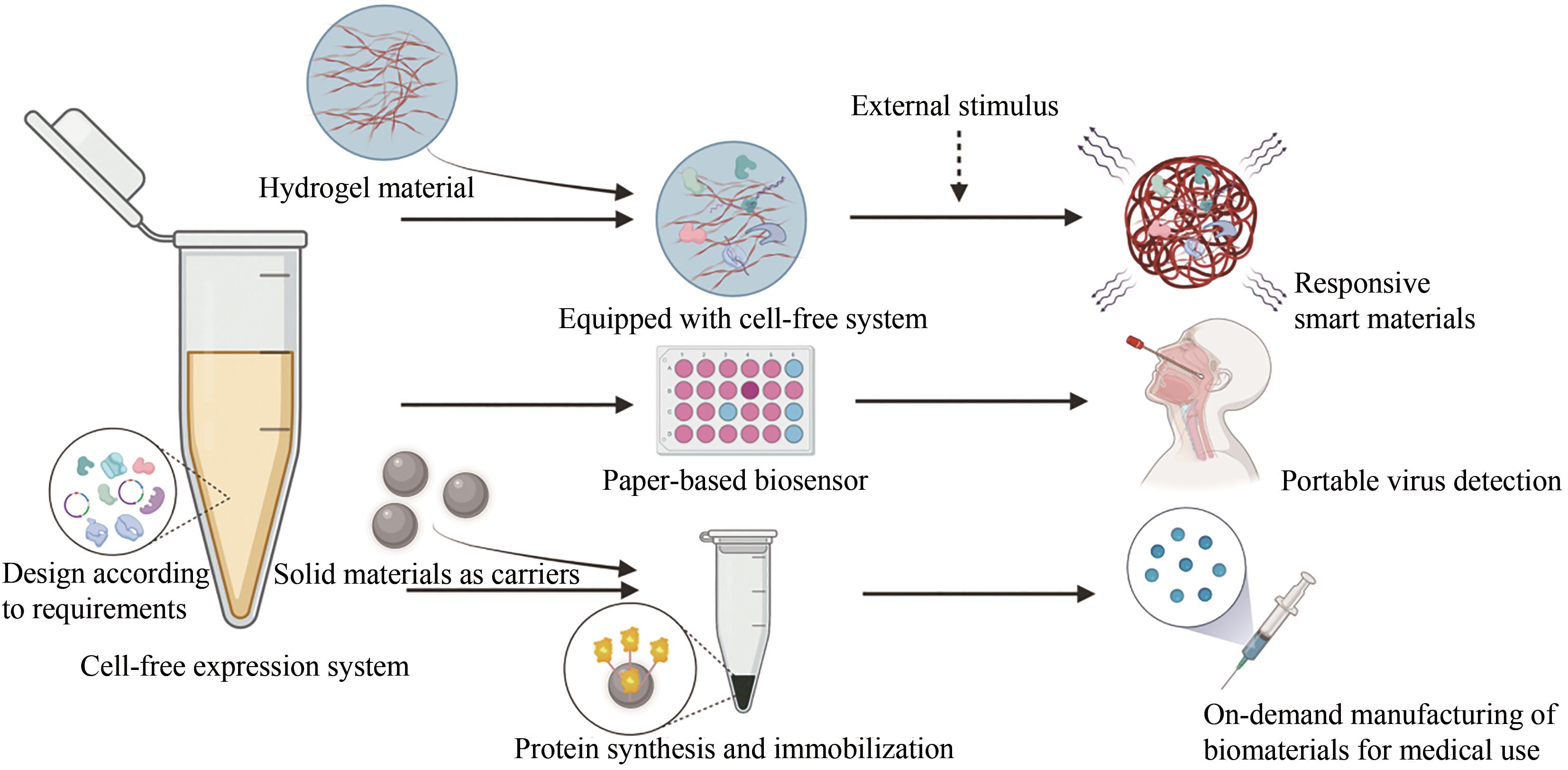
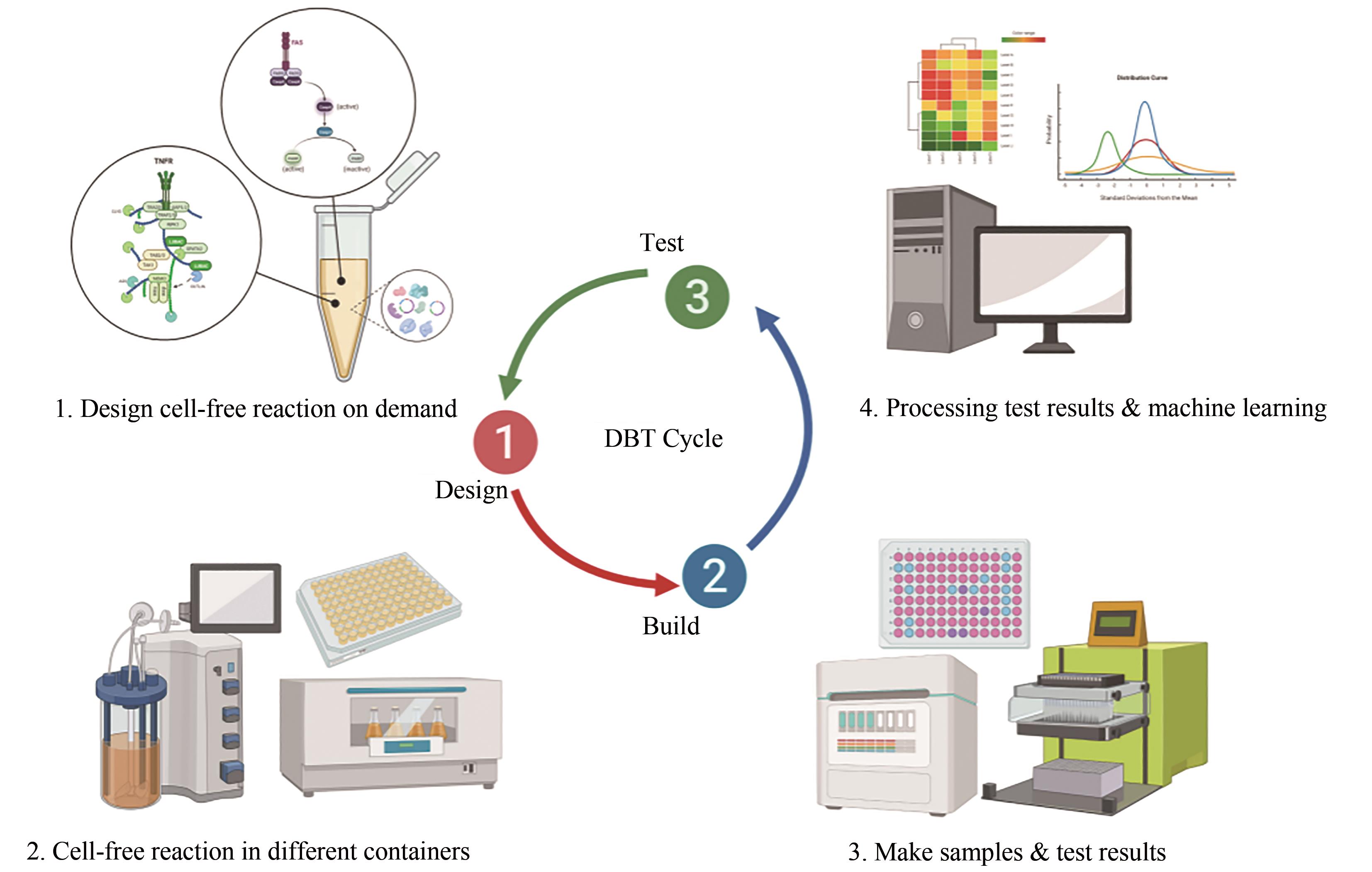
生物材料凭借其可再生、可降解及生物相容性好等诸多优点,在生物技术及生物医学应用领域备受关注。基于“自下而上”设计理念的无细胞表达系统,作为细胞合成体系的有益补充,大力加速了生物材料的开发生产及其应用范围,并成为了材料合成生物学发展的前沿。本文介绍了无细胞表达系统的独特优势,并列举了其用于生物材料可持续生产的策略、与材料结合后的创新设计、赋予材料功能化和智能化方法,以及通过加速设计-构建-测试(DBT)循环来促进新型生物材料开发的各项应用,而这些例证都充分表明无细胞策略在生物材料设计与合成中的创新优势。尽管无细胞表达系统用于生物材料合成仍然存在诸如成本、翻译后修饰较弱、亟需交叉领域合作等方面的挑战,但相信随着合成生物学的发展及多学科交叉研究的深入,无细胞表达系统将为新型生物材料研发及产业化提供强大的助力,并为共同解决全球环境问题、保护人类生命健康等方面作出不可忽视的贡献。
中图分类号:
引用本文
吉博涛, 钱志刚, 夏小霞. 无细胞合成策略在生物材料研究中的应用[J]. 合成生物学, 2022, 3(4): 658-675.
JI Botao, QIAN Zhigang, XIA Xiaoxia. Application of cell-free synthesis strategy in biomaterial research[J]. Synthetic Biology Journal, 2022, 3(4): 658-675.
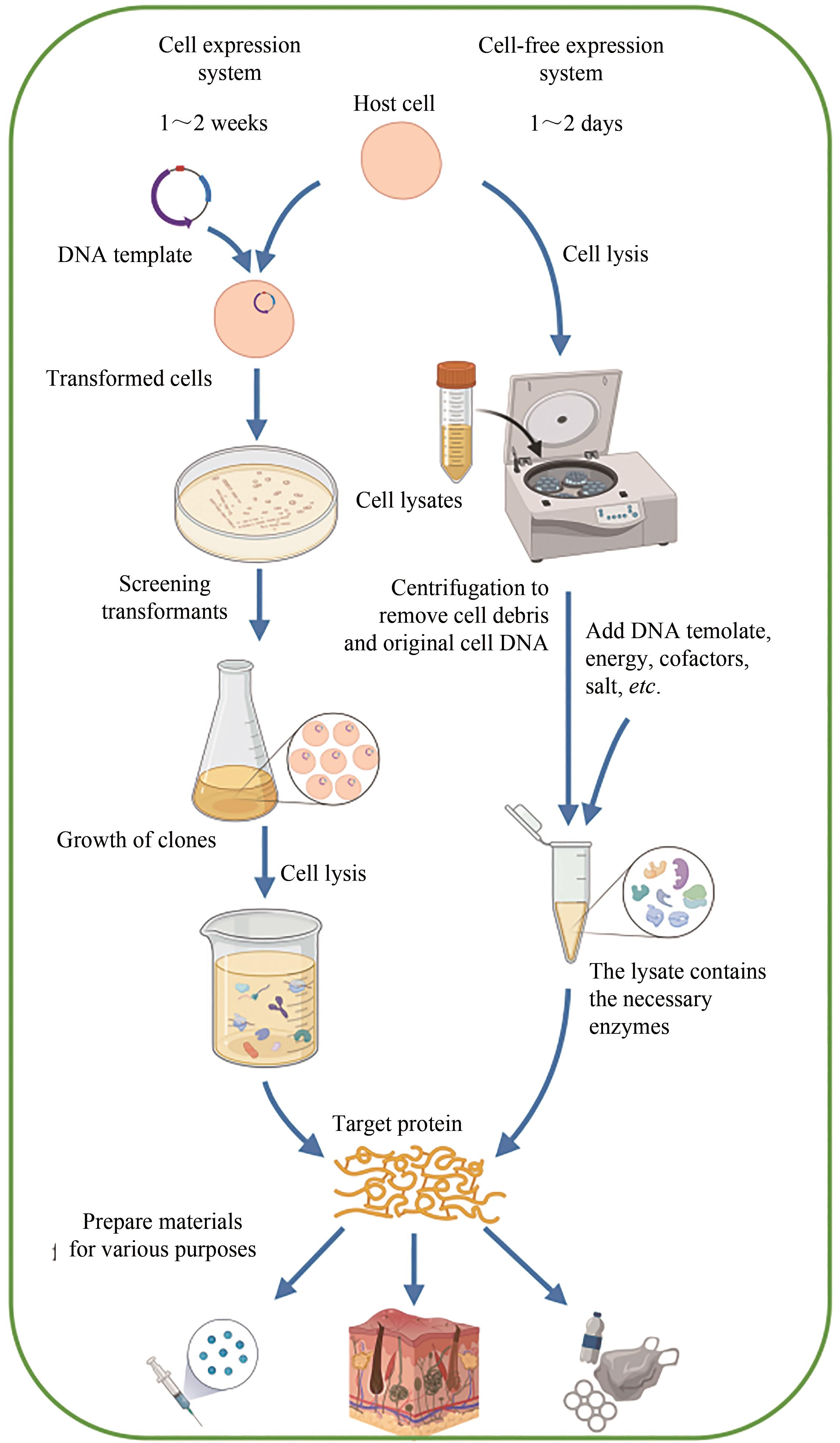
| 特征 | 细胞表达系统 | 无细胞表达系统 |
|---|---|---|
| 转录和翻译 | -具有细胞膜,细胞内表征和产物释放可能具有挑战性 -内源性调控难以预测和修改 +能够使用定向进化技术 | +开放的环境易于实时操作和控制 +直接添加底物或收获产物 +易于使用来自多种生物体的酶途径 |
| 翻译后修饰 | +简单 | -困难 |
| 自我复制 | +简单 | -困难 |
| DNA模板 | -质粒(或者整合到基因组的DNA) | +质粒或 PCR 产物 |
| 引入非天然氨基酸 | -困难 | +简单 |
| 膜蛋白和复合蛋白的合成 | -由于细胞内环境有限而难以合成 | +通过添加表面活性剂或调整系统环境易于合成 |
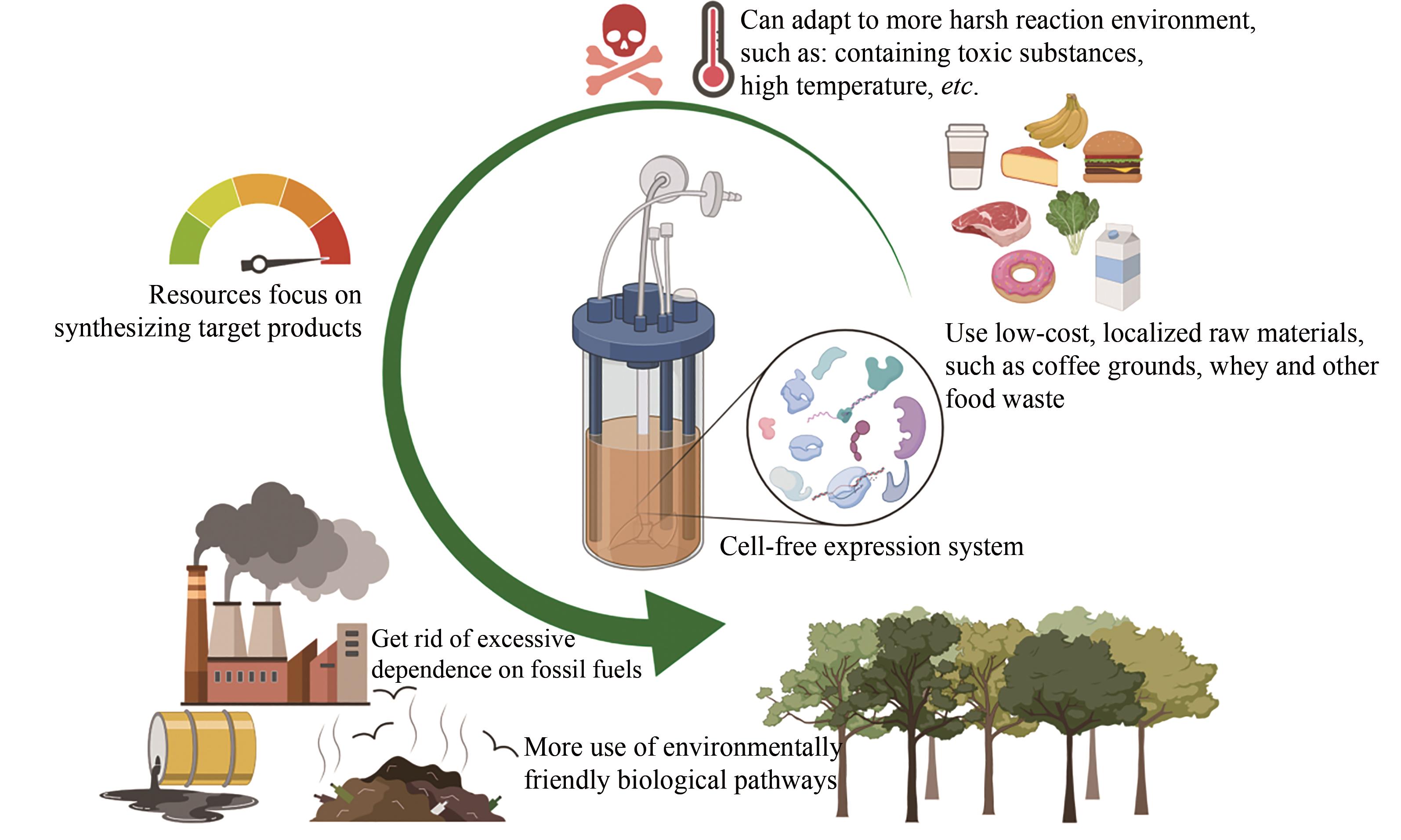
| 对毒性的耐受力 | -低,难以合成毒性蛋白 -若中间产物有毒性,则可行性受到限制 | +高,可以合成毒性蛋白 +无限制(如某些途径酶使用体积分数为12%异丁醇) |
| 设计-构建-测试(DBT)循环 | -慢,1~2周 | +快,1~2天 |
| 资源利用率 | -较低,由于复杂的细胞代谢而将资源转向细胞维护和副产品 | +较高,所有资源都可以直接用于生产产品 |
| 与材料整合的能力 | -困难 | +简单 |
| 生物制造 | -适中的产率和产量 -纯化前需再次裂解细胞 -不易放大生产,工业规模发酵条件是不同的 -菌种污染可能会是灾难性的 +多年实践经验;完善的方法 | +较高的产率和产量 +无须细胞裂解的简单的纯化过程 +易于放大生产,具有线性放大的可能 -较为年轻,最近成立 |
| 成本 | +低到适中 | -中高,主要为酶和辅因子成本 |
| 稳定性 | -通过发酵条件影响细胞环境 | +由工程师直接控制反应条件 |
表1 无细胞表达系统的优缺点[32-34]
Tab. 1 The advantages and disadvantages of cell-free expression systems
| 特征 | 细胞表达系统 | 无细胞表达系统 |
|---|---|---|
| 转录和翻译 | -具有细胞膜,细胞内表征和产物释放可能具有挑战性 -内源性调控难以预测和修改 +能够使用定向进化技术 | +开放的环境易于实时操作和控制 +直接添加底物或收获产物 +易于使用来自多种生物体的酶途径 |
| 翻译后修饰 | +简单 | -困难 |
| 自我复制 | +简单 | -困难 |
| DNA模板 | -质粒(或者整合到基因组的DNA) | +质粒或 PCR 产物 |
| 引入非天然氨基酸 | -困难 | +简单 |
| 膜蛋白和复合蛋白的合成 | -由于细胞内环境有限而难以合成 | +通过添加表面活性剂或调整系统环境易于合成 |
| 对毒性的耐受力 | -低,难以合成毒性蛋白 -若中间产物有毒性,则可行性受到限制 | +高,可以合成毒性蛋白 +无限制(如某些途径酶使用体积分数为12%异丁醇) |
| 设计-构建-测试(DBT)循环 | -慢,1~2周 | +快,1~2天 |
| 资源利用率 | -较低,由于复杂的细胞代谢而将资源转向细胞维护和副产品 | +较高,所有资源都可以直接用于生产产品 |
| 与材料整合的能力 | -困难 | +简单 |
| 生物制造 | -适中的产率和产量 -纯化前需再次裂解细胞 -不易放大生产,工业规模发酵条件是不同的 -菌种污染可能会是灾难性的 +多年实践经验;完善的方法 | +较高的产率和产量 +无须细胞裂解的简单的纯化过程 +易于放大生产,具有线性放大的可能 -较为年轻,最近成立 |
| 成本 | +低到适中 | -中高,主要为酶和辅因子成本 |
| 稳定性 | -通过发酵条件影响细胞环境 | +由工程师直接控制反应条件 |
| 组成生物材料的特殊蛋白质 | 使CFE系统提高生产力和/或生物活性的方法 |
|---|---|
| 对生产宿主有毒的蛋白质 | 额外的tRNA补充[ 选择不同的提取物(例如昆虫)[ 连续交换无细胞表达系统[ 纯化标签以提高溶解度[ |
| 具有氨基酸组成偏好的蛋白质 | 稀有tRNA的添加[ 选择不同的提取物(例如小麦胚芽)[ 用同义密码子替换初始密码子[ |
| 需要分子伴侣或特定化学环境合成的蛋白质 | 伴侣蛋白,蛋白质二硫键异构酶(PDI),还原和氧化的谷胱甘肽(glutathione)调整[ 前导肽和微粒体囊泡[ 对于膜蛋白,加入洗涤剂/表面活性剂[ |
| 含有非天然氨基酸的蛋白质 | 通过基因组工程消除提取物中的负面效应物,如释放因子1(RF1)的竞争限制[ 去除同源tRNA[ |
| 需要糖基化修饰的蛋白质 | 内切糖苷酶介导的N-糖基化蛋白的制备[ 原核寡糖基转移酶(OST)介导的无细胞糖蛋白生物合成[ 糖基转移酶(GT)介导的蛋白质糖基化和聚糖加工[ |
| 需要磷酸化的蛋白质 | 基因组重新编码的大肠杆菌菌株[ |
表2 CFE系统针对特殊蛋白质合成的应对方法
Tab. 2 Corresponding methods for special protein synthesis using CFE system
| 组成生物材料的特殊蛋白质 | 使CFE系统提高生产力和/或生物活性的方法 |
|---|---|
| 对生产宿主有毒的蛋白质 | 额外的tRNA补充[ 选择不同的提取物(例如昆虫)[ 连续交换无细胞表达系统[ 纯化标签以提高溶解度[ |
| 具有氨基酸组成偏好的蛋白质 | 稀有tRNA的添加[ 选择不同的提取物(例如小麦胚芽)[ 用同义密码子替换初始密码子[ |
| 需要分子伴侣或特定化学环境合成的蛋白质 | 伴侣蛋白,蛋白质二硫键异构酶(PDI),还原和氧化的谷胱甘肽(glutathione)调整[ 前导肽和微粒体囊泡[ 对于膜蛋白,加入洗涤剂/表面活性剂[ |
| 含有非天然氨基酸的蛋白质 | 通过基因组工程消除提取物中的负面效应物,如释放因子1(RF1)的竞争限制[ 去除同源tRNA[ |
| 需要糖基化修饰的蛋白质 | 内切糖苷酶介导的N-糖基化蛋白的制备[ 原核寡糖基转移酶(OST)介导的无细胞糖蛋白生物合成[ 糖基转移酶(GT)介导的蛋白质糖基化和聚糖加工[ |
| 需要磷酸化的蛋白质 | 基因组重新编码的大肠杆菌菌株[ |
| 73 | TSUBOI T, TAKEO S, ARUMUGAM T U, et al. The wheat germ cell-free protein synthesis system: a key tool for novel malaria vaccine candidate discovery[J]. Acta Tropica, 2010, 114(3): 171-176. |
| 74 | AHN J H, KEUM J W, KIM D M. High-throughput, combinatorial engineering of initial codons for tunable expression of recombinant proteins[J]. Journal of Proteome Research, 2008, 7(5): 2107-2113. |
| 75 | PARK Y J, LEE K H, BAEK M S, et al. High-throughput engineering of initial coding regions for maximized production of recombinant proteins[J]. Biotechnology and Bioprocess Engineering, 2017, 22(5): 497-503. |
| 76 | STECH M, KUBICK S. Cell-free synthesis meets antibody production: a review[J]. Antibodies, 2015, 4(1): 12-33. |
| 77 | STECH M, NIKOLAEVA O, THORING L, et al. Cell-free synthesis of functional antibodies using a coupled in vitro transcription-translation system based on CHO cell lysates[J]. Scientific Reports, 2017, 7: 12030. |
| 78 | SACHSE R, DONDAPATI S K, FENZ S F, et al. Membrane protein synthesis in cell-free systems: from bio-mimetic systems to bio-membranes[J]. FEBS Letters, 2014, 588(17): 2774-2781. |
| 79 | BÄCKLUND E, IGNATUSHCHENKO M, LARSSON G. Suppressing glucose uptake and acetic acid production increases membrane protein overexpression in Escherichia coli [J]. Microbial Cell Factories, 2011, 10: 35. |
| 80 | MICHEL-REYDELLET N, CALHOUN K, SWARTZ J. Amino acid stabilization for cell-free protein synthesis by modification of the Escherichia coli genome[J]. Metabolic Engineering, 2004, 6(3): 197-203. |
| 81 | GAN Q L, FAN C G. Increasing the fidelity of noncanonical amino acid incorporation in cell-free protein synthesis[J]. Biochimica et Biophysica Acta (BBA)-General Subjects, 2017, 1861(11): 3047-3052. |
| 82 | GIDDENS J P, LOMINO J V, AMIN M N, et al. Endo-F3 glycosynthase mutants enable chemoenzymatic synthesis of core-fucosylated triantennary complex type glycopeptides and glycoproteins[J]. Journal of Biological Chemistry, 2016, 291(17): 9356-9370. |
| 83 | FAN S Q, HUANG W, WANG L X. Remarkable transglycosylation activity of glycosynthase mutants of endo-D, an endo-β-N-acetylglucosaminidase from Streptococcus pneumoniae [J]. Journal of Biological Chemistry, 2012, 287(14): 11272-11281. |
| 84 | NATARAJAN A, JAROENTOMEECHAI T, CABRERA-SÁNCHEZ M, et al. Engineering orthogonal human O-linked glycoprotein biosynthesis in bacteria[J]. Nature Chemical Biology, 2020, 16(10): 1062-1070. |
| 85 | KIGHTLINGER W, DUNCKER K E, RAMESH A, et al. A cell-free biosynthesis platform for modular construction of protein glycosylation pathways[J]. Nature Communications, 2019, 10: 5404. |
| 86 | LIU F, VIJAYAKRISHNAN B, FARIDMOAYER A, et al. Rationally designed short polyisoprenol-linked PglB substrates for engineered polypeptide and protein N-glycosylation[J]. Journal of the American Chemical Society, 2014, 136(2): 566-569. |
| 87 | OZA J P, AERNI H R, PIRMAN N L, et al. Robust production of recombinant phosphoproteins using cell-free protein synthesis[J]. Nature Communications, 2015, 6: 8168. |
| 88 | JIN X, HONG S H. Cell-free protein synthesis for producing 'difficult-to-express' proteins[J]. Biochemical Engineering Journal, 2018, 138: 156-164. |
| 89 | DONDAPATI S K, STECH M, ZEMELLA A, et al. Cell-free protein synthesis: a promising option for future drug development[J]. BioDrugs, 2020, 34(3): 327-348. |
| 90 | BENCI J L, XU B H, QIU Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade[J]. Cell, 2016, 167(6): 1540-1554.e12. |
| 91 | XIE Q H, MATSUNAGA S, WEN Z S, et al. In vitro system for high-throughput screening of random peptide libraries for antimicrobial peptides that recognize bacterial membranes[J]. Journal of Peptide Science: an Official Publication of the European Peptide Society, 2006, 12(10): 643-652. |
| 92 | RAGHUNATH P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in vibrio parahaemolyticus[J]. Frontiers in Microbiology, 2015, 5: 805. |
| 93 | PARK N, KAHN J S, RICE E J, et al. High-yield cell-free protein production from P-gel[J]. Nature Protocols, 2009, 4(12): 1759-1770. |
| 94 | UM S H, LEE J B, PARK N, et al. Enzyme-catalysed assembly of DNA hydrogel[J]. Nature Materials, 2006, 5(10): 797-801. |
| 95 | PARK N, UM S H, FUNABASHI H, et al. A cell-free protein-producing gel[J]. Nature Materials, 2009, 8(5): 432-437. |
| 96 | RUIZ R C H, KIATWUTHINON P, KAHN J S, et al. Cell-free protein expression from DNA-based hydrogel (P-gel) droplets for scale-up production[J]. Industrial Biotechnology, 2012, 8(6): 372-377. |
| 1 | MARTIN J D. What's in a name change? [J]. Physics in Perspective, 2015, 17(1): 3-32. |
| 2 | CALLISTER JR W D, RETHWISCH D G. Fundamentals of materials science and engineering: an integrated approach[M]. John Wiley & Sons, 2020. |
| 3 | CROWTHER T W, GLICK H B, COVEY K R, et al. Mapping tree density at a global scale[J]. Nature, 2015, 525(7568): 201-205. |
| 4 | ARMSTRONG E. Voluntary greenhouse gas reporting[J]. Environmental Quality Management, 2011, 20(4): 29-42. |
| 5 | ERIKSEN M, LEBRETON L C M, CARSON H S, et al. Plastic pollution in the world's oceans: more than 5 trillion plastic pieces weighing over 250, 000 tons afloat at sea[J]. PLoS One, 2014, 9(12): e111913. |
| 6 | LENTON T M, ROCKSTRÖM J, GAFFNEY O, et al. Climate tipping points—too risky to bet against[J]. Nature, 2019, 575(7784): 592-595. |
| 7 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11: 5174. |
| 8 | MAPLESTON P. New technologies for a greener industry[J]. Plastics Engineering, 2008, 64(1): 10-15. |
| 9 | COLWILL J, RAHIMIFARD S, CLEGG A. Eco-design tool to support the use of renewable polymers within packaging applications[C]// Glocalized Solutions for Sustainability in Manufacturing. Springer, 2011: 160-165. |
| 10 | AKSAKAL R, MERTENS C, SOETE M, et al. Applications of discrete synthetic macromolecules in life and materials science: recent and future trends[J]. Advanced Science, 2021, 8(6): 2004038. |
| 11 | YADAV P, YADAV H, SHAH V G, et al. Biomedical biopolymers, their origin and evolution in biomedical sciences: a systematic review[J]. Journal of Clinical and Diagnostic Research: JCDR, 2015, 9(9): ZE21-ZE25. |
| 12 | WEINER S, ADDADI L, WAGNER H D. Materials design in biology[J]. Materials Science and Engineering: C, 2000, 11(1): 1-8. |
| 97 | KAHN J S, RUIZ R C H, SUREKA S, et al. DNA microgels as a platform for cell-free protein expression and display[J]. Biomacromolecules, 2016, 17(6): 2019-2026. |
| 98 | CUI J H, WU D, SUN Q, et al. A PEGDA/DNA hybrid hydrogel for cell-free protein synthesis[J]. Frontiers in Chemistry, 2020, 8: 28. |
| 99 | YANG D Y, PENG S M, HARTMAN M R, et al. Enhanced transcription and translation in clay hydrogel and implications for early life evolution[J]. Scientific Reports, 2013, 3: 3165. |
| 100 | JIAO Y, LIU Y, LUO D, et al. Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis[J]. ACS Applied Materials & Interfaces, 2018, 10(35): 29308-29313. |
| 101 | LEE M S, HUNG C S, PHILLIPS D A, et al. Silk fibroin as an additive for cell-free protein synthesis[J]. Synthetic and Systems Biotechnology, 2020, 5(3): 145-154. |
| 102 | PARDEE K, GREEN A A, FERRANTE T, et al. Paper-based synthetic gene networks[J]. Cell, 2014, 159(4): 940-954. |
| 103 | PARDEE K. Perspective: solidifying the impact of cell-free synthetic biology through lyophilization[J]. Biochemical Engineering Journal, 2018, 138: 91-97. |
| 104 | BLUM S M, LEE M S, MGBOJI G E, et al. Impact of porous matrices and concentration by lyophilization on cell-free expression[J]. ACS Synthetic Biology, 2021, 10(5): 1116-1131. |
| 105 | CHO E, LU Y. Compartmentalizing cell-free systems: toward creating life-like artificial cells and beyond[J]. ACS Synthetic Biology, 2020, 9(11): 2881-2901. |
| 106 | PATRICK J F, ROBB M J, SOTTOS N R, et al. Polymers with autonomous life-cycle control[J]. Nature, 2016, 540(7633): 363-370. |
| 107 | SUCH G K, EVANS R A, DAVIS T P. Rapid photochromic switching in a rigid polymer matrix using living radical polymerization[J]. Macromolecules, 2006, 39(4): 1391-1396. |
| 108 | CASTANO L M, FLATAU A B. Smart fabric sensors and e-textile technologies: a review[J]. Smart Materials and Structures, 2014, 23(5): 053001. |
| 13 | SCHMALZ G. Determination of biocompatibility[M]//Biocompatibility of dental materials. Berlin, Heidelberg: Springer, 2009: 13-43. |
| 14 | GAD S C, GAD-MCDONALD S. Biomaterials, medical devices, and combination products: biocompatibility testing and safety assessment[M]. CRC Press, 2015. |
| 15 | ANDERSON J, STRELKOWA N, STAN G B, et al. Engineering and ethical perspectives in synthetic biology. Rigorous, robust and predictable designs, public engagement and a modern ethical framework are vital to the continued success of synthetic biology[J]. EMBO Reports, 2012, 13(7): 584-590. |
| 16 | CHURCH G M, ELOWITZ M B, SMOLKE C D, et al. Realizing the potential of synthetic biology[J]. Nature Reviews Molecular Cell Biology, 2014, 15(4): 289-294. |
| 17 | KELWICK R, MACDONALD J T, WEBB A J, et al. Developments in the tools and methodologies of synthetic biology[J]. Frontiers in Bioengineering and Biotechnology, 2014, 2: 60. |
| 18 | KHAMBHATI K, BHATTACHARJEE G, GOHIL N, et al. Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 248. |
| 19 | MOORE S J, MACDONALD J T, WIENECKE S, et al. Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(19): E4340-E4349. |
| 20 | MARTIN R W, DES SOYE B J, KWON Y C, et al. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids[J]. Nature Communications, 2018, 9(1): 1203. |
| 21 | BOWIE J U, SHERKHANOV S, KORMAN T P, et al. Synthetic biochemistry: the bio-inspired cell-free approach to commodity chemical production[J]. Trends in Biotechnology, 2020, 38(7): 766-778. |
| 22 | HODGMAN C E, JEWETT M C. Cell-free synthetic biology: thinking outside the cell[J]. Metabolic Engineering, 2012, 14(3): 261-269. |
| 23 | SMOLKE C D, SILVER P A. Informing biological design by integration of systems and synthetic biology[J]. Cell, 2011, 144(6): 855-859. |
| 24 | YUE K, ZHU Y Y, KAI L. Cell-free protein synthesis: chassis toward the minimal cell[J]. Cells, 2019, 8(4): 315. |
| 109 | HOARE T R, KOHANE D S. Hydrogels in drug delivery: progress and challenges[J]. Polymer, 2008, 49(8): 1993-2007. |
| 110 | AHN S K, KASI R M, KIM S C, et al. Stimuli-responsive polymer gels[J]. Soft Matter, 2008, 4(6): 1151. |
| 111 | QIAN Z G, ZHOU M L, SONG W W, et al. Dual thermosensitive hydrogels assembled from the conserved C‑terminal domain of spider dragline silk[J]. Biomacromolecules, 2015, 16(11): 3704-3711. |
| 112 | SONG W W, QIAN Z G, LIU H, et al. On-demand regulation of dual thermosensitive protein hydrogels[J]. ACS Macro Letters, 2021, 10(4): 395-400. |
| 113 | HU X, XIA X X, HUANG S C, et al. Development of adhesive and conductive resilin-based hydrogels for wearable sensors[J]. Biomacromolecules, 2019, 20(9): 3283-3293. |
| 114 | SARIKAYA M, TAMERLER C, JEN A K Y, et al. Molecular biomimetics: nanotechnology through biology[J]. Nature Materials, 2003, 2(9): 577-585. |
| 115 | WHITFIELD C J, BANKS A M, DURA G, et al. Cell-free genetic devices confer autonomic and adaptive properties to hydrogels[EB/OL]. bioRxiv, 2019. |
| 116 | KENNE L, GOHIL S, NILSSON E M, et al. Modification and cross-linking parameters in hyaluronic acid hydrogels — definitions and analytical methods[J]. Carbohydrate Polymers, 2013, 91(1): 410-418. |
| 117 | SMITH M T, BERKHEIMER S D, WERNER C J, et al. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage[J]. BioTechniques, 2014, 56(4): 186-193. |
| 118 | LEE K H, KIM D M. In vitro use of cellular synthetic machinery for biosensing applications[J]. Frontiers in Pharmacology, 2019, 10: 1166. |
| 119 | PARDEE K, GREEN A A, TAKAHASHI M K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components[J]. Cell, 2016, 165(5): 1255-1266. |
| 120 | AHN D G, JEON I J, KIM J D, et al. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein[J]. The Analyst, 2009, 134(9): 1896-1901. |
| 25 | BUCHNER E. Alkoholische gährung ohne hefezellen[J]. Berichte Der Deutschen Chemischen Gesellschaft, 1897, 30(1): 117-124. |
| 26 | NIRENBERG M W, MATTHAEI J H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 1961, 47(10): 1588-1602. |
| 27 | GREGORIO N E, LEVINE M Z, OZA J P. A user's guide to cell-free protein synthesis[J]. Methods and Protocols, 2019, 2(1): 24. |
| 28 | LIU W Q, ZHANG L K, CHEN M Z, et al. Cell-free protein synthesis: recent advances in bacterial extract sources and expanded applications[J]. Biochemical Engineering Journal, 2019, 141: 182-189. |
| 29 | GARENNE D, NOIREAUX V. Cell-free transcription-translation: engineering biology from the nanometer to the millimeter scale[J]. Current Opinion in Biotechnology, 2019, 58: 19-27. |
| 30 | SWARTZ J R. Expanding biological applications using cell-free metabolic engineering: an overview[J]. Metabolic Engineering, 2018, 50: 156-172. |
| 31 | WILDING K M, SCHINN S M, LONG E A, et al. The emerging impact of cell-free chemical biosynthesis[J]. Current Opinion in Biotechnology, 2018, 53: 115-121. |
| 32 | LU Y. Cell-free synthetic biology: engineering in an open world[J]. Synthetic and Systems Biotechnology, 2017, 2(1): 23-27. |
| 33 | DUDLEY Q M, KARIM A S, JEWETT M C. Cell-free metabolic engineering: biomanufacturing beyond the cell[J]. Biotechnology Journal, 2015, 10(1): 69-82. |
| 34 | GUTERL J K, GARBE D, CARSTEN J, et al. Cell-free metabolic engineering: production of chemicals by minimized reaction cascades[J]. ChemSusChem, 2012, 5(11): 2165-2172. |
| 35 | CARLSON E D, GAN R, HODGMAN C E, et al. Cell-free protein synthesis: applications come of age[J]. Biotechnology Advances, 2012, 30(5): 1185-1194. |
| 36 | SUN Z Z, HAYES C A, SHIN J, et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology[J]. Journal of Visualized Experiments: JoVE, 2013(79): e50762. |
| 37 | CALHOUN K A, SWARTZ J R. An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates[J]. Biotechnology Progress, 2005, 21(4): 1146-1153. |
| 38 | LIU D V, ZAWADA J F, SWARTZ J R. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis[J]. Biotechnology Progress, 2005, 21(2): 460-465. |
| 39 | JEWETT M C, CALHOUN K A, VOLOSHIN A, et al. An integrated cell-free metabolic platform for protein production and synthetic biology[J]. Molecular Systems Biology, 2008, 4(1): 220. |
| 40 | SMOLSKAYA S, LOGASHINA Y A, ANDREEV Y A. Escherichia coli extract-based cell-free expression system as an alternative for difficult-to-obtain protein biosynthesis[J]. International Journal of Molecular Sciences, 2020, 21(3): 928. |
| 41 | KWON Y C, JEWETT M C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis[J]. Scientific Reports, 2015, 5: 8663. |
| 42 | DIDOVYK A, TONOOKA T, TSIMRING L, et al. Rapid and scalable preparation of bacterial lysates for cell-free gene expression[J]. ACS Synthetic Biology, 2017, 6(12): 2198-2208. |
| 43 | SILVERMAN A D, KARIM A S, JEWETT M C. Cell-free gene expression: an expanded repertoire of applications[J]. Nature Reviews Genetics, 2020, 21(3): 151-170. |
| 44 | GÜTTEL R. Chemical process technology. von J. A. Moulijn, M. Makkee, A. E. van Diepen[J]. Chemie Ingenieur Technik, 2014, 86(5): 585. |
| 45 | HÖÖK M, TANG X. Depletion of fossil fuels and anthropogenic climate change—a review[J]. Energy Policy, 2013, 52: 797-809. |
| 46 | CHAE T U, CHOI S Y, KIM J W, et al. Recent advances in systems metabolic engineering tools and strategies[J]. Current Opinion in Biotechnology, 2017, 47: 67-82. |
| 47 | CHUBUKOV V, MUKHOPADHYAY A, PETZOLD C J, et al. Synthetic and systems biology for microbial production of commodity chemicals[J]. NPJ Systems Biology and Applications, 2016, 2: 16009. |
| 48 | STEPHANOPOULOS G. Challenges in engineering microbes for biofuels production[J]. Science, 2007, 315(5813): 801-804. |
| 121 | LIU R, FU A S, DENG Z X, et al. Promising methods for detection of novel coronavirus SARS-CoV-2[J]. View, 2020, 1(1): e4. |
| 122 | RAMACHANDRAN N, RAPHAEL J V, HAINSWORTH E, et al. Next-generation high-density self-assembling functional protein arrays[J]. Nature Methods, 2008, 5(6): 535-538. |
| 123 | HE M Y, TAUSSIG M J. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method)[J]. Nucleic Acids Research, 2001, 29(15): e73. |
| 124 | BHIDE M, NATARAJAN S, HRESKO S, et al. Rapid in vitro protein synthesis pipeline: a promising tool for cost-effective protein array design[J]. Molecular BioSystems, 2014, 10(6): 1236-1245. |
| 125 | HE M Y, LIU H, TURNER M, et al. Detection of protein-protein interactions by ribosome display and protein in situ immobilisation[J]. New Biotechnology, 2009, 26(6): 277-281. |
| 126 | HE M Y, STOEVESANDT O, TAUSSIG M J. In situ synthesis of protein arrays[J]. Current Opinion in Biotechnology, 2008, 19(1): 4-9. |
| 127 | LEE K H, KWON Y C, YOO S J, et al. Ribosomal synthesis and in situ isolation of peptide molecules in a cell-free translation system[J]. Protein Expression and Purification, 2010, 71(1): 16-20. |
| 128 | LEE K H, LEE K Y, BYUN J Y, et al. On-bead expression of recombinant proteins in an agarose gel matrix coated on a glass slide[J]. Lab on a Chip, 2012, 12(9): 1605-1610. |
| 129 | BYUN J Y, LEE K H, LEE K Y, et al. In-gel expression and in situ immobilization of proteins for generation of three dimensional protein arrays in a hydrogel matrix[J]. Lab on a Chip, 2013, 13(5): 886-891. |
| 130 | KARIG D K, BESSLING S, THIELEN P, et al. Preservation of protein expression systems at elevated temperatures for portable therapeutic production[J]. Journal of the Royal Society, Interface, 2017, 14(129): 20161039. |
| 131 | BENÍTEZ-MATEOS A I, LLARENA I, SÁNCHEZ-IGLESIAS A, et al. Expanding one-pot cell-free protein synthesis and immobilization for on-demand manufacturing of biomaterials[J]. ACS Synthetic Biology, 2018, 7(3): 875-884. |
| 132 | MISIRLI G, NGUYEN T, MCLAUGHLIN J A, et al. A computational workflow for the automated generation of models of genetic designs[J]. ACS Synthetic Biology, 2019, 8(7): 1548-1559. |
| 49 | HASUNUMA T, OKAZAKI F, OKAI N, et al. A review of enzymes and microbes for lignocellulosic biorefinery and the possibility of their application to consolidated bioprocessing technology[J]. Bioresource Technology, 2013, 135: 513-522. |
| 50 | NIELSEN C, RAHMAN A, REHMAN A U, et al. Food waste conversion to microbial polyhydroxyalkanoates[J]. Microbial Biotechnology, 2017, 10(6): 1338-1352. |
| 51 | STRONG P J, LAYCOCK B, MAHAMUD S N S, et al. The opportunity for high-performance biomaterials from methane[J]. Microorganisms, 2016, 4(1): 11. |
| 52 | KELWICK R J R, WEBB A J, FREEMONT P S. Biological materials: the next frontier for cell-free synthetic biology[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 399. |
| 53 | KELWICK R, RICCI L, CHEE S M, et al. Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics[J]. Synthetic Biology, 2018, 3(1): ysy016. |
| 54 | PETROLL K, KOPP D, CARE A, et al. Tools and strategies for constructing cell-free enzyme pathways[J]. Biotechnology Advances, 2019, 37(1): 91-108. |
| 55 | KOPP D, WILLOWS R D, SUNNA A. Cell-free enzymatic conversion of spent coffee grounds into the platform chemical lactic acid[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 389. |
| 56 | CASTRO-AGUIRRE E, IÑIGUEZ-FRANCO F, SAMSUDIN H, et al. Poly(lactic acid)—mass production, processing, industrial applications, and end of life[J]. Advanced Drug Delivery Reviews, 2016, 107: 333-366. |
| 57 | OPGENORTH P H, KORMAN T P, BOWIE J U. A synthetic biochemistry module for production of bio-based chemicals from glucose[J]. Nature Chemical Biology, 2016, 12(6): 393-395. |
| 58 | ULLAH M W, UL-ISLAM M, KHAN S, et al. Innovative production of bio-cellulose using a cell-free system derived from a single cell line[J]. Carbohydrate Polymers, 2015, 132: 286-294. |
| 59 | GREEN E M. Fermentative production of butanol—the industrial perspective[J]. Current Opinion in Biotechnology, 2011, 22(3): 337-343. |
| 60 | ENDOH T, KANAI T, SATO Y T, et al. Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile[J]. Journal of Biotechnology, 2006, 126(2): 186-195. |
| 133 | LIAN J Z, JIN R, ZHAO H M. Construction of plasmids with tunable copy numbers in Saccharomyces cerevisiae and their applications in pathway optimization and multiplex genome integration[J]. Biotechnology and Bioengineering, 2016, 113(11): 2462-2473. |
| 134 | DU J, YUAN Y B, SI T, et al. Customized optimization of metabolic pathways by combinatorial transcriptional engineering[J]. Nucleic Acids Research, 2012, 40(18): e142. |
| 135 | JIANG L H, ZHAO J R, LIAN J Z, et al. Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology[J]. Synthetic and Systems Biotechnology, 2018, 3(2): 90-96. |
| 136 | MOORE S J, MACDONALD J T, WIENECKE S, et al. Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(19): E4340-E4349. |
| 137 | KOPNICZKY M B, CANAVAN C, MCCLYMONT D W, et al. Cell-free protein synthesis as a prototyping platform for mammalian synthetic biology[J]. ACS Synthetic Biology, 2020, 9(1): 144-156. |
| 138 | SWANK Z, LAOHAKUNAKORN N, MAERKL S J. Cell-free gene-regulatory network engineering with synthetic transcription factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(13): 5892-5901. |
| 139 | ZHANG Y, MINAGAWA Y, KIZOE H, et al. Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array[J]. Science Advances, 2019, 5(8): eaav8185. |
| 140 | SAWASAKI T, OGASAWARA T, MORISHITA R, et al. A cell-free protein synthesis system for high-throughput proteomics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(23): 14652-14657. |
| 141 | KANTER G, YANG J H, VOLOSHIN A, et al. Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines[J]. Blood, 2007, 109(8): 3393-3399. |
| 142 | FAN J Z, VILLARREAL F, WEYERS B, et al. Multi-dimensional studies of synthetic genetic promoters enabled by microfluidic impact printing[J]. Lab on a Chip, 2017, 17(13): 2198-2207. |
| 143 | CONTRERAS-LLANO L E, TAN C. High-throughput screening of biomolecules using cell-free gene expression systems[J]. Synthetic Biology, 2018, 3(1): ysy012. |
| 144 | GUBERNATIS J E, LOOKMAN T. Machine learning in materials design and discovery: examples from the present and suggestions for the future[J]. Physical Review Materials, 2018, 2(12): 120301. |
| 61 | KRUTSAKORN B, HONDA K, YE X T, et al. In vitro production of n-butanol from glucose[J]. Metabolic Engineering, 2013, 20: 84-91. |
| 62 | YE X T, HONDA K, MORIMOTO Y, et al. Direct conversion of glucose to malate by synthetic metabolic engineering[J]. Journal of Biotechnology, 2013, 164(1): 34-40. |
| 63 | YOUNT N Y, YEAMAN M R. Immunocontinuum: perspectives in antimicrobial peptide mechanisms of action and resistance[J]. Protein and Peptide Letters, 2005, 12(1): 49-67. |
| 64 | XU M, LEWIS R V. Structure of a protein superfiber: spider dragline silk[J]. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(18): 7120-7124. |
| 65 | SAHIN KEHRIBAR E, ISILAK M E, BOZKURT E U, et al. Engineering of biofilms with a glycosylation circuit for biomaterial applications[J]. Biomaterials Science, 2021, 9(10): 3650-3661. |
| 66 | PEREZ J G, STARK J C, JEWETT M C. Cell-free synthetic biology: engineering beyond the cell[J]. Cold Spring Harbor Perspectives in Biology, 2016, 8(12): a023853. |
| 67 | CHONG S R. Overview of cell-free protein synthesis: historic landmarks, commercial systems, and expanding applications[J]. Current Protocols in Molecular Biology, 2014, 108: 16.30.1-16.3011. |
| 68 | SALEHI A S M, SMITH M T, BENNETT A M, et al. Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system[J]. Biotechnology Journal, 2016, 11(2): 274-281. |
| 69 | ORTH J H C, SCHORCH B, BOUNDY S, et al. Cell-free synthesis and characterization of a novel cytotoxic pierisin-like protein from the cabbage butterfly Pieris rapae [J]. Toxicon, 2011, 57(2): 199-207. |
| 70 | MARTEMYANOV K A, SHIROKOV V A, KURNASOV O V, et al. Cell-free production of biologically active polypeptides: application to the synthesis of antibacterial peptide cecropin[J]. Protein Expression and Purification, 2001, 21(3): 456-461. |
| 71 | CHEN H Q, FAN L M, XU Z N, et al. Efficient production of soluble human beta-defensin-3-4 fusion proteins in Escherichia coli cell-free system[J]. Process Biochemistry, 2007, 42(3): 423-428. |
| 72 | MU J B, AWADALLA P, DUAN J H, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome[J]. Nature Genetics, 2007, 39(1): 126-130. |
| 145 | GAO W, CHO E, LIU Y Y, et al. Advances and challenges in cell-free incorporation of unnatural amino acids into proteins[J]. Frontiers in Pharmacology, 2019, 10: 611. |
| 146 | HONG S H, NTAI I, HAIMOVICH A D, et al. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation[J]. ACS Synthetic Biology, 2014, 3(6): 398-409. |
| 147 | MANKOWSKA S A, GATTI-LAFRANCONI P, CHODORGE M, et al. A shorter route to antibody binders via quantitative in vitro bead-display screening and consensus analysis[J]. Scientific Reports, 2016, 6: 36391. |
| 148 | ALBAYRAK C, SWARTZ J R. Direct polymerization of proteins[J]. ACS Synthetic Biology, 2014, 3(6): 353-362. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 李石开, 曾东鳌, 杜方舟, 张京钟, 余爽. 血管化类器官的构建方法及生物材料[J]. 合成生物学, 2024, 5(4): 851-866. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||