合成生物学 ›› 2024, Vol. 5 ›› Issue (3): 447-473.DOI: 10.12211/2096-8280.2023-086
基因组挖掘指导天然药物分子的发现
奚萌宇1,2, 胡逸灵1, 顾玉诚3, 戈惠明1
- 1.南京大学生命科学学院,医药生物技术全国重点实验室,江苏 南京 210023
2.南京大学化学化工学院,江苏 南京 210023
3.先正达Jealott’s Hill国际研发中心,英国,伯克郡,布拉克内尔 RG42 6EY
-
收稿日期:2023-11-28修回日期:2024-02-20出版日期:2024-06-30发布日期:2024-07-12 -
通讯作者:戈惠明 -
作者简介:奚萌宇 (1995—),女,博士研究生。研究方向为解析放线菌来源天然产物生物合成途径和机制;基因组挖掘发现新颖天然产物。E-mail:dg20240120@smail.nju.edu.cn胡逸灵 (1989—),男,博士,博士后。研究方向为天然产物的基因组挖掘和人工智能在天然产物发现中的应用。E-mail:huyiling10@163.com戈惠明 (1980—),男,教授,博士生导师。研究方向为挖掘微生物中新型药源分子;解析重要微生物活性分子的生物合成途径和机制;工程改造新型生物催化剂;合成生物学智造高值化学品。E-mail:hmge@nju.edu.cn -
基金资助:国家重点研发计划(2018YFA0902000);国家自然科学基金(81925033)
Genome mining-directed discovery for natural medicinal products
XI Mengyu1,2, HU Yiling1, GU Yucheng3, GE Huiming1
- 1.State Key Laboratory of Pharmaceutical Biotechnology,School of Life Sciences,Nanjing University,Nanjing 210023,Jiangsu,China
2.School of Chemistry and Chemical Engineering,Nanjing University,Nanjing 210023,Jiangsu,China
3.Syngenta Jealott’s Hill International Research Centre,Bracknell RG42 6EY,Berkshire,UK
-
Received:2023-11-28Revised:2024-02-20Online:2024-06-30Published:2024-07-12 -
Contact:GE Huiming
摘要:
天然产物是临床药物的主要来源,也是新药研发过程中先导化合物结构设计和优化的灵感源泉。但传统策略天然药源分子的发现却遭遇了瓶颈,新颖天然产物的数量逐渐无法满足现代药物开发的需求和应对全球多药耐药的威胁。随着测序技术的快速迭代,生物学的研究进入了基因组时代,基因组挖掘指导天然产物定向发现的策略得以确立,成功摆脱了传统天然产物发现策略对于生物样本生物量的依赖,极大提高了活性天然产物发现的特异性和成功率。本文简述了基因组挖掘以及相关数据库和生物信息学工具的发展,详细介绍了包括基于核心基因或后修饰基因的经典挖掘手段,自抗性机制、进化理论指导的基因组挖掘和人工智能在活性天然产物发现中的具体应用,并对基因组挖掘在药物发现和多学科交叉领域的影响和发展进行了展望。基因组信息中蕴藏着无可估量的化学潜能,促进基因组挖掘与其他学科间的交叉融合,提升对遗传信息的处理和分析能力,增强下游基因簇表达通量和产物结构预测能力,可实现天然小分子高通量、高新颖性和高效率的发现,为开发具有自主知识产权的新药物、新化学品和新型酶催化剂服务。
中图分类号:
引用本文
奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473.
XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products[J]. Synthetic Biology Journal, 2024, 5(3): 447-473.
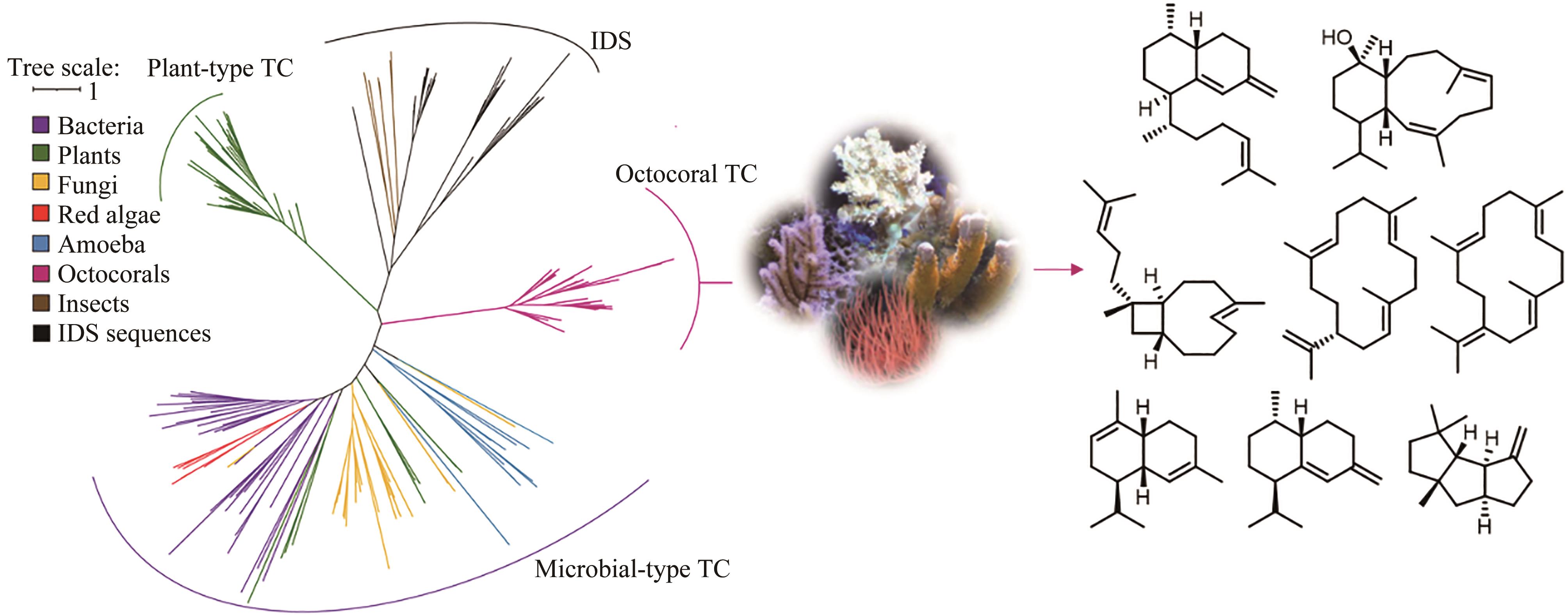
图4 针对萜类合酶的基因组挖掘发现八放珊瑚来源的萜类[74]
Fig. 4 Discovery of octocoral terpene cyclases and natural products synthesized by the enzymes through the genome mining[74]
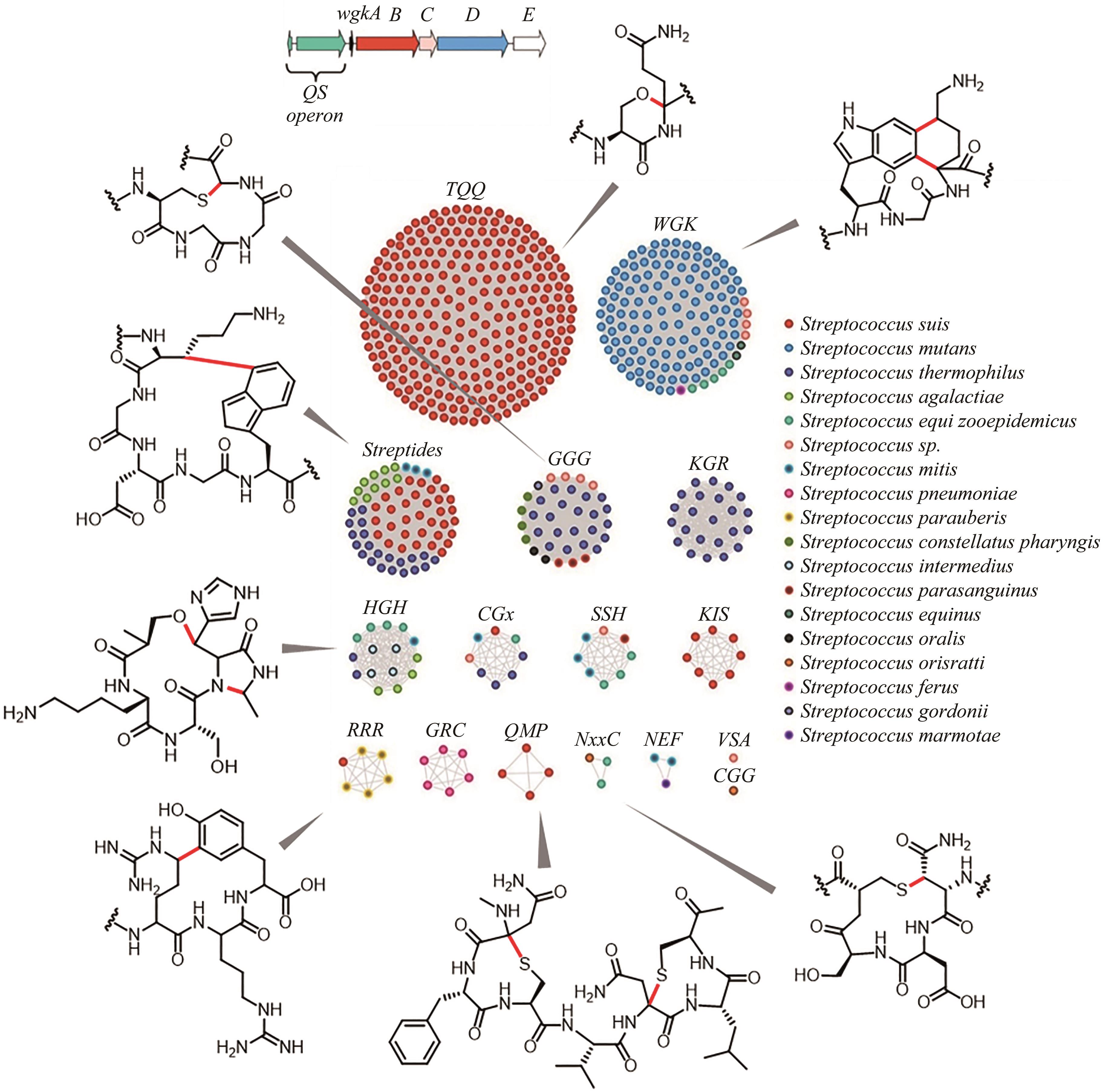
图8 RaS-RiPPs序列相似性网络和对应的交联产物[111-115](已表征类群的rSAM酶催化形成的交联由红色标注)
Fig. 8 Sequence similarity network analysis of RaS-RiPPs and the discovery of the cross-linked products [111-115](red: new bonds formed by rSAM enzymes)
| 1 | MEDEMA M H, DE ROND T, MOORE B S. Mining genomes to illuminate the specialized chemistry of life[J]. Nature Reviews Genetics, 2021, 22(9): 553-571. |
| 2 | BUCAR F, WUBE A, SCHMID M. Natural product isolation: how to get from biological material to pure compounds[J]. Natural Product Reports, 2013, 30(4): 525-545. |
| 3 | KATZ L, BALTZ R H. Natural product discovery: past, present, and future[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(2-3): 155-176. |
| 4 | THOMFORD N E, SENTHEBANE D A, ROWE A, et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery[J]. International Journal of Molecular Sciences, 2018, 19(6): 1578. |
| 5 | SMITH D J, BURNHAM M K, EDWARDS J, et al. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillium-Chrysogenum [J]. Bio/technology, 1990, 8(1): 39-41. |
| 6 | TOBERT J A. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors[J]. Nature Reviews Drug Discovery, 2003, 2(7): 517-526. |
| 7 | ROWINSKY E K, DONEHOWER R C. Drug-Therapy-Paclitaxel (taxol)[J]. The New England Journal of Medicine, 1995, 332(15): 1004-1014. |
| 8 | TU Y Y. Artemisinin - a gift from traditional Chinese medicine to the world (Nobel lecture)[J]. Angewandte Chemie International Edition, 2016, 55(35): 10210-10226. |
| 9 | PANTER F, BADER C D, MÜLLER R. Synergizing the potential of bacterial genomics and metabolomics to find novel antibiotics[J]. Chemical Science, 2021, 12(17): 5994-6010. |
| 10 | CHALLIS G L, HOPWOOD D A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(): 14555-14561. |
| 11 | MALPARTIDA F, HOPWOOD D A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host[J]. Nature, 1984, 309(5967): 462-464. |
| 12 | KEATINGE-CLAY A T. The structures of type I polyketide synthases[J]. Natural Product Reports, 2012, 29(10): 1050-1073. |
| 13 | SÜSSMUTH R D, MAINZ A. Nonribosomal peptide synthesis — principles and prospects[J]. Angewandte Chemie International Edition, 2017, 56(14): 3770-3821. |
| 14 | ZIEMERT N, ALANJARY M, WEBER T. The evolution of genome mining in microbes - a review[J]. Natural Product Reports, 2016, 33(8): 988-1005. |
| 15 | GAVRIILIDOU A, KAUTSAR S A, ZABURANNYI N, et al. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes[J]. Nature Microbiology, 2022, 7(5): 726-735. |
| 16 | BENTLEY S D, CHATER K F, CERDEÑO-TÁRRAGA A M, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2)[J]. Nature, 2002, 417(6885): 141-147. |
| 17 | IKEDA H, ISHIKAWA J, HANAMOTO A, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis [J]. Nature Biotechnology, 2003, 21(5): 526-531. |
| 18 | SANGER F, COULSON A R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase[J]. Journal of Molecular Biology, 1975, 94(3): 441-448. |
| 19 | International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome[J]. Nature, 2001, 409(6822): 860-921. |
| 20 | METZKER M L. Sequencing technologies—the next generation[J]. Nature Reviews Genetics, 2010, 11: 31-46. |
| 21 | DE COSTER W, WEISSENSTEINER M H, SEDLAZECK F J. Towards population-scale long-read sequencing[J]. Nature Reviews Genetics, 2021, 22(9): 572-587. |
| 22 | SAYERS E W, CAVANAUGH M, CLARK K, et al. GenBank 2023 update[J]. Nucleic Acids Research, 2023, 51(D1): D141-D144. |
| 23 | O’LEARY N A, WRIGHT M W, BRISTER J R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation[J]. Nucleic Acids Research, 2016, 44(D1): D733-D745. |
| 24 | NORDBERG H, CANTOR M, DUSHEYKO S, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates[J]. Nucleic Acids Research, 2014, 42(D1): D26-D31. |
| 25 | KITTS P A, CHURCH D M, THIBAUD-NISSEN F, et al. Assembly: a resource for assembled genomes at NCBI[J]. Nucleic Acids Research, 2016, 44(D1): D73-D80. |
| 26 | TURNBAUGH P J, LEY R E, HAMADY M, et al. The human microbiome project[J]. Nature, 2007, 449(7164): 804-810. |
| 27 | The Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project[J]. Nature, 2019, 569(7758): 641-648. |
| 28 | DONIA M S, CIMERMANCIC P, SCHULZE C J, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics[J]. Cell, 2014, 158(6): 1402-1414. |
| 29 | GUO C J, CHANG F Y, WYCHE T P, et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases[J]. Cell, 2017, 168(3): 517-526. e18. |
| 30 | PAOLI L, RUSCHEWEYH H J, FORNERIS C C, et al. Biosynthetic potential of the global ocean microbiome[J]. Nature, 2022, 607(7917): 111-118. |
| 31 | The UniProt Consortium. UniProt: the universal protein knowledgebase in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D480-D489. |
| 32 | FINN R D, COGGILL P, EBERHARDT R Y, et al. The Pfam protein families database: towards a more sustainable future[J]. Nucleic Acids Research, 2016, 44(D1): D279-D285. |
| 33 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D412-D419. |
| 34 | BLUM M, CHANG H Y, CHUGURANSKY S, et al. The InterPro protein families and domains database: 20 years on[J]. Nucleic Acids Research, 2021, 49(D1): D344-D354. |
| 35 | KAUTSAR S A, BLIN K, SHAW S, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function[J]. Nucleic Acids Research, 2020, 48(D1): D454-D458. |
| 36 | BLIN K, SHAW S, AUGUSTIJN H E, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation[J]. Nucleic Acids Research, 2023, 51(W1): W46-W50. |
| 37 | BLIN K, SHAW S, MEDEMA M H, et al. The antiSMASH database version 4: additional genomes and BGCs, new sequence-based searches and more[J]. Nucleic Acids Research, 2024, 52(D1): D586-D589. |
| 38 | CAMACHO C, COULOURIS G, AVAGYAN V, et al. BLAST+: architecture and applications[J]. BMC Bioinformatics, 2009, 10: 421. |
| 39 | ALTSCHUL S F, MADDEN T L, SCHÄFFER A A, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389-3402. |
| 40 | LI W Z, GODZIK A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences[J]. Bioinformatics, 2006, 22(13): 1658-1659. |
| 41 | STARCEVIC A, ZUCKO J, SIMUNKOVIC J, et al. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures[J]. Nucleic Acids Research, 2008, 36(21): 6882-6892. |
| 42 | WEBER T, RAUSCH C, LOPEZ P, et al. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters[J]. Journal of Biotechnology, 2009, 140(1-2): 13-17. |
| 43 | LI M H T, UNG P M U, ZAJKOWSKI J, et al. Automated genome mining for natural products[J]. BMC Bioinformatics, 2009, 10: 185. |
| 44 | SKINNIDER M A, DEJONG C A, REES P N, et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM)[J]. Nucleic Acids Research, 2015, 43(20): 9645-9662. |
| 45 | TIETZ J I, SCHWALEN C J, PATEL P S, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape[J]. Nature Chemical Biology, 2017, 13(5): 470-478. |
| 46 | KLOOSTERMAN A M, SHELTON K E, VAN WEZEL G P, et al. RRE-Finder: a genome-mining tool for class-independent RiPP discovery[J]. mSystems, 2020, 5(5): e00267-20. |
| 47 | MERWIN N J, MOUSA W K, DEJONG C A, et al. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(1): 371-380. |
| 48 | SUGIMOTO Y, CAMACHO F R, WANG S, et al. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome[J]. Science, 2019, 366(6471): eaax9176. |
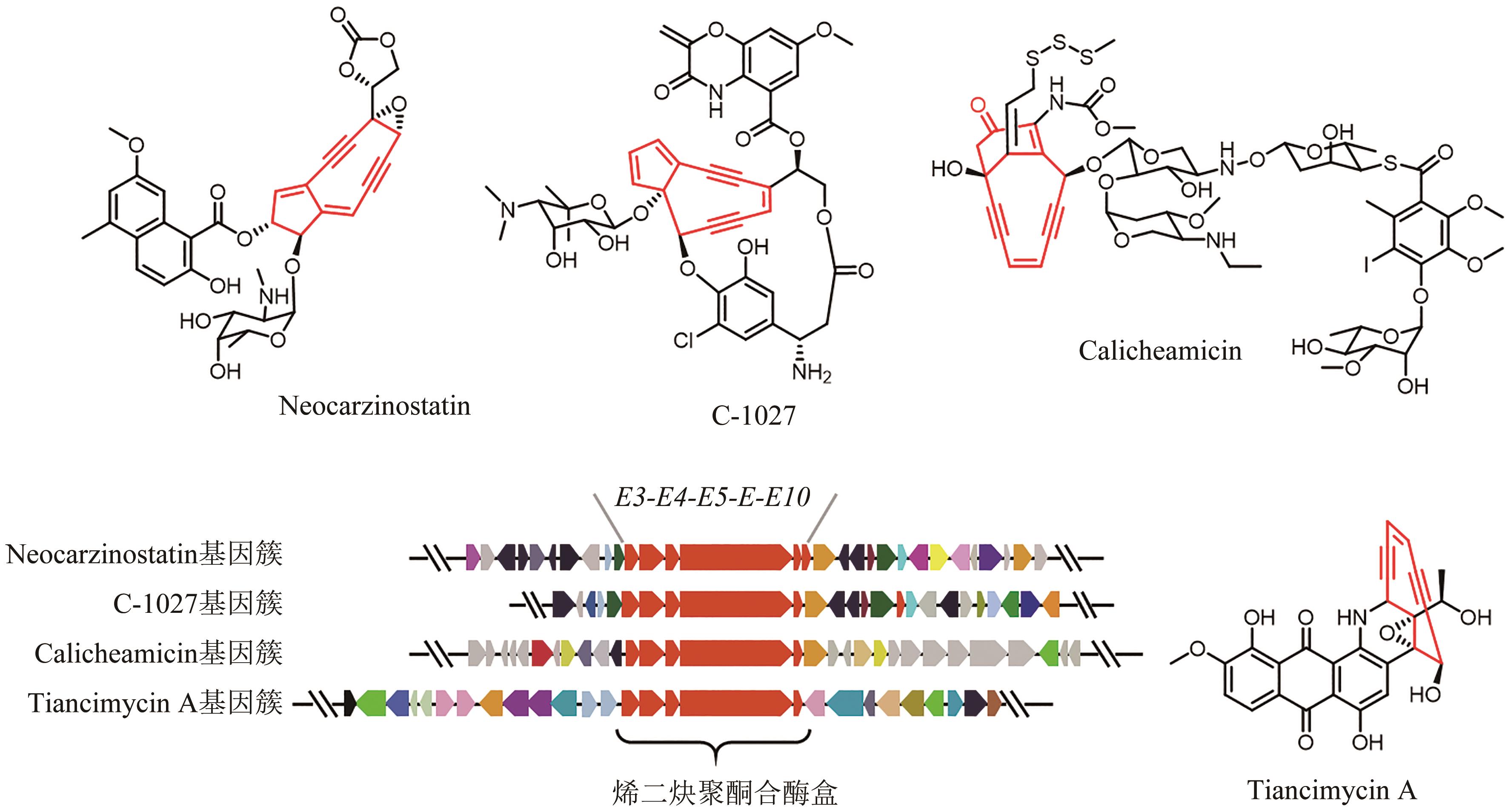
| 49 | SHEN B, HINDRA, YAN X H, et al. Enediynes: exploration of microbial genomics to discover new anticancer drug leads[J]. Bioorganic & Medicinal Chemistry Letters, 2015, 25(1): 9-15. |
| 50 | ADHIKARI A, SHEN B, RADER C. Challenges and opportunities to develop enediyne natural products as payloads for antibody-drug conjugates[J]. Antibody Therapeutics, 2021, 4(1): 1-15. |
| 51 | RUDOLF J D, YAN X H, SHEN B. Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(2-3): 261-276. |
| 52 | HINDRA, HUANG T T, YANG D, et al. Strain prioritization for natural product discovery by a high-throughput real-time PCR method[J]. Journal of Natural Products, 2014, 77(10): 2296-2303. |
| 53 | YAN X H, GE H M, HUANG T T, et al. Strain prioritization and genome mining for enediyne natural products[J]. mBio, 2016, 7(6): e02104-e02116. |
| 54 | GUTIÉRREZ-CHÁVEZ C, BENAUD N, FERRARI B C. The ecological roles of microbial lipopeptides: where are we going?[J]. Computational and Structural Biotechnology Journal, 2021, 19: 1400-1413. |
| 55 | ZHANG S B, MUKHERJI R, CHOWDHURY S, et al. Lipopeptide-mediated bacterial interaction enables cooperative predator defense[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(6): e2013759118. |
| 56 | CHU J, VILA-FARRES X, INOYAMA D, et al. Discovery of MRSA active antibiotics using primary sequence from the human microbiome[J]. Nature Chemical Biology, 2016, 12(12): 1004-1006. |
| 57 | BISWAS S, BRUNEL J M, DUBUS J C, et al. Colistin: an update on the antibiotic of the 21st century[J]. Expert Review of Anti-Infective Therapy, 2012, 10(8): 917-934. |
| 58 | LIU Y Y, WANG Y, WALSH T R, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study[J]. The Lancet Infectious Diseases, 2016, 16(2): 161-168. |
| 59 | WANG Z Q, KOIRALA B, HERNANDEZ Y, et al. A naturally inspired antibiotic to target multidrug-resistant pathogens[J]. Nature, 2022, 601(7894): 606-611. |
| 60 | VILA-FARRES X, CHU J, INOYAMA D, et al. Antimicrobials inspired by nonribosomal peptide synthetase gene clusters[J]. Journal of the American Chemical Society, 2017, 139(4): 1404-1407. |
| 61 | CHU J, VILA-FARRES X, BRADY S F. Bioactive synthetic-bioinformatic natural product cyclic peptides inspired by nonribosomal peptide synthetase gene clusters from the human microbiome[J]. Journal of the American Chemical Society, 2019, 141(40): 15737-15741. |
| 62 | CHU J, KOIRALA B, FORELLI N, et al. Synthetic-bioinformatic natural product antibiotics with diverse modes of action[J]. Journal of the American Chemical Society, 2020, 142(33): 14158-14168. |
| 63 | PECK S C, VAN DER DONK W A. Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology[J]. Current Opinion in Chemical Biology, 2013, 17(4): 580-588. |
| 64 | PETKOWSKI J J, BAINS W, SEAGER S. Natural products containing “rare” organophosphorus functional groups[J]. Molecules, 2019, 24(5): 866. |
| 65 | CHIN J P, MCGRATH J W, QUINN J P. Microbial transformations in phosphonate biosynthesis and catabolism, and their importance in nutrient cycling[J]. Current Opinion in Chemical Biology, 2016, 31: 50-57. |
| 66 | SHIRAISHI T, KUZUYAMA T. Biosynthetic pathways and enzymes involved in the production of phosphonic acid natural products[J]. Bioscience, Biotechnology, and Biochemistry, 2021, 85(1): 42-52. |
| 67 | JU K S, GAO J T, DOROGHAZI J R, et al. Discovery of phosphonic acid natural products by mining the genomes of 10 000 actinomycetes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(39): 12175-12180. |
| 68 | DICKSCHAT J S. Bacterial diterpene biosynthesis[J]. Angewandte Chemie International Edition, 2019, 58(45): 15964-15976. |
| 69 | DONG L B, RUDOLF J D, DENG M R, et al. Discovery of the tiancilactone antibiotics by genome mining of atypical bacterial Type Ⅱ diterpene synthases[J]. ChemBioChem, 2018: 19(16): 1727-1733. |
| 70 | YAMADA Y, KUZUYAMA T, KOMATSU M, et al. Terpene synthases are widely distributed in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(3): 857-862. |
| 71 | HU Y L, ZHANG Q, LIU S H, et al. Building Streptomyces albus as a chassis for synthesis of bacterial terpenoids[J]. Chemical Science, 2023, 14(13): 3661-3667. |
| 72 | LÜ C Y, CHEN T, QIANG B, et al. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean[J]. Nucleic Acids Research, 2021, 49(D1): D509-D515. |
| 73 | BARSBY T, KUBANEK J. Isolation and structure elucidation of feeding deterrent diterpenoids from the sea pansy, Renilla reniformis [J]. Journal of Natural Products, 2005, 68(4): 511-516. |
| 74 | BURKHARDT I, DE ROND T, CHEN P Y T, et al. Ancient plant-like terpene biosynthesis in corals[J]. Nature Chemical Biology, 2022, 18(6): 664-669. |
| 75 | SCESA P D, LIN Z J, SCHMIDT E W. Ancient defensive terpene biosynthetic gene clusters in the soft corals[J]. Nature Chemical Biology, 2023, 19(6): 790. |
| 76 | LEELANANDA S P, LINDERT S. Computational methods in drug discovery[J]. Beilstein Journal of Organic Chemistry, 2016, 12: 2694-2718. |
| 77 | GIORDANO D, BIANCANIELLO C, ARGENIO M A, et al. Drug design by pharmacophore and virtual screening approach[J]. Pharmaceuticals, 2022, 15(5): 646. |
| 78 | ZHU H J, ZHANG B, WANG L, et al. Redox modifications in the biosynthesis of alchivemycin A enable the formation of its key pharmacophore[J]. Journal of the American Chemical Society, 2021, 143(12): 4751-4757. |
| 79 | HOWAT S, PARK B, OH I S, et al. Paclitaxel: biosynthesis, production and future prospects[J]. New Biotechnology, 2014, 31(3): 242-245. |
| 80 | WALSH C T. The chemical versatility of natural-product assembly lines[J]. Accounts of Chemical Research, 2008, 41(1): 4-10. |
| 81 | WALSH C T, WENCEWICZ T A. Flavoenzymes: versatile catalysts in biosynthetic pathways[J]. Natural Product Reports, 2013, 30(1): 175-200. |
| 82 | WANG P, GAO X, TANG Y. Complexity generation during natural product biosynthesis using redox enzymes[J]. Current Opinion in Chemical Biology, 2012, 16(3-4): 362-369. |
| 83 | HERNANDES M Z, CAVALCANTI S M, MOREIRA D R, et al. Halogen atoms in the modern medicinal chemistry: hints for the drug design[J]. Current Drug Targets, 2010, 11(3): 303-314. |
| 84 | MONDAL S, RAJA K, SCHWEIZER U, et al. Chemistry and biology in the biosynthesis and action of thyroid hormones[J]. Angewandte Chemie International Edition, 2016, 55(27): 7606-7630. |
| 85 | HARRIS C M, KANNAN R, KOPECKA H, et al. The role of the chlorine substituents in the antibiotic vancomycin: preparation and characterization of mono- and didechlorovancomycin[J]. Journal of the American Chemical Society, 1985, 107(23): 6652-6658. |
| 86 | GROLL M, HUBER R, POTTS B C M. Crystal structures of Salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding[J]. Journal of the American Chemical Society, 2006, 128(15): 5136-5141. |
| 87 | LATHAM J, BRANDENBURGER E, SHEPHERD S A, et al. Development of halogenase enzymes for use in synthesis[J]. Chemical Reviews, 2018, 118(1): 232-269. |
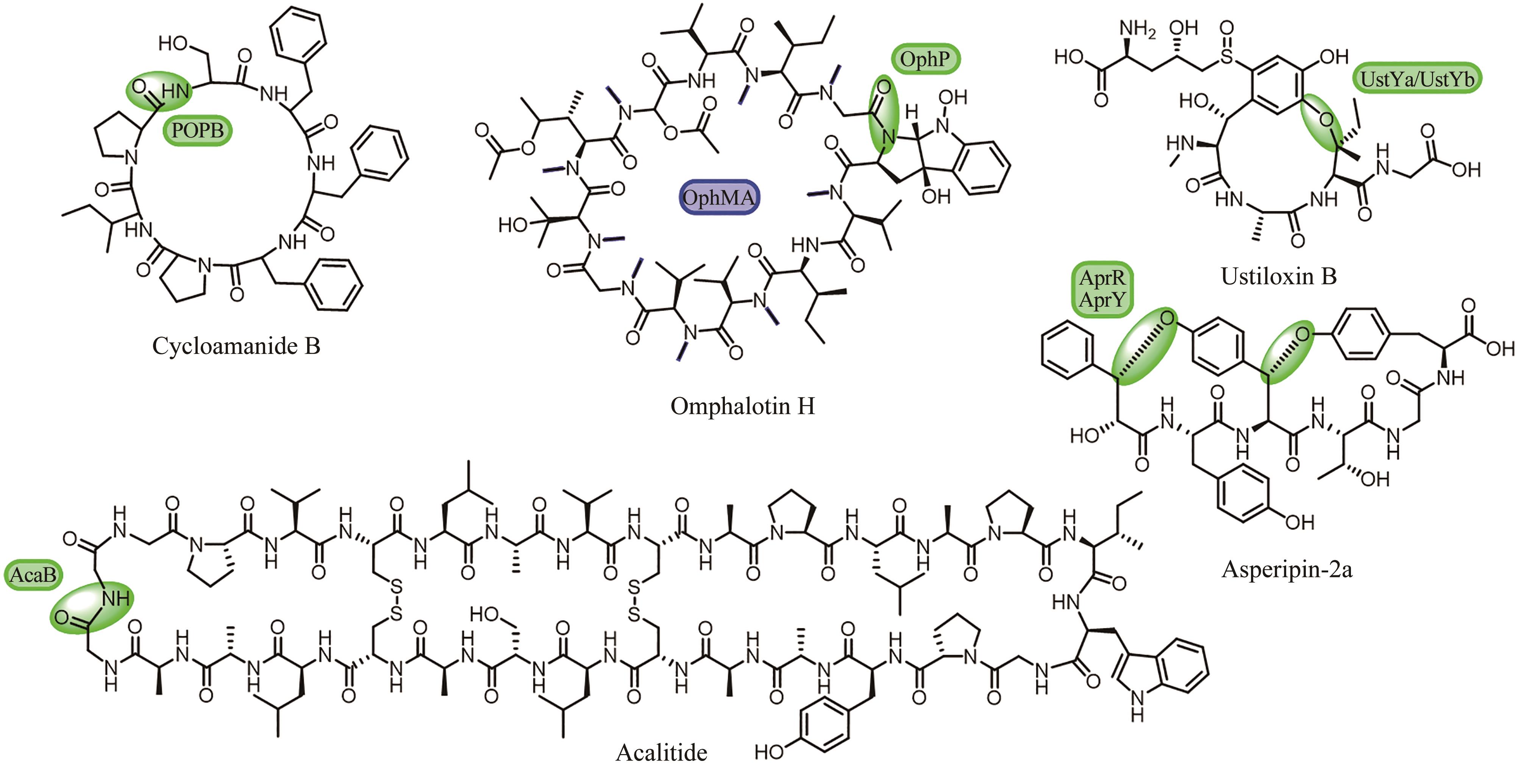
| 88 | HORNUNG A, BERTAZZO M, DZIARNOWSKI A, et al. A genomic screening approach to the structure-guided identification of drug candidates from natural sources[J]. ChemBioChem, 2007, 8(7): 757-766. |
| 89 | LUO M N, WANG M Y, CHANG S S, et al. Halogenase-targeted genome mining leads to the discovery of (±) pestalachlorides A1a, A2a, and their atropisomers[J]. Antibiotics, 2022, 11(10): 1304. |
| 90 | DENG H, MA L, BANDARANAYAKA N, et al. Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining[J]. ChemBioChem, 2014, 15(3): 364-368. |
| 91 | REICH H J, HONDAL R J. Why nature chose selenium[J]. ACS Chemical Biology, 2016, 11(4): 821-841. |
| 92 | KAYROUZ C M, HUANG J, HAUSER N, et al. Biosynthesis of selenium-containing small molecules in diverse microorganisms[J]. Nature, 2022, 610(7930): 199-204. |
| 93 | WOLFE M D, AHMED F, LACOURCIERE G M, et al. Functional diversity of the rhodanese homology domain: the Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase[J]. The Journal of Biological Chemistry, 2004, 279(3): 1801-1809. |
| 94 | FORCHHAMMER K, BÖCK A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence[J]. Journal of Biological Chemistry, 1991, 266(10): 6324-6328. |
| 95 | EHRENREICH A, FORCHHAMMER K, TORMAY P, et al. Selenoprotein synthesis in E. coli. Purification and characterisation of the enzyme catalysing selenium activation[J]. European Journal of Biochemistry, 1992, 206(3): 767-773. |
| 96 | SEEBECK F P. In vitro reconstitution of mycobacterial ergothioneine biosynthesis[J]. Journal of the American Chemical Society, 2010, 132(19): 6632-6633. |
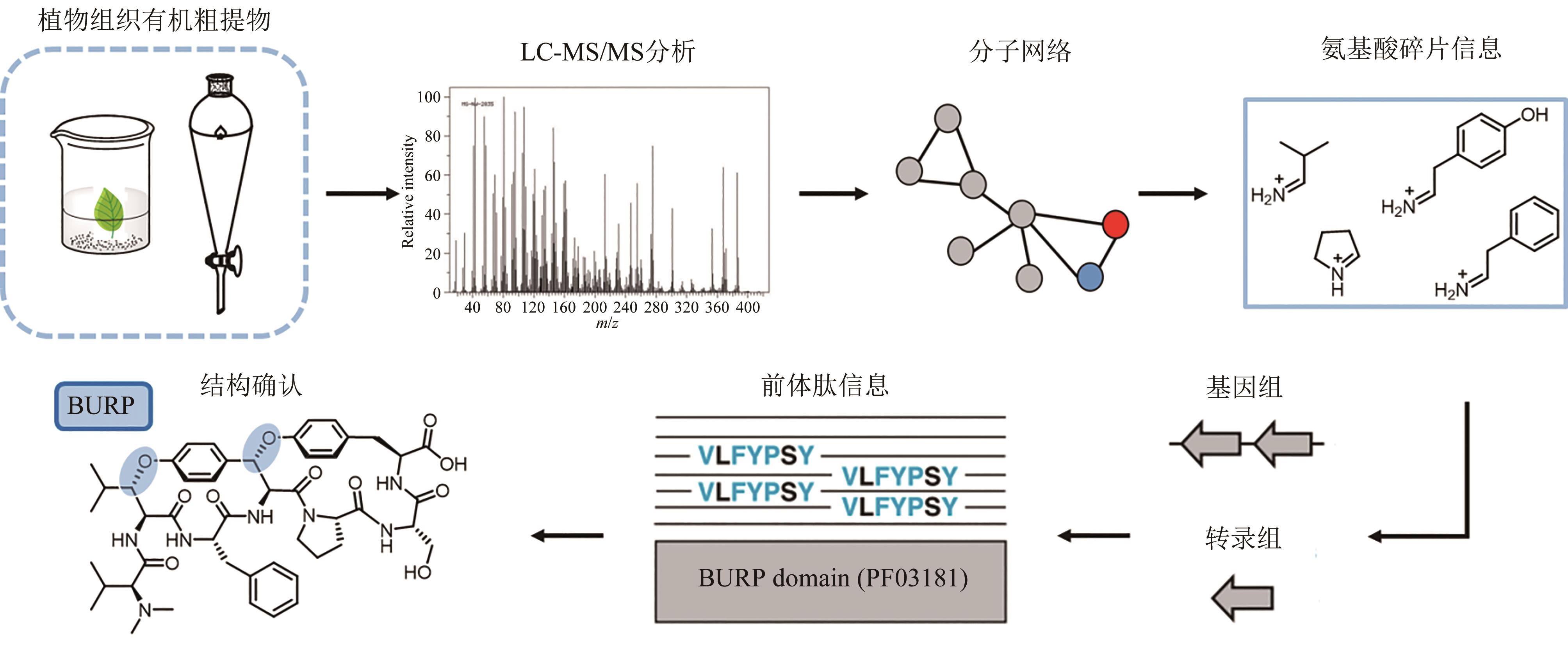
| 97 | PAUL N P, GALVÁN A E, YOSHINAGA-SAKURAI K, et al. Arsenic in medicine: past, present and future[J]. Biometals, 2023, 36(2): 283-301. |
| 98 | CHEN J, ROSEN B P. The arsenic methylation cycle: how microbial communities adapted methylarsenicals for use as weapons in the continuing war for dominance[J]. Frontiers in Environmental Science, 2020, 8: 43. |
| 99 | HOSHINO S, IJICHI S, ASAMIZU S, et al. Insights into arsenic secondary metabolism in actinomycetes from the structure and biosynthesis of bisenarsan[J]. Journal of the American Chemical Society, 2023, 145(32): 17863-17871. |
| 100 | HE H Y, NIIKURA H, DU Y L, et al. Synthetic and biosynthetic routes to nitrogen-nitrogen bonds[J]. Chemical Society Reviews, 2022, 51(8): 2991-3046. |
| 101 | FU D, CALVO J A, SAMSON L D. Balancing repair and tolerance of DNA damage caused by alkylating agents[J]. Nature Reviews Cancer, 2012, 12(2): 104-120. |
| 102 | WANG M H, NIIKURA H, HE H Y, et al. Biosynthesis of the N-N-bond-containing compound L-alanosine[J]. Angewandte Chemie International Edition, 2020, 59(10): 3881-3885. |
| 103 | BROBERG A, MENKIS A, VASILIAUSKAS R. Kutznerides 1-4, depsipeptides from the actinomycete Kutzneria sp. 744 inhabiting mycorrhizal roots of Picea abies seedlings[J]. Journal of Natural Products, 2006, 69(1): 97-102. |
| 104 | DU Y L, HE H Y, HIGGINS M A, et al. A heme-dependent enzyme forms the nitrogen-nitrogen bond in piperazate[J]. Nature Chemical Biology, 2017, 13(8): 836-838. |
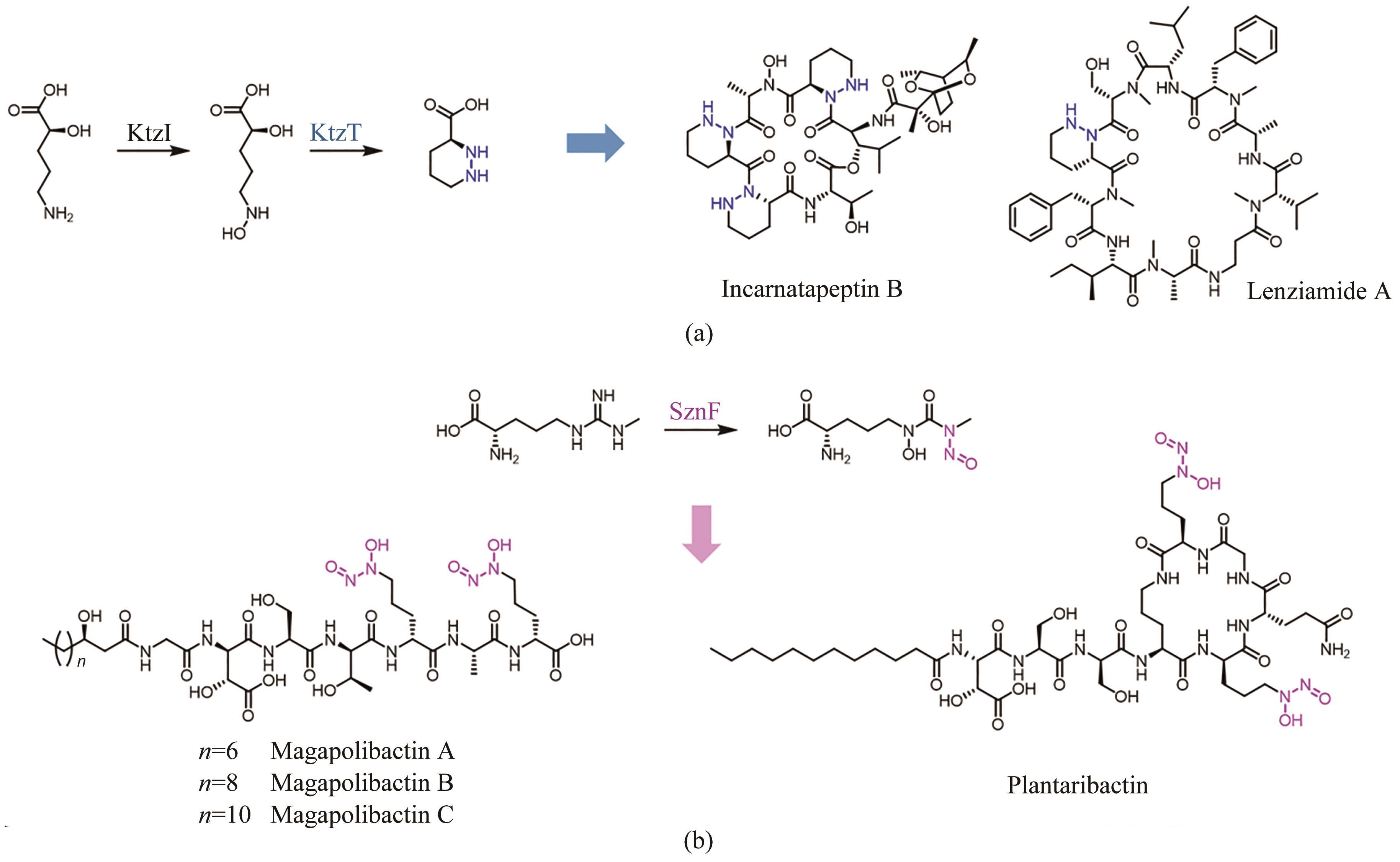
| 105 | MORGAN K D, WILLIAMS D E, PATRICK B O, et al. Incarnatapeptins A and B, nonribosomal peptides discovered using genome mining and 1H/15N HSQC-TOCSY[J]. Organic Letters, 2020, 22(11): 4053-4057. |
| 106 | SHIN D, BYUN W S, KANG S, et al. Targeted and logical discovery of piperazic acid-bearing natural products based on genomic and spectroscopic signatures[J]. Journal of the American Chemical Society, 2023, 145(36): 19676-19690. |
| 107 | NG T L, ROHAC R, MITCHELL A J, et al. An N-nitrosating metalloenzyme constructs the pharmacophore of streptozotocin[J]. Nature, 2019, 566(7742): 94-99. |
| 108 | HERMENAU R, MEHL J L, ISHIDA K, et al. Genomics-driven discovery of NO-donating diazeniumdiolate siderophores in diverse plant-associated bacteria[J]. Angewandte Chemie International Edition, 2019, 58(37): 13024-13029. |
| 109 | BRODERICK J B, DUFFUS B R, DUSCHENE K S, et al. Radical S-adenosylmethionine enzymes[J]. Chemical Reviews, 2014, 114(8): 4229-4317. |
| 110 | HUDSON G A, BURKHART B J, DICAPRIO A J, et al. Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides[J]. Journal of the American Chemical Society, 2019, 141(20): 8228-8238. |
| 111 | BUSHIN L B, CLARK K A, PELCZER I, et al. Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif[J]. Journal of the American Chemical Society, 2018, 140(50): 17674-17684. |
| 112 | CARUSO A, MARTINIE R J, BUSHIN L B, et al. Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes[J]. Journal of the American Chemical Society, 2019, 141(42): 16610-16614. |
| 113 | CLARK K A, BUSHIN L B, SEYEDSAYAMDOST M R. Aliphatic ether bond formation expands the scope of radical SAM enzymes in natural product biosynthesis[J]. Journal of the American Chemical Society, 2019, 141(27): 10610-10615. |
| 114 | CARUSO A, BUSHIN L B, CLARK K A, et al. Radical approach to enzymatic β-thioether bond formation[J]. Journal of the American Chemical Society, 2019, 141(2): 990-997. |
| 115 | BUSHIN L B, COVINGTON B C, RUED B E, et al. Discovery and biosynthesis of streptosactin, a sactipeptide with an alternative topology encoded by commensal bacteria in the human microbiome[J]. Journal of the American Chemical Society, 2020, 142(38): 16265-16275. |
| 116 | KESSLER S C, CHOOI Y H. Out for a RiPP: challenges and advances in genome mining of ribosomal peptides from fungi[J]. Natural Product Reports, 2022, 39(2): 222-230. |
| 117 | HALLEN H E, LUO H, SCOTT-CRAIG J S, et al. Gene family encoding the major toxins of lethal Amanita mushrooms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(48): 19097-19101. |
| 118 | PULMAN J A, CHILDS K L, SGAMBELLURI R M, et al. Expansion and diversification of the MSDIN family of cyclic peptide genes in the poisonous agarics Amanita phalloides and A. bisporigera [J]. BMC Genomics, 2016, 17(1): 1038. |
| 119 | QUIJANO M R, ZACH C, MILLER F S, et al. Distinct autocatalytic α-N-methylating precursors expand the borosin RiPP family of peptide natural products[J]. Journal of the American Chemical Society, 2019, 141(24): 9637-9644. |
| 120 | YE Y, MINAMI A, IGARASHI Y, et al. Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi[J]. Angewandte Chemie International Edition, 2016, 55(28): 8072-8075. |
| 121 | NAGANO N, UMEMURA M, IZUMIKAWA M, et al. Class of cyclic ribosomal peptide synthetic genes in filamentous fungi[J]. Fungal Genetics and Biology, 2016, 86: 58-70. |
| 122 | TONG Z W, XIE X H, GE H M, et al. Disulfide bridge-targeted metabolome mining unravels an antiparkinsonian peptide[J]. Acta Pharmaceutica Sinica B, 2024, 14(2): 881-892. |
| 123 | CRAIK D J, DALY N L, BOND T, et al. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif[J]. Journal of Molecular Biology, 1999, 294(5): 1327-1336. |
| 124 | BARBER C J, PUJARA P T, REED D W, et al. The two-step biosynthesis of cyclic peptides from linear precursors in a member of the plant family Caryophyllaceae involves cyclization by a serine protease-like enzyme[J]. The Journal of Biological Chemistry, 2013, 288(18): 12500-12510. |
| 125 | KERSTEN R D, WENG J K. Gene-guided discovery and engineering of branched cyclic peptides in plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(46): E10961-E10969. |
| 126 | KERSTEN R D, MYDY L S, FALLON T R, et al. Gene-guided discovery and ribosomal biosynthesis of moroidin peptides[J]. Journal of the American Chemical Society, 2022, 144(17): 7686-7692. |
| 127 | HATTORI J, BOUTILIER K A, VAN LOOKEREN CAMPAGNE M M, et al. A conserved BURP domain defines a novel group of plant proteins with unusual primary structures[J]. Molecular & General Genetics, 1998, 259(4): 424-428. |
| 128 | CHIGUMBA D N, MYDY L S, DE WAAL F, et al. Discovery and biosynthesis of cyclic plant peptides via autocatalytic cyclases[J]. Nature Chemical Biology, 2022, 18(1): 18-28. |
| 129 | KAUTSAR S A, SUAREZ DURAN H G, BLIN K, et al. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters[J]. Nucleic Acids Research, 2017, 45(W1): W55-W63. |
| 130 | MOUSA J J, BRUNER S D. Structural and mechanistic diversity of multidrug transporters[J]. Natural Product Reports, 2016, 33(11): 1255-1267. |
| 131 | TENCONI E, RIGALI S. Self-resistance mechanisms to DNA-damaging antitumor antibiotics in Actinobacteria[J]. Current Opinion in Microbiology, 2018, 45: 100-108. |
| 132 | TOOKE C L, HINCHLIFFE P, BRAGGINTON E C, et al. β-Lactamases and β-lactamase inhibitors in the 21st century[J]. Journal of Molecular Biology, 2019, 431(18): 3472-3500. |
| 133 | WEISBLUM B. Insights into erythromycin action from studies of its activity as inducer of resistance[J]. Antimicrobial Agents and Chemotherapy, 1995, 39(4): 797-805. |
| 134 | YAN Y, LIU N, TANG Y. Recent developments in self-resistance gene directed natural product discovery[J]. Natural Product Reports, 2020, 37(7): 879-892. |
| 135 | THAKER M N, WANG W L, SPANOGIANNOPOULOS P, et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance[J]. Nature Biotechnology, 2013, 31(10): 922-927. |
| 136 | TANG X Y, LI J, MILLÁN-AGUIÑAGA N, et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining[J]. ACS Chemical Biology, 2015, 10(12): 2841-2849. |
| 137 | CULP E J, SYCHANTHA D, HOBSON C, et al. ClpP inhibitors are produced by a widespread family of bacterial gene clusters[J]. Nature Microbiology, 2022, 7(3): 451-462. |
| 138 | LI L Y, HU Y L, SUN J L, et al. Resistance and phylogeny guided discovery reveals structural novelty of tetracycline antibiotics[J]. Chemical Science, 2022, 13(43): 12892-12898. |
| 139 | ZHANG W, ZHANG X, FENG D D, et al. Discovery of a unique flavonoid biosynthesis mechanism in fungi by genome mining[J]. Angewandte Chemie International Edition, 2023, 62(12): e202215529. |
| 140 | ALANJARY M, KRONMILLER B, ADAMEK M, et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery[J]. Nucleic Acids Research, 2017, 45(W1): W42-W48. |
| 141 | MUNGAN M D, ALANJARY M, BLIN K, et al. ARTS 2.0: feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining[J]. Nucleic Acids Research, 2020, 48(W1): W546-W552. |
| 142 | MUNGAN M D, BLIN K, ZIEMERT N. ARTS-DB: a database for antibiotic resistant targets[J]. Nucleic Acids Research, 2022, 50(D1): D736-D740. |
| 143 | KANG H S. Phylogeny-guided (meta)genome mining approach for the targeted discovery of new microbial natural products[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(2): 285-293. |
| 144 | CHEN S C, ZHANG C, ZHANG L H. Investigation of the molecular landscape of bacterial aromatic polyketides by global analysis of type II polyketide synthases[J]. Angewandte Chemie International Edition, 2022, 61(24): e202202286. |
| 145 | HELFRICH E J N, PIEL J. Biosynthesis of polyketides by trans-AT polyketide synthases[J]. Natural Product Reports, 2016, 33(2): 231-316. |
| 146 | TETA R, GURGUI M, HELFRICH E J N, et al. Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial Phylum bacteroidetes[J]. ChemBioChem, 2010, 11(18): 2506-2512. |
| 147 | NAKABACHI A, UEOKA R, OSHIMA K, et al. Defensive bacteriome symbiont with a drastically reduced genome[J]. Current Biology, 2013, 23(15): 1478-1484. |
| 148 | KAMPA A, GAGUNASHVILI A N, GULDER T A M, et al. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(33): E3129-E3137. |
| 149 | HELFRICH E J N, UEOKA R, DOLEV A, et al. Automated structure prediction of trans-acyltransferase polyketide synthase products[J]. Nature Chemical Biology, 2019, 15(8): 813-821. |
| 150 | LAZZARO B P, ZASLOFF M, ROLFF J. Antimicrobial peptides: application informed by evolution[J]. Science, 2020, 368(6490): eaau5480. |
| 151 | MA Y, GUO Z Y, XIA B B, et al. Identification of antimicrobial peptides from the human gut microbiome using deep learning[J]. Nature Biotechnology, 2022, 40(6): 921-931. |
| 152 | ARNISON P G, BIBB M J, BIERBAUM G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature[J]. Natural Product Reports, 2013, 30(1): 108-160. |
| 153 | SHEN B. A new golden age of natural products drug discovery[J]. Cell, 2015, 163(6): 1297-1300. |
| 154 | LI J W H, VEDERAS J C. Drug discovery and natural products: end of an era or an endless frontier?[J]. Science, 2009, 325(5937): 161-165. |
| 155 | BLIN K, SHAW S, STEINKE K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline[J]. Nucleic Acids Research, 2019, 47(W1): W81-W87. |
| 156 | The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease[J]. Cell Host & Microbe, 2014, 16(3): 276-289. |
| 157 | TREVATHAN-TACKETT S M, SHERMAN C D H, HUGGETT M J, et al. A horizon scan of priorities for coastal marine microbiome research[J]. Nature Ecology & Evolution, 2019, 3(11): 1509-1520. |
| 158 | XIONG X Y, GOU J B, LIAO Q G, et al. The Taxus genome provides insights into paclitaxel biosynthesis[J]. Nature Plants, 2021, 7(8): 1026-1036. |
| 159 | ZHANG Y J, WIESE L, FANG H, et al. Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis[J]. Molecular Plant, 2023, 16(12): 1951-1961. |
| 160 | ZHAO Y, LIANG F Y, XIE Y M, et al. Oxetane ring formation in taxol biosynthesis is catalyzed by a bifunctional cytochrome P450 enzyme[J]. Journal of the American Chemical Society, 2024, 146(1): 801-810. |
| 161 | JIANG B, GAO L, WANG H J, et al. Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin Ⅲ[J]. Science, 2024, 383(6683): 622-629. |
| 162 | LI P, YAN M X, LIU P, et al. Multiomics analyses of two Leonurus species illuminate leonurine biosynthesis and its evolution[J]. Molecular Plant, 2024, 17(1): 158-177. |
| 163 | NETT R S, DHO Y, TSAI C, et al. Plant carbonic anhydrase-like enzymes in neuroactive alkaloid biosynthesis[J]. Nature, 2023, 624(7990): 182-191. |
| 164 | MICHELLOD D, BIEN T, BIRGEL D, et al. De novo phytosterol synthesis in animals[J]. Science, 2023, 380(6644): 520-526. |
| 165 | COOKE T F, FISCHER C R, WU P, et al. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars[J]. Cell, 2017, 171(2): 427-439. e21. |
| 166 | GIZZI A S, GROVE T L, ARNOLD J J, et al. A naturally occurring antiviral ribonucleotide encoded by the human genome[J]. Nature, 2018, 558(7711): 610-614. |
| 167 | KIM E, MOORE B S, YOON Y J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology[J]. Nature Chemical Biology, 2015, 11(9): 649-659. |
| 168 | VAN DER HELM E, GENEE H J, SOMMER M O A. The evolving interface between synthetic biology and functional metagenomics[J]. Nature Chemical Biology, 2018, 14(8): 752-759. |
| 169 | SMANSKI M J, ZHOU H, CLAESEN J, et al. Synthetic biology to access and expand nature’s chemical diversity[J]. Nature Reviews Microbiology, 2016, 14(3): 135-149. |
| 170 | FISCHBACH M A, SEGRE J A. Signaling in host-associated microbial communities[J]. Cell, 2016, 164(6): 1288-1300. |
| 171 | FISCHBACH M A. Microbiome: focus on causation and mechanism[J]. Cell, 2018, 174(4): 785-790. |
| 172 | SILPE J E, BALSKUS E P. Deciphering human microbiota-host chemical interactions[J]. ACS Central Science, 2021, 7(1): 20-29. |
| [1] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [2] | 仲泉周, 单依怡, 裴清云, 金艳芸, 王艺涵, 孟璐远, 王歆韵, 张雨鑫, 刘坤媛, 王慧中, 冯尚国. 生物合成法生产α-熊果苷的研究进展[J]. 合成生物学, 2025, 6(1): 118-135. |
| [3] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [4] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [5] | 李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517. |
| [6] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [7] | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| [8] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| [9] | 刘子健, 穆柏杨, 段志强, 王璇, 陆晓杰. 与核酸兼容的化学反应开发进展[J]. 合成生物学, 2024, 5(5): 1102-1124. |
| [10] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| [11] | 谢向前, 郭雯, 王欢, 李进. 含氨基乙烯半胱氨酸核糖体肽的生物合成与化学合成[J]. 合成生物学, 2024, 5(5): 981-996. |
| [12] | 汤志军, 胡友财, 刘文. 酶促4+2和2+2环加成反应:区域与立体选择性的理解与应用[J]. 合成生物学, 2024, 5(3): 401-407. |
| [13] | 张俊, 金诗雪, 云倩, 瞿旭东. 聚酮化合物非天然延伸单元的生物合成与结构改造应用[J]. 合成生物学, 2024, 5(3): 561-570. |
| [14] | 陈锡玮, 张华然, 邹懿. 真菌源非核糖体肽类药物生物合成及代谢工程[J]. 合成生物学, 2024, 5(3): 571-592. |
| [15] | 虞旭昶, 吴辉, 李雷. 文库构建与基因簇靶向筛选驱动的微生物天然产物高效发现[J]. 合成生物学, 2024, 5(3): 492-506. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||