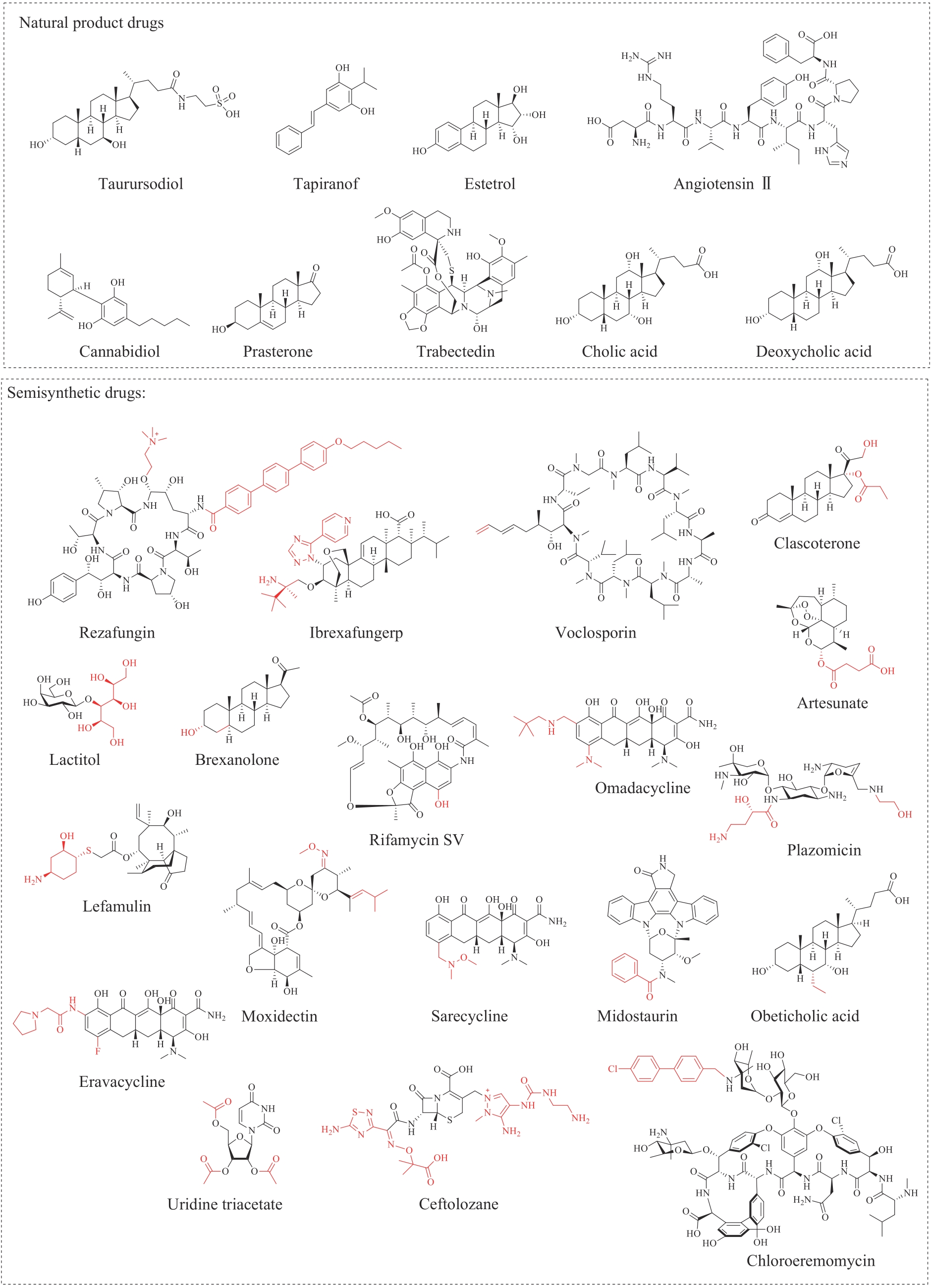
合成生物学 ›› 2024, Vol. 5 ›› Issue (3): 408-446.DOI: 10.12211/2096-8280.2023-092
近十年天然产物药物的生物合成研究进展
冯金1, 潘海学1,2, 唐功利1,2
- 1.中国科学院大学杭州高等研究院,化学与材料科学学院,浙江 杭州 310024
2.中国科学院上海有机化学研究所,生命过程小分子调控全国重点实验室,上海 200032
-
收稿日期:2023-11-30修回日期:2023-12-22出版日期:2024-06-30发布日期:2024-07-12 -
通讯作者:唐功利 -
作者简介:冯金 (1988—),男,博士,博士后。研究方向为活性天然产物的生物合成。E-mail:fengjin@ucas.ac.cn唐功利 (1971—),男,博士,研究员。研究方向为天然产物生物合成和化学生物学。E-mail:gltang@sioc.ac.cn -
基金资助:国家自然科学基金(32070065)
Research advances in biosynthesis of natural product drugs within the past decade
FENG Jin1, PAN Haixue1,2, TANG Gongli1,2
- 1.School of Chemistry and Materials Science,Hangzhou Institute for Advanced Study,University of Chinese Academy of Sciences,Hangzhou 310024,Zhejiang,China
2.State Key Laboratory of Chemical Biology,Shanghai Institute of Organic Chemistry,Chinese Academy of Sciences,Shanghai 200032,China
-
Received:2023-11-30Revised:2023-12-22Online:2024-06-30Published:2024-07-12 -
Contact:TANG Gongli
摘要:
天然产物一直是潜在的先导药物的重要来源,天然产物及其结构类似物在历史上对疾病治疗做出了重大贡献,特别是对癌症和传染病的治疗。在过去两百年的时间里,天然产物的发现和研究经历了巨大的变化,由传统的分离鉴定为主的经典研究方法转为了基因组时代的多学科组合研究。虽然近二十年发现和挖掘了丰富的活性天然产物,但与自然界中巨大的天然产物合成潜力相比仍有不足,庞大的陆地和海洋天然产物资源尚待开发。同时,与传统的化学合成分子相比,天然产物具有丰富的骨架多样性和结构复杂性,在新药发现中展现了巨大的优势。虽然在天然产物的新药创新方面仍面临着种种挑战,但新的分析技术和挖掘策略的出现有望迎来天然产物发现的新阶段。本文总结了近十年(2014年1月—2023年10月)美国食品药品监督管理局批准成药的天然产物及源自天然产物的半合成药物,并对其中纯天然产物来源分子、重要的半合成天然产物前体的生物合成研究进展进行了详细总结。此外还简要总结了一些FDA批准的老药在过去十年中取得的重要生物合成研究进展。期望通过对成药天然产物生物合成途径及机制的深入理解,为更多天然产物创新药物的发现和研究提供借鉴。
中图分类号:
引用本文
冯金, 潘海学, 唐功利. 近十年天然产物药物的生物合成研究进展[J]. 合成生物学, 2024, 5(3): 408-446.
FENG Jin, PAN Haixue, TANG Gongli. Research advances in biosynthesis of natural product drugs within the past decade[J]. Synthetic Biology Journal, 2024, 5(3): 408-446.
| 批准药物 | 药物前体 | 来源 | 主要功效 | 结构类型 | 批准时间 | 商品名 | 参考文献 |
|---|---|---|---|---|---|---|---|
| Natural product drugs: | |||||||
| 牛磺酸二醇(taurursodiol)* | — | Ursusthibetanus | antiapoptotic | steroid | 2022 | Relyvrio | [ |
| tapinarof* | — | Photorhabdusluminescens | AhR agonist | polyketide | 2022 | Vtama | [ |
| 雌四醇(estetrol)* | — | humans | hormone regulation | steroid | 2021 | Nextstellis | [ |
| 大麻二酚(cannabidiol)* | — | Cannabissativa L. | analgesic, anticonvulsant | polyketide | 2018 | Epidiolex | [ |
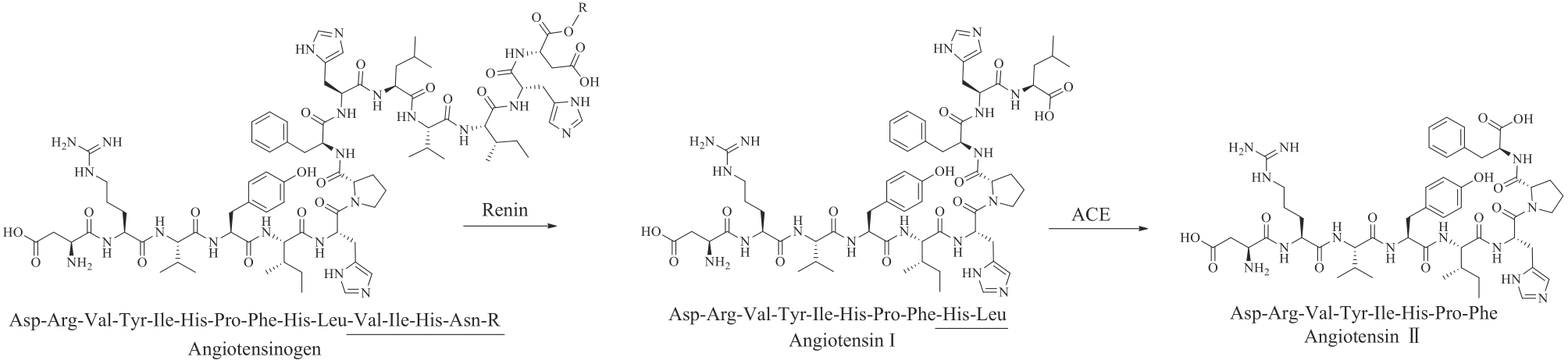
| 血管紧张素(angiotensin)Ⅱ* | — | humans | blood pressure regulation | peptide | 2017 | Giapreza | [ |
| 普拉睾酮(prasterone)* | — | humans | hormone regulation | steroid | 2016 | Intrarosa | [ |
| 曲贝替定(trabectedin)* | — | CandidatusEndoecteinascidiafrumentensis | antitumor | non-ribosomal peptide | 2015 | Yondelis | [ |
| 胆酸(cholic acid)* | — | animals | facilitating fat absorption | steroid | 2015 | Cholbam | [ |
| 去氧胆酸 (deoxycholic acid)* | — | animals | cytolytic agent | steroid | 2015 | Kybella | [ |
| Semisynthetic drugs: | |||||||
| 瑞扎芬净(rezafungin)* | echinocandin | Aspergillusdelacroxii | antifungal | non-ribosomal peptide | 2023 | Rezzayo | [ |
| 艾瑞芬净(ibrexafungerp) | enfumafungin | Hormonemacarpetanum | antifungal | terpene | 2021 | Brexafemme | [ |
| 伏环孢素(voclosporin) | cyclosporine | Beauverianivea | calcineurin inhibitor | non-ribosomal peptide | 2021 | Lupkynis | [ |
| 克拉司酮(clascoterone) | progesterone | humans | topical androgen antagonist | steroid | 2020 | Winlevi | [ |
| 青蒿琥酯(artesunate) | artemisinin | Artemisiaannua | antimalarial | terpene | 2020 | Artesunate | [ |
| 乳糖醇(lactitol) | lactose | animals | osmotic laxative | sugar alcohol | 2020 | Pizensy | [ |
| 来法莫林(lefamulin)* | pleuromutilin | Clitopiluspasseckerianus | antibacterial | terpene | 2019 | Xenleta | [ |
| 布瑞诺龙(brexanolone) | pregnanolone | humans | antidepressant | steroid | 2019 | Zulresso | [ |
| 利福霉素 (rifamycin)SV | rifamycin B | Amycolatopsisrifamycinica | antibacterial | polyketide | 2018 | Aemcolo | [ |
| 奥玛环素(omadacycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Nuzyra | [ |
| 普拉佐米星(plazomicin) | sisomicin | Micromonosporainositola | antibacterial | aminoglycoside | 2018 | Zemdri | [ |
| 莫昔克丁(Moxidectin) | avermectin | Streptomycesavermitilis | antiparasitic | polyketide | 2018 | Moxidectin | [ |
| 艾拉环素(eravacycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Xerava | [ |
| 萨瑞环素(sarecycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Seysara | [ |
| 米哚妥林(midostaurin) | staurosporine | Streptomycesstaurosporeus | antineoplastic | alkaloid | 2017 | Rydapt | [ |
| 奥贝胆酸 (obeticholic acid) | cholic acid | animals | FXR agonist | steroid | 2016 | Ocaliva | [ |
| 尿苷三乙酸酯 (uridine triacetate) | uridine | animals | antidote | nucleoside | 2015 | Xuriden | [ |
| 头孢洛扎(ceftolozane) | cephalosporin | Acremonium | antibacterial | beta-lactam | 2014 | Zerbaxa | [ |
| 奥利万星(oritavancin) | chloroeremomycin | Amycolatopsisorientalis | antibacterial | glycopeptide | 2014 | Orbactiv | [ |
| Earlier FDA-approved drugs: | |||||||
| 奥利司他(orlistat)* | lipstatin | Streptomycestoxytricini | lipase inhibitor | fatty acid | 1999 | Xenical | [ |
| 依托泊苷(etoposide)* | podophyllotoxin | Podophyllumhexandrum | antitumor | lignan | 1983 | Vepesid | [ |
| 长春花碱(vinblastine)* | — | Catharanthusroseus | antitumor | alkaloid | 1965 | Velban | [ |
| 长春新碱(vincristine)* | — | Catharanthusroseus | antitumor | alkaloid | 1963 | Oncovin | [ |
| 秋水仙碱(colchicine)* | — | Gloriosasuperba | anti-inflammatory | alkaloid | 1961 | Colbenemid | [ |
| 吗啡(morphine)* | — | Papaversomniferum | analgesic | alkaloid | 1941 | Morphine sulfate | [ |
| 可待因(codeine)* | — | Papaversomniferum | analgesic | alkaloid | 1950 | Codeine sulfate | [ |
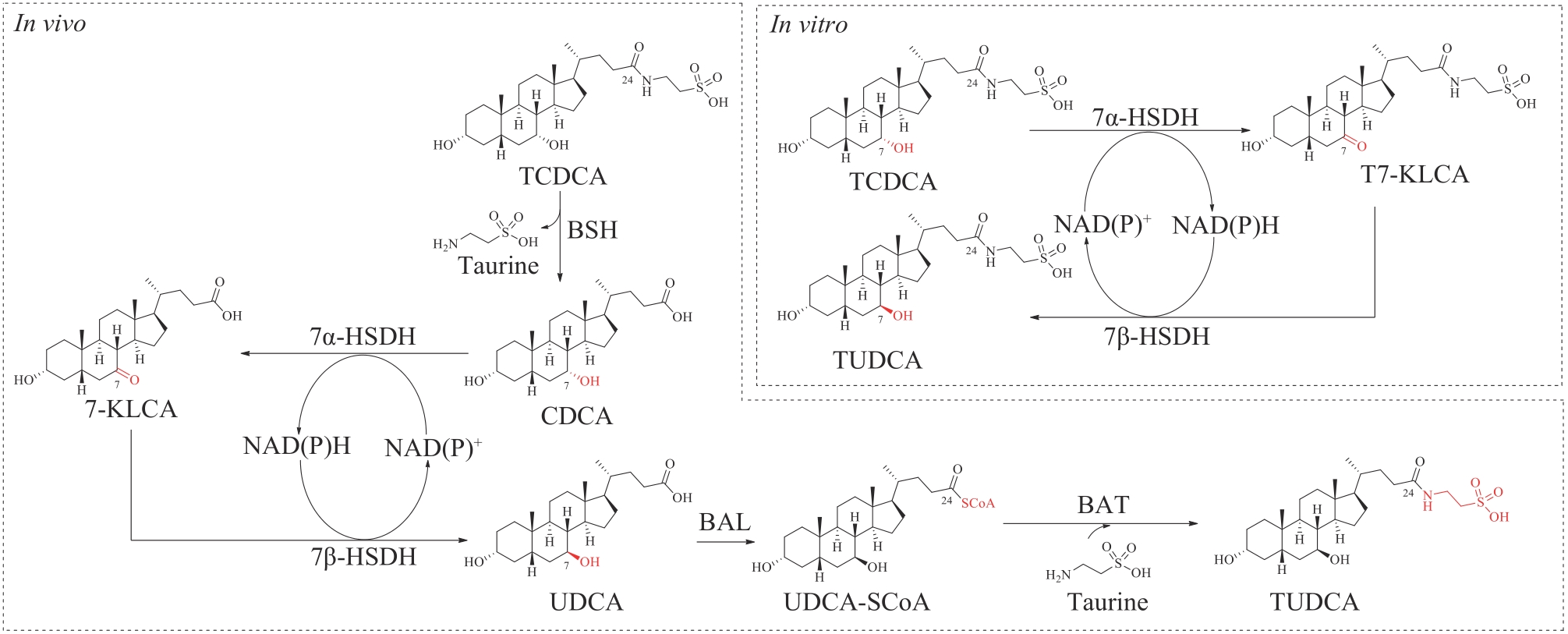
| 东莨菪碱(scopolamine)* | — | Solanaceae | anticholinergic | alkaloid | 1979 | Transderm Scop | [ |
| 维生素(vitamin)B12* | — | Archaea, bacteria | supplement | alkaloid | 1942 | Cyanocobalamin | [ |
| 紫杉醇(paclitaxel)* | — | Taxusbrevifolia | anticancer | terpene | 1992 | Taxol | [ |
表1 FDA批准的天然产物药物和半合成药物总结
Table 1 Summary of FDA-approved natural product drugs and semisynthetic drugs
| 批准药物 | 药物前体 | 来源 | 主要功效 | 结构类型 | 批准时间 | 商品名 | 参考文献 |
|---|---|---|---|---|---|---|---|
| Natural product drugs: | |||||||
| 牛磺酸二醇(taurursodiol)* | — | Ursusthibetanus | antiapoptotic | steroid | 2022 | Relyvrio | [ |
| tapinarof* | — | Photorhabdusluminescens | AhR agonist | polyketide | 2022 | Vtama | [ |
| 雌四醇(estetrol)* | — | humans | hormone regulation | steroid | 2021 | Nextstellis | [ |
| 大麻二酚(cannabidiol)* | — | Cannabissativa L. | analgesic, anticonvulsant | polyketide | 2018 | Epidiolex | [ |
| 血管紧张素(angiotensin)Ⅱ* | — | humans | blood pressure regulation | peptide | 2017 | Giapreza | [ |
| 普拉睾酮(prasterone)* | — | humans | hormone regulation | steroid | 2016 | Intrarosa | [ |
| 曲贝替定(trabectedin)* | — | CandidatusEndoecteinascidiafrumentensis | antitumor | non-ribosomal peptide | 2015 | Yondelis | [ |
| 胆酸(cholic acid)* | — | animals | facilitating fat absorption | steroid | 2015 | Cholbam | [ |
| 去氧胆酸 (deoxycholic acid)* | — | animals | cytolytic agent | steroid | 2015 | Kybella | [ |
| Semisynthetic drugs: | |||||||
| 瑞扎芬净(rezafungin)* | echinocandin | Aspergillusdelacroxii | antifungal | non-ribosomal peptide | 2023 | Rezzayo | [ |
| 艾瑞芬净(ibrexafungerp) | enfumafungin | Hormonemacarpetanum | antifungal | terpene | 2021 | Brexafemme | [ |
| 伏环孢素(voclosporin) | cyclosporine | Beauverianivea | calcineurin inhibitor | non-ribosomal peptide | 2021 | Lupkynis | [ |
| 克拉司酮(clascoterone) | progesterone | humans | topical androgen antagonist | steroid | 2020 | Winlevi | [ |
| 青蒿琥酯(artesunate) | artemisinin | Artemisiaannua | antimalarial | terpene | 2020 | Artesunate | [ |
| 乳糖醇(lactitol) | lactose | animals | osmotic laxative | sugar alcohol | 2020 | Pizensy | [ |
| 来法莫林(lefamulin)* | pleuromutilin | Clitopiluspasseckerianus | antibacterial | terpene | 2019 | Xenleta | [ |
| 布瑞诺龙(brexanolone) | pregnanolone | humans | antidepressant | steroid | 2019 | Zulresso | [ |
| 利福霉素 (rifamycin)SV | rifamycin B | Amycolatopsisrifamycinica | antibacterial | polyketide | 2018 | Aemcolo | [ |
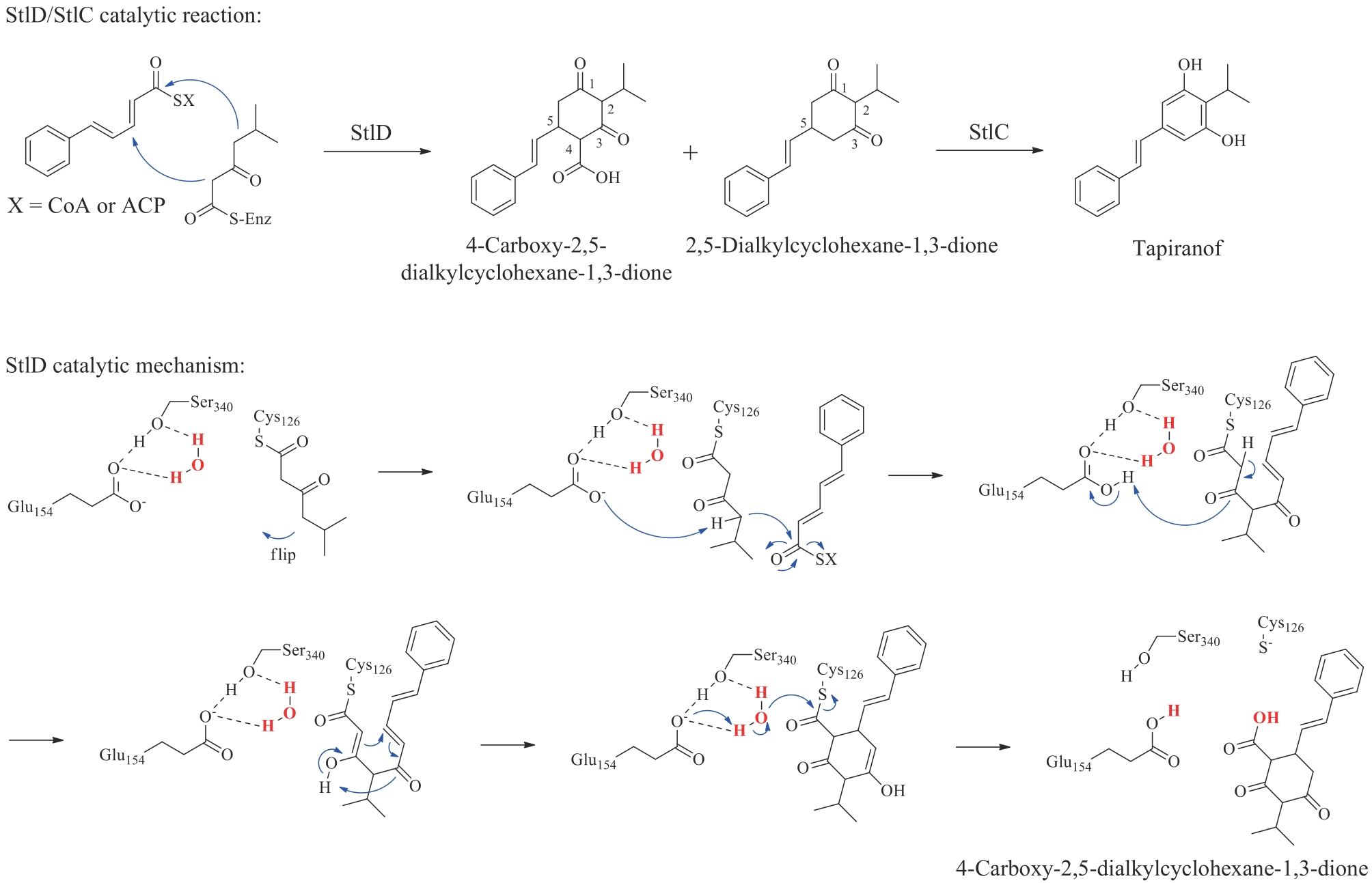
| 奥玛环素(omadacycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Nuzyra | [ |
| 普拉佐米星(plazomicin) | sisomicin | Micromonosporainositola | antibacterial | aminoglycoside | 2018 | Zemdri | [ |
| 莫昔克丁(Moxidectin) | avermectin | Streptomycesavermitilis | antiparasitic | polyketide | 2018 | Moxidectin | [ |
| 艾拉环素(eravacycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Xerava | [ |
| 萨瑞环素(sarecycline) | tetracyclines | Streptomyces | antibacterial | polyketide | 2018 | Seysara | [ |
| 米哚妥林(midostaurin) | staurosporine | Streptomycesstaurosporeus | antineoplastic | alkaloid | 2017 | Rydapt | [ |
| 奥贝胆酸 (obeticholic acid) | cholic acid | animals | FXR agonist | steroid | 2016 | Ocaliva | [ |
| 尿苷三乙酸酯 (uridine triacetate) | uridine | animals | antidote | nucleoside | 2015 | Xuriden | [ |
| 头孢洛扎(ceftolozane) | cephalosporin | Acremonium | antibacterial | beta-lactam | 2014 | Zerbaxa | [ |
| 奥利万星(oritavancin) | chloroeremomycin | Amycolatopsisorientalis | antibacterial | glycopeptide | 2014 | Orbactiv | [ |
| Earlier FDA-approved drugs: | |||||||
| 奥利司他(orlistat)* | lipstatin | Streptomycestoxytricini | lipase inhibitor | fatty acid | 1999 | Xenical | [ |
| 依托泊苷(etoposide)* | podophyllotoxin | Podophyllumhexandrum | antitumor | lignan | 1983 | Vepesid | [ |
| 长春花碱(vinblastine)* | — | Catharanthusroseus | antitumor | alkaloid | 1965 | Velban | [ |
| 长春新碱(vincristine)* | — | Catharanthusroseus | antitumor | alkaloid | 1963 | Oncovin | [ |
| 秋水仙碱(colchicine)* | — | Gloriosasuperba | anti-inflammatory | alkaloid | 1961 | Colbenemid | [ |
| 吗啡(morphine)* | — | Papaversomniferum | analgesic | alkaloid | 1941 | Morphine sulfate | [ |
| 可待因(codeine)* | — | Papaversomniferum | analgesic | alkaloid | 1950 | Codeine sulfate | [ |
| 东莨菪碱(scopolamine)* | — | Solanaceae | anticholinergic | alkaloid | 1979 | Transderm Scop | [ |
| 维生素(vitamin)B12* | — | Archaea, bacteria | supplement | alkaloid | 1942 | Cyanocobalamin | [ |
| 紫杉醇(paclitaxel)* | — | Taxusbrevifolia | anticancer | terpene | 1992 | Taxol | [ |
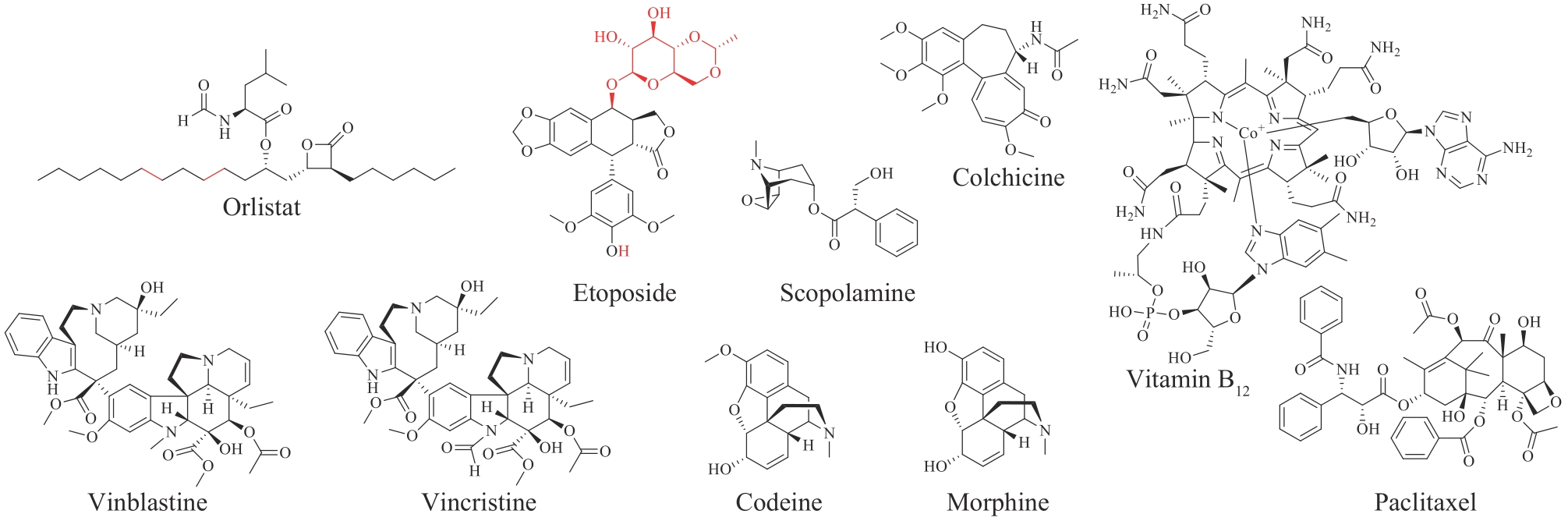
图2 近十年取得重要生物合成进展的FDA批准的老药的化学结构
Fig. 2 Chemical structures of drugs approved earlier by FDA and significant progress in their biosynthesis achieved within the past decade
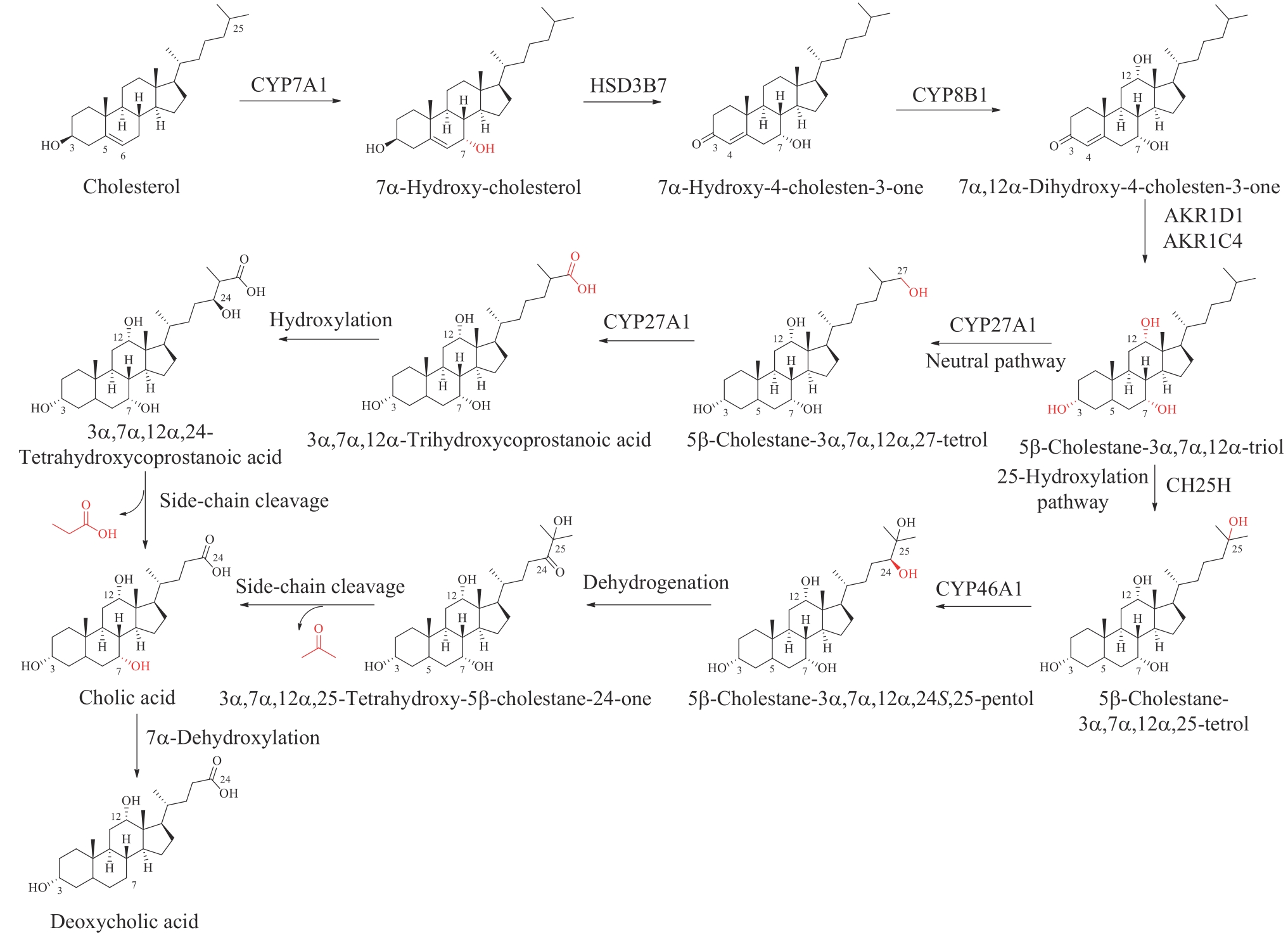
图3 胆酸和去氧胆酸的生物合成CYP7A1—胆固醇7α-羟化酶;HSD3B7—3β-羟基-Δ5-C27-类固醇氧化还原酶;CYP8B1—12α-羟化酶;AKR1D1—Δ5-3-氧代类固醇5β-还原酶;AKR1C4—3α-羟基类固醇脱氢酶;CYP27A1—甾醇27-羟化酶;CH25H—甾醇25-羟化酶;CYP46A1—24β-羟化酶
Fig. 3 Biosynthesis of cholic and deoxycholic acidsCYP7A1—cholesterol 7α-hydroxylase; HSD3B7—3β-hydroxy-Δ5-C27-steroid oxidoreductase; CYP8B1—12α-hydroxylase; AKR1D1—Δ5-3-oxosteroid 5β-reductase; AKR1C4—3α-hydroxysteroid dehydrogenase; CYP27A1—sterol 27-hydroxylase; CH25H—sterol 25-hydroxylase; CYP46A1—24β-hydroxylase
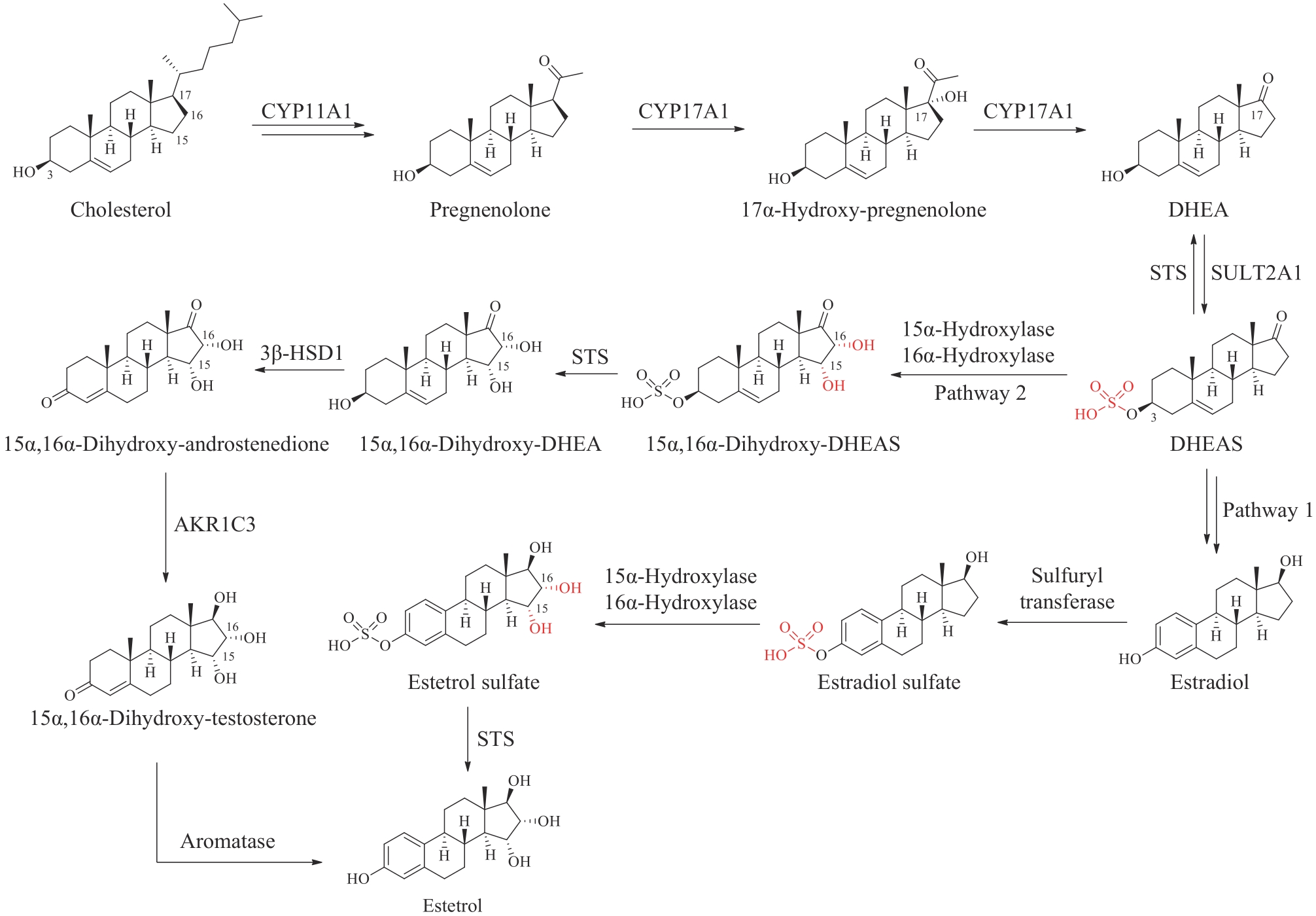
图6 普拉睾酮和雌四醇在人体内的生物合成路径CYP11A1—细胞色素P450胆固醇侧链裂解酶;CYP17A1—17α-羟化酶/17,20-裂解酶;SULT2A1—磺基转移酶;STS—类固醇硫酸酯酶;AKR1C3—醛酮还原酶
Fig. 6 Biosynthetic pathways of prasterone and estetrol in humans CYP11A1—cytochrome P450 cholesterol side chain cleavage enzyme; CYP17A1—17α-hydroxylase/17,20-lyase; SULT2A1—sulfotransferase; STS—steroid sulfatase; AKR1C3—aldo/keto reductase
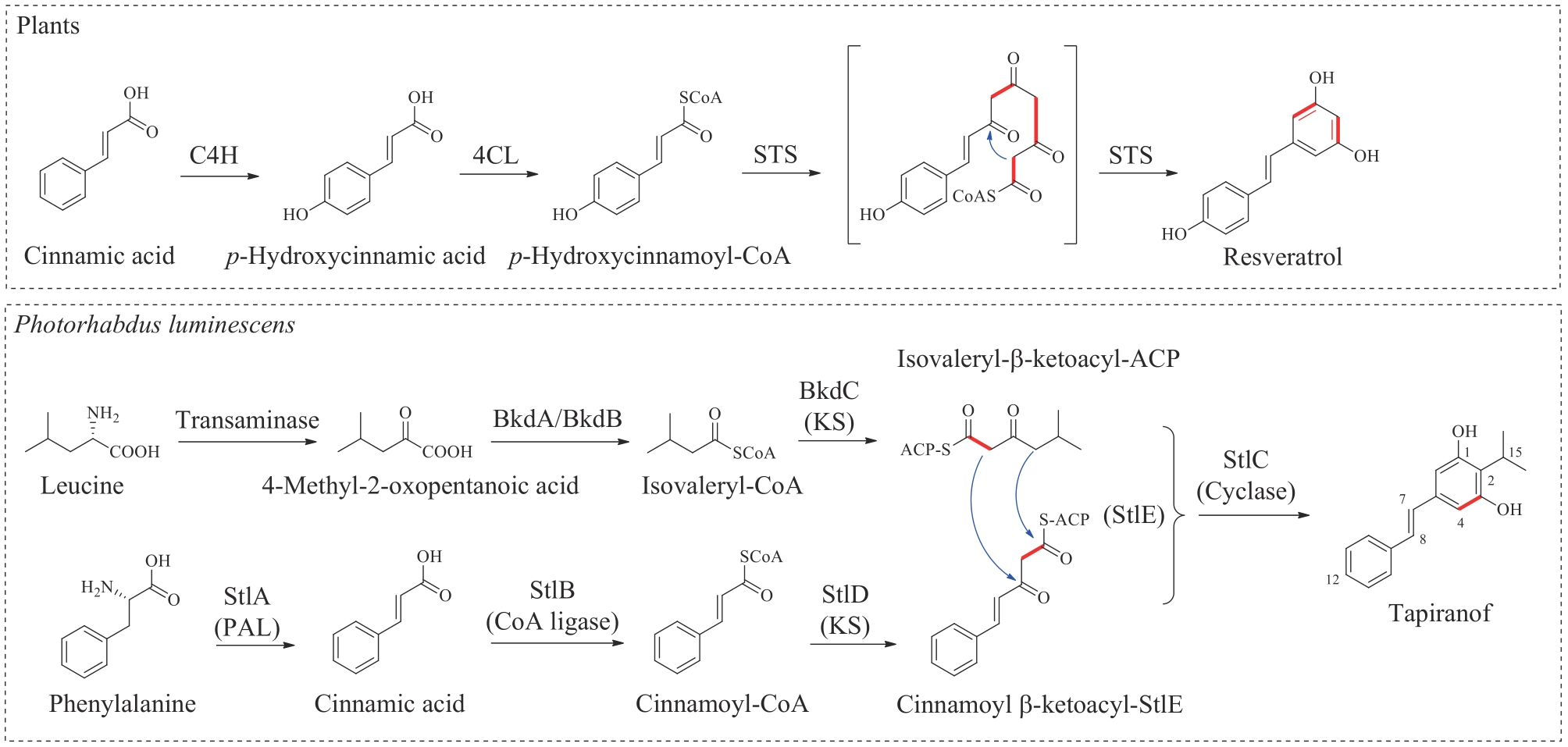
图7 二苯乙烯类天然产物在植物和发光杆菌中的生物合成途径C4H—肉桂酸-4-羟化酶;4CL—4-香豆酰-CoA连接酶;STS—二苯乙烯合酶;KS—酮基合酶;PAL—苯丙氨酸解氨酶
Fig. 7 Biosynthesis of stilbene natural products in plants and PhotorhabdusluminescensC4H—cinnamate-4-hydroxylase; 4CL—4-coumaroyl-CoA ligase; STS—stilbene synthase; KS—ketosynthase; PAL—phenylalanine ammonia-lyase
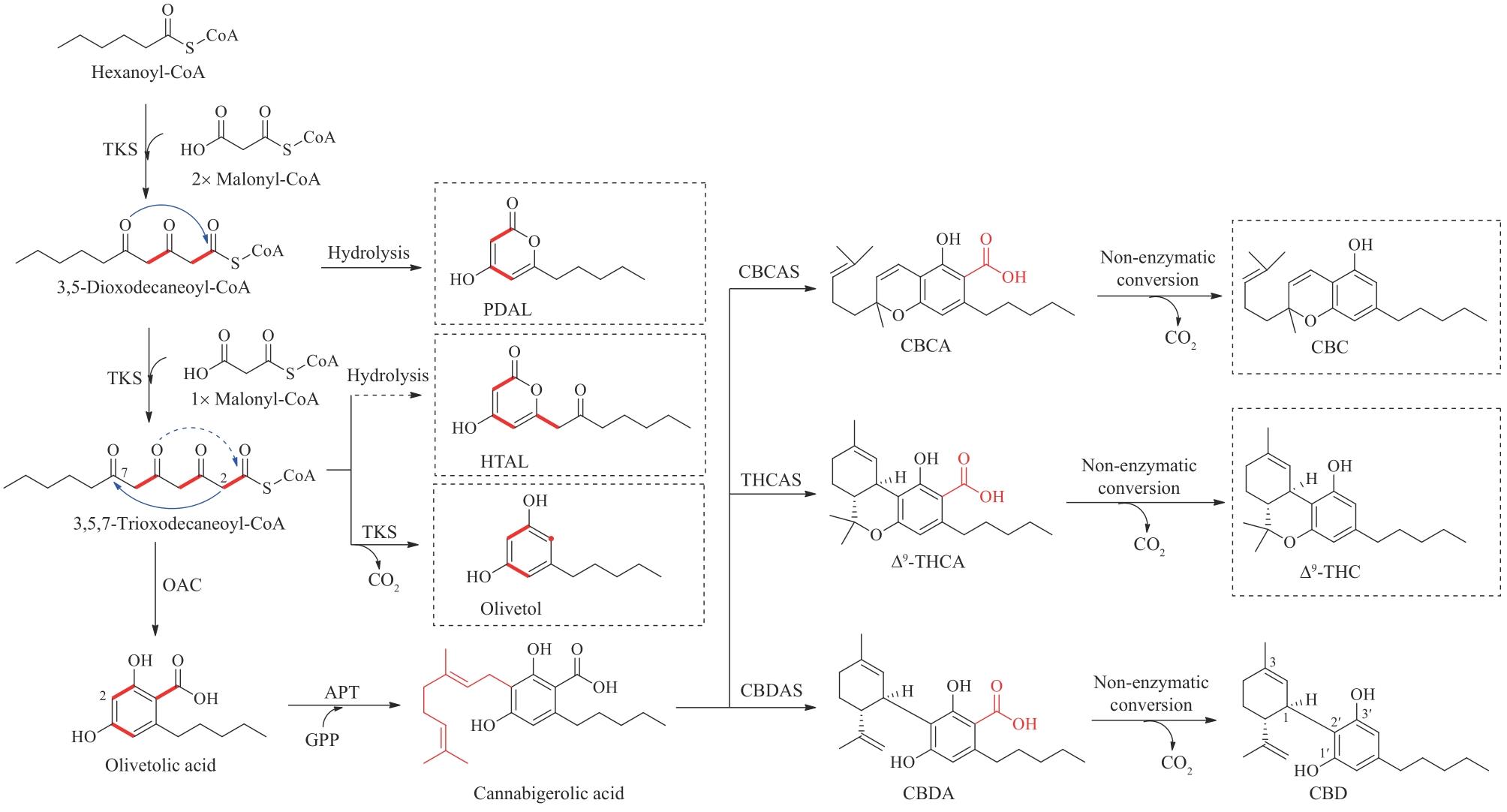
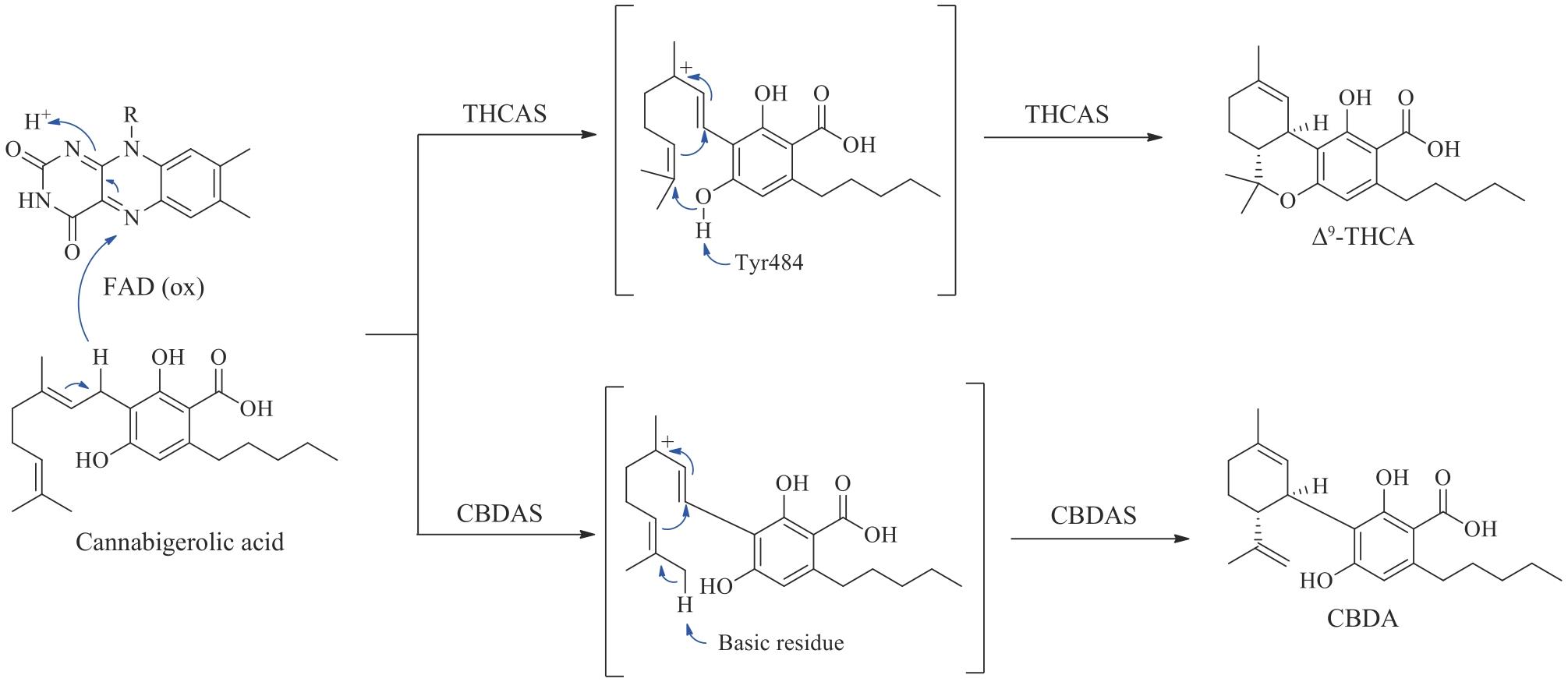
图9 大麻二酚及其他大麻素类似物的生物合成APT—芳香异戊烯基转移酶;CBDAS—大麻二酚酸合酶;THCAS—四氢大麻酚酸合酶;CBCAS—大麻色烯酸合酶;Δ9-THCA—Δ9-四氢大麻酚酸;CBCA—大麻色烯酸;Δ9-THC—Δ9-四氢大麻酚;CBC—大麻色原烯;PDAL—戊二乙酸内酯;HTAL—己酰三乙酸内酯;GPP—牻牛儿基焦磷酸
Fig. 9 Biosynthesis of CBD and other cannabinoid analoguesAPT—aromatic prenyltransferase; CBDAS—cannabidiolic acid synthase; THCAS—tetrahydrocannabinolic acid synthase; CBCAS—cannabichromenic acid synthase; Δ9-THCA—Δ9-tetrahydrocannabinolic acid; CBCA—cannabichromenic acid; Δ9-THC—Δ9-tetrahydrocannabinol; CBC—cannabichromene; PDAL—pentyl diacetic lactone; HTAL—hexanoyl triacetic acid lactone; GPP—geranyl pyrophosphate
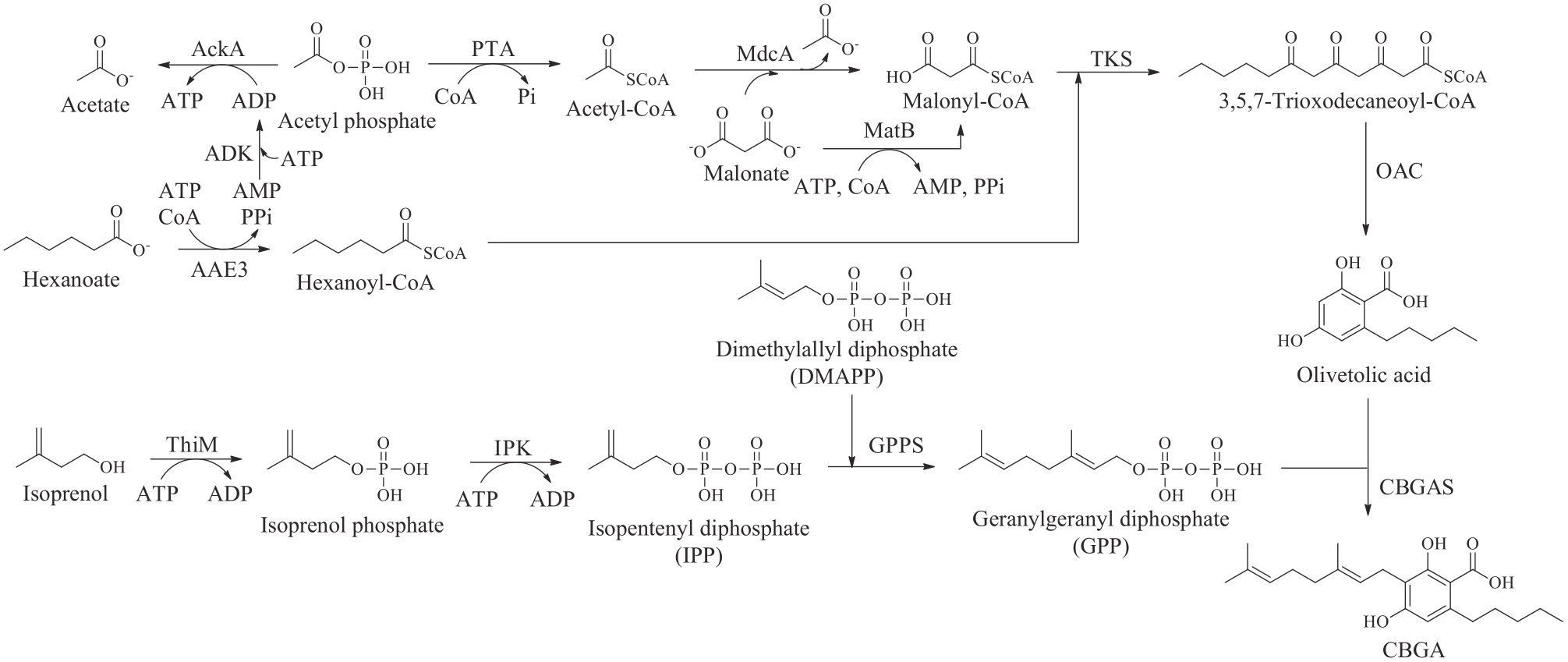
图11 CBGA在体外的酶法生物合成AckA—乙酸激酶;PTA—磷酸乙酰基转移酶;MdcA—丙二酸脱羧酶;MatB—丙二酰辅酶A合成酶;ADK—腺苷酸激酶;AAE3—酰基激活酶;ThiM—羟乙基噻唑激酶;IPK—磷酸激酶;GPPS—牻牛儿基焦磷酸合酶
Fig. 11 Enzymatic biosynthesis of CBGA invitroAckA—acetate kinase; PTA—phosphate acetyltransferase; MdcA—malonate decarboxylase; MatB—malonyl-CoA synthetase; ADK—adenylate kinase; AAE3—acyl-activating enzyme; ThiM—hydroxyethylthiazole kinase; IPK—phosphate kinase; GPPS—geranyl pyrophosphate synthase
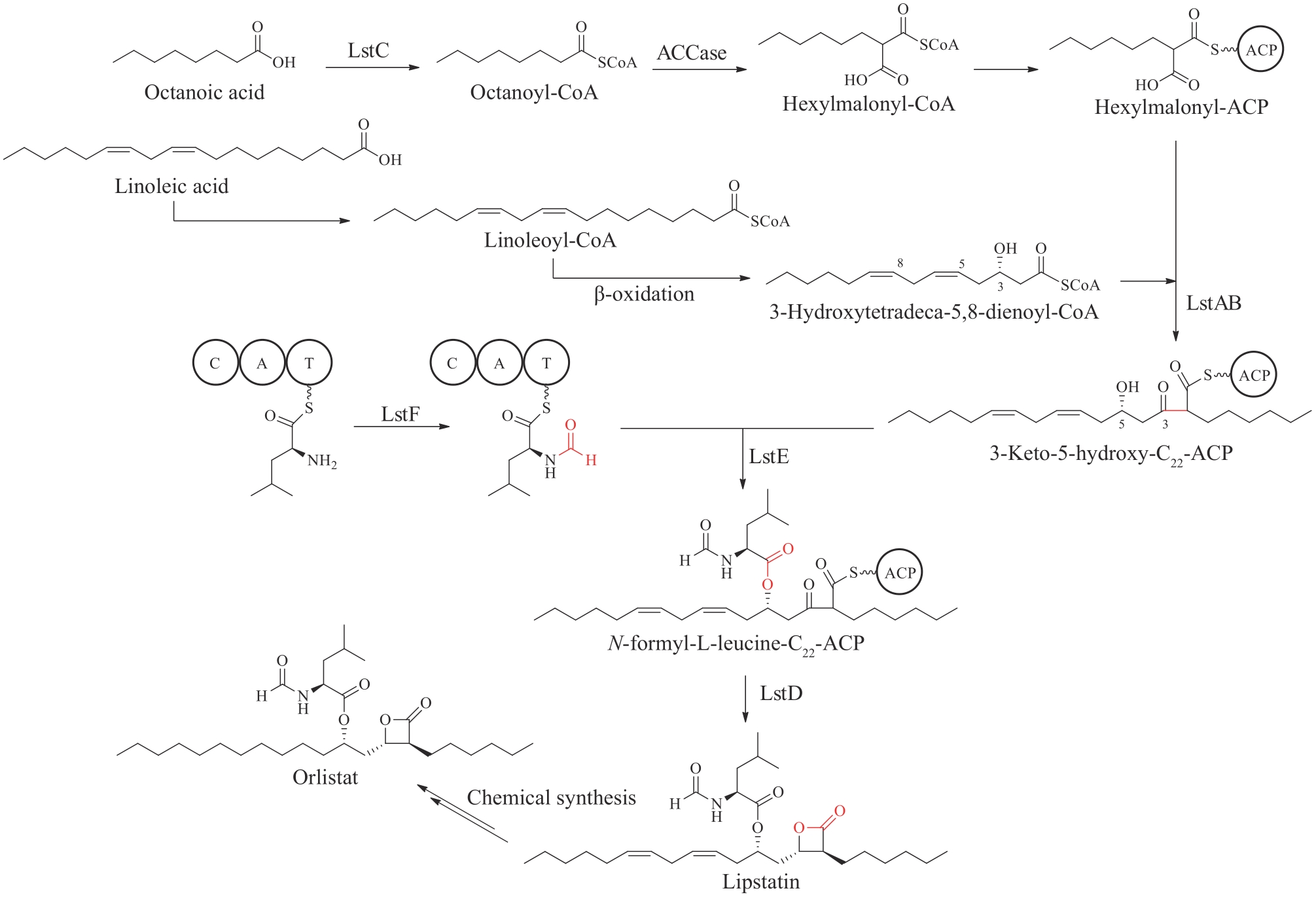
图12 奥利司他化学结构及推测的利普司他汀生物合成途径C—缩合结构域;A—腺苷化结构域;T—硫酯化结构域
Fig. 12 Chemical structure of orlistat and proposed biosynthetic pathway of lipstatinC—condensation domain; A—adenylation domain; T—thiolation domain
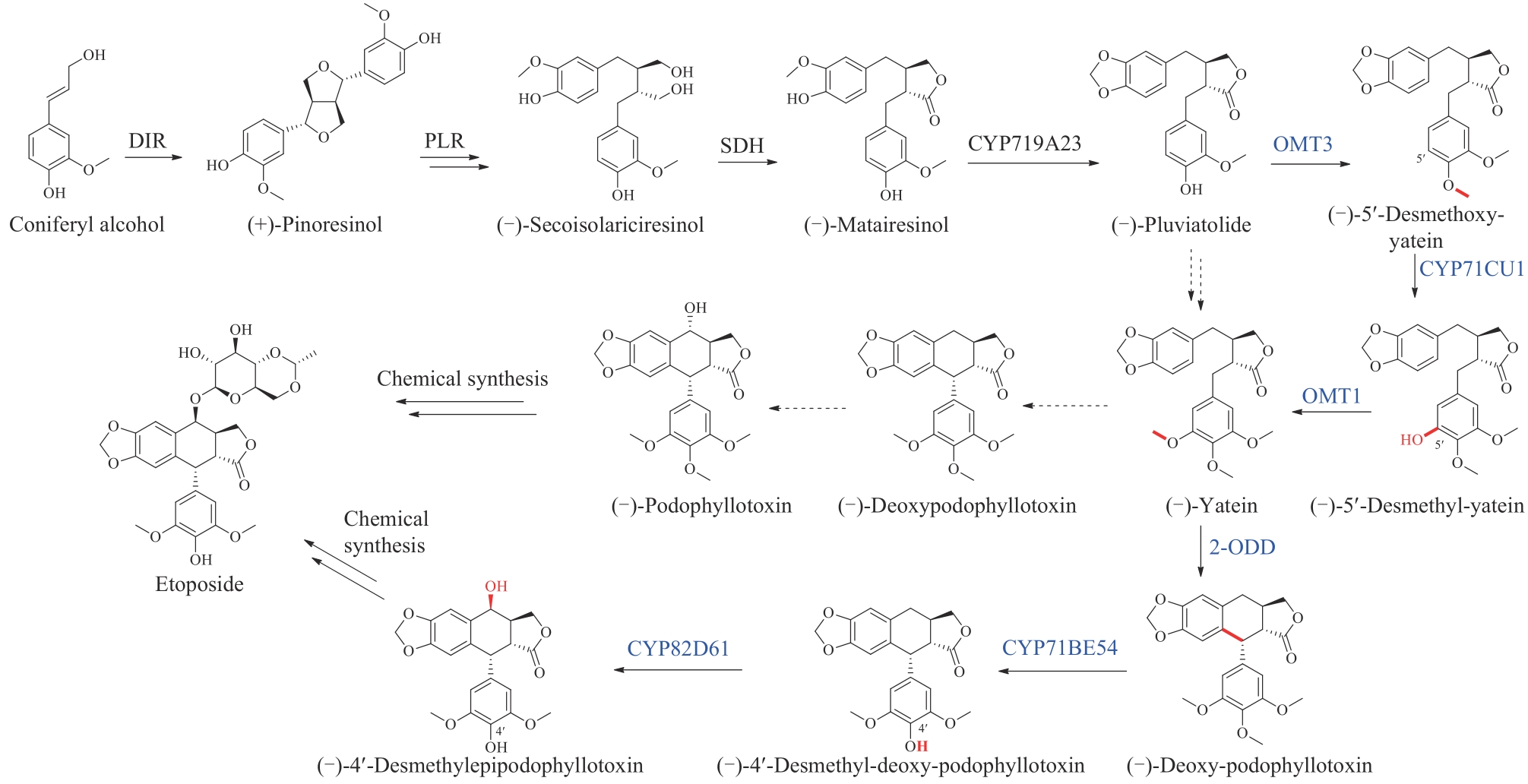
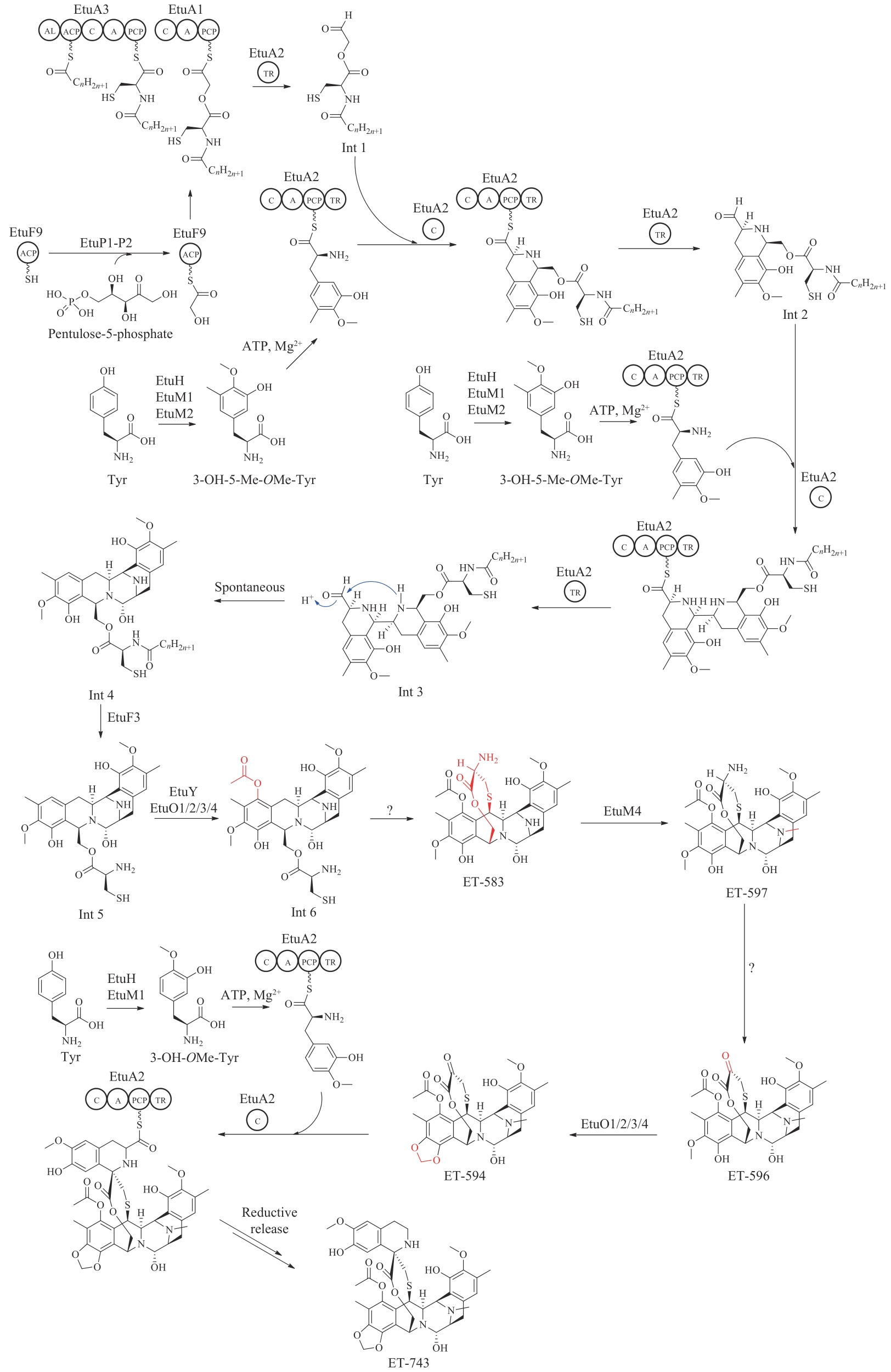
图16 依托泊苷苷元的生物合成途径(蓝色为新表征的酶)DIR—定向蛋白;PLR—松脂醇-落叶松脂醇还原酶;SDH—开环异落叶松脂酚脱氢酶;CYP719A23/71CU1/71BE54/82D61—细胞色素P450;OMT1/3—O-甲基转移酶;2-ODD—2-酮戊二酸/Fe(Ⅱ)依赖双加氧酶
Fig. 16 Biosynthetic pathway of etoposide aglycone(Newly characterized enzymes are highlighted in blue) DIR—dirigent protein; PLR—pinoresinol-lariciresinol reductase; SDH—secoisolariciresinol dehydrogenase; CYP719A23/71CU1/71BE54/82D61—cytochromes P450; OMT1/3—O-methyltransferases; 2-ODD—2-oxoglutarate/Fe(Ⅱ)-dependent dioxygenase
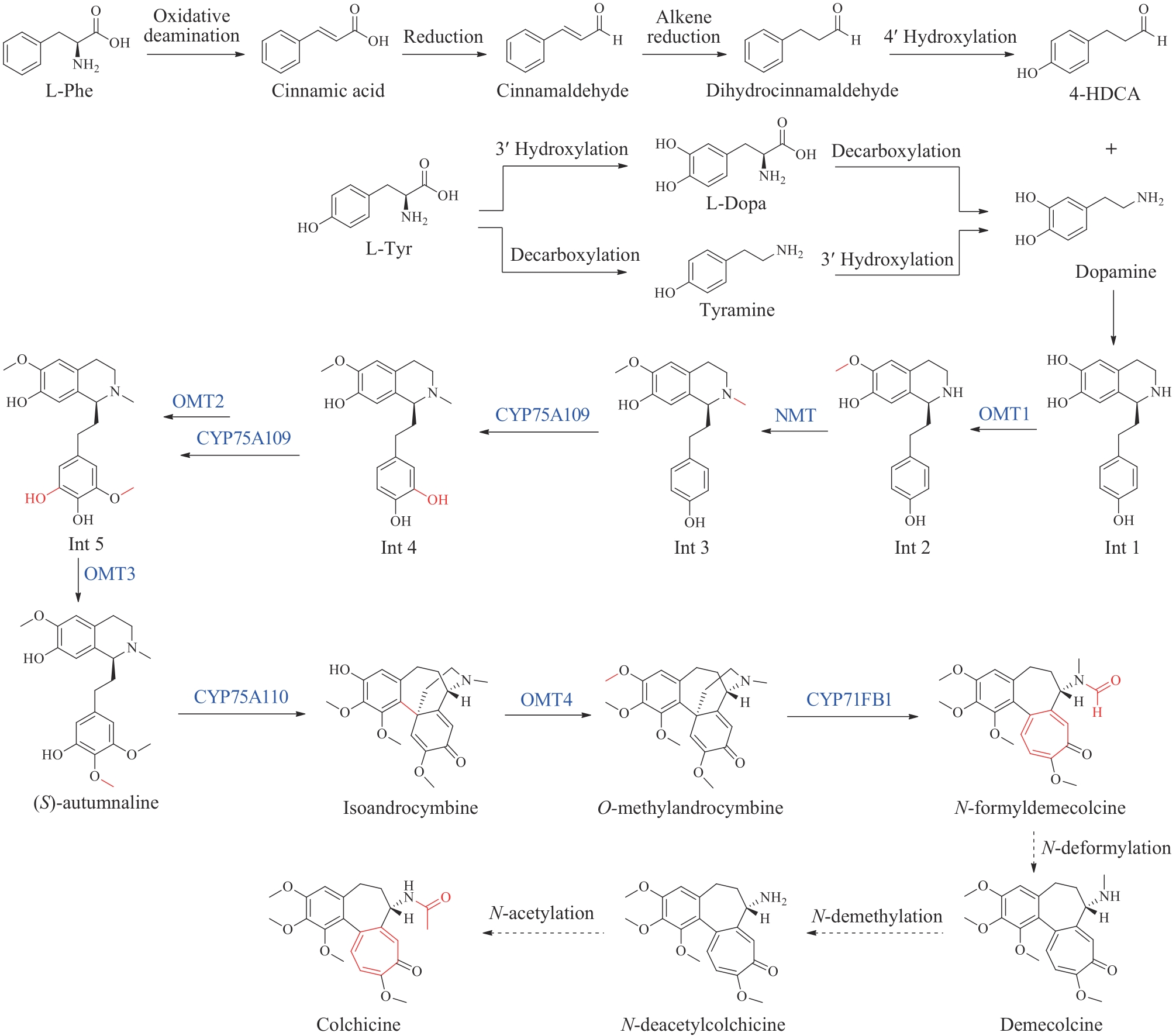
图18 秋水仙碱的生物合成途径(蓝色为新表征的酶)OMT1/2/3/4—O-甲基转移酶;NMT—N-甲基转移酶;CYP75A109/75A110/71FB1—细胞色素P450
Fig. 18 Biosynthetic pathway of colchicine(Newly characterized enzymes are highlighted in blue) OMT1/2/3/4—O-methyltransferases; NMT—N-methyltransferase; CYP75A109/75A110/71FB1—cytochromes P450
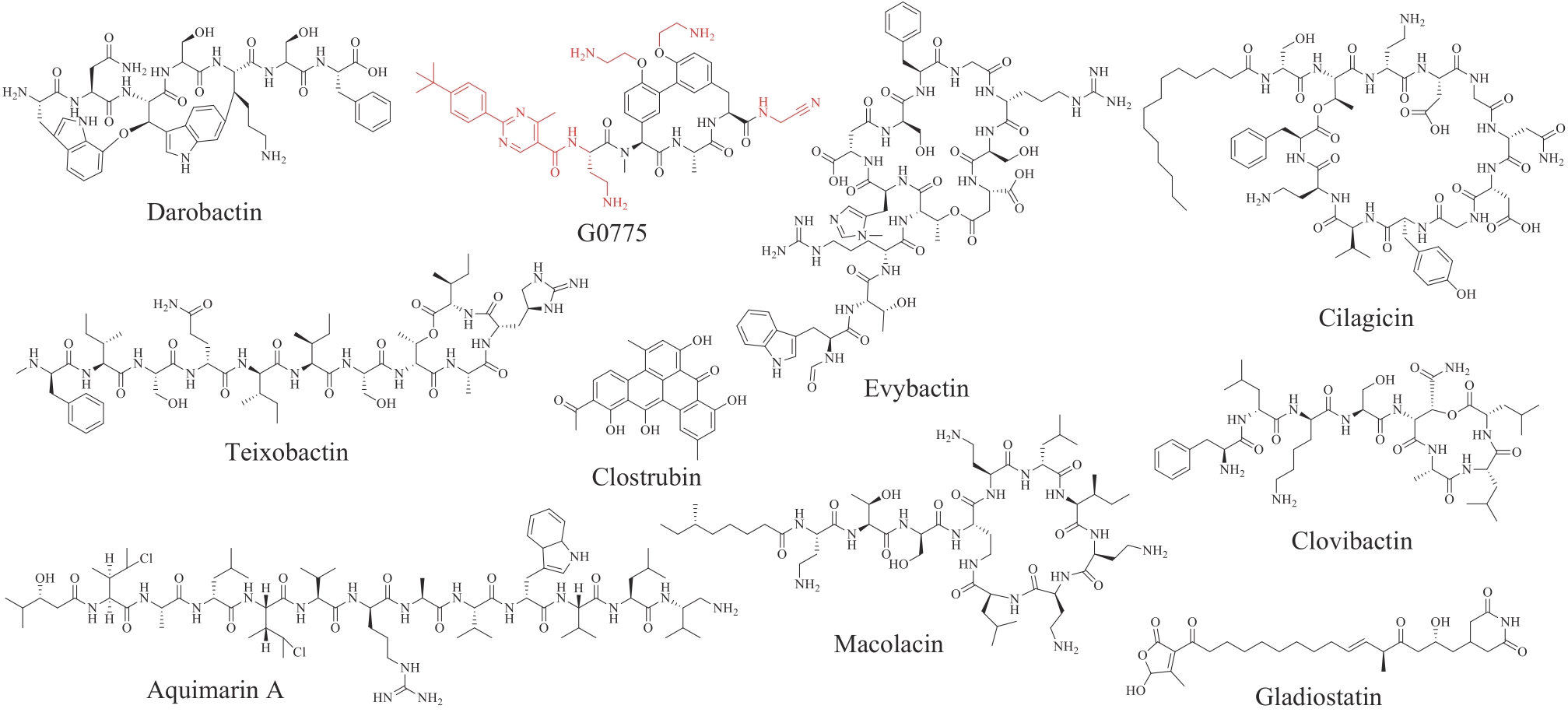
图23 近十年新发现的涉及生物合成且有发展为抗生素药物潜力的代表性天然产物
Fig. 23 Representative natural products biosynthesied within the past decade with potentials for being developed as antibiotics
| 1 | DIAS D A, URBAN S, ROESSNER U. A historical overview of natural products in drug discovery[J]. Metabolites, 2012, 2(2): 303-336. |
| 2 | VEERESHAM C. Natural products derived from plants as a source of drugs[J]. Journal of Advanced Pharmaceutical Technology & Research, 2012, 3(4): 200-201. |
| 3 | HUTCHINGS M I, TRUMAN A W, WILKINSON B. Antibiotics: past, present and future[J]. Current Opinion in Microbiology, 2019, 51: 72-80. |
| 4 | 饶聪, 云轩, 虞沂, 等. 微生物药物的合成生物学研究进展[J]. 合成生物学, 2020, 1(1): 92-102. |
| RAO C, YUN X, YU Y, et al. Recent progress of synthetic biology applications in microbial pharmaceuticals research[J]. Synthetic Biology Journal, 2020, 1(1): 92-102. | |
| 5 | 张博, 马永硕, 尚轶, 等. 植物合成生物学研究进展[J]. 合成生物学, 2020, 1(2): 121-140. |
| ZHANG B, MA Y S, SHANG Y, et al. Recent advances in plant synthetic biology[J]. Synthetic Biology Journal, 2020, 1(2): 121-140. | |
| 6 | 赖奇龙, 姚帅, 查毓国, 等. 微生物组生物合成基因簇发掘方法及应用前景[J]. 合成生物学, 2023, 4(3): 611-627. |
| LAI Q L, YAO S, ZHA Y G, et al. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials[J]. Synthetic Biology Journal, 2023, 4(3): 611-627. | |
| 7 | KENSHOLE E, HERISSE M, MICHAEL M, et al. Natural product discovery through microbial genome mining[J]. Current Opinion in Chemical Biology, 2021, 60: 47-54. |
| 8 | LI L. Accessing hidden microbial biosynthetic potential from underexplored sources for novel drug discovery[J]. Biotechnology Advances, 2023, 66: 108176. |
| 9 | CRAGG G M, NEWMAN D J. Biodiversity: a continuing source of novel drug leads[J]. Pure and Applied Chemistry, 2005, 77(1): 7-24. |
| 10 | ATANASOV A G, WALTENBERGER B, PFERSCHY-WENZIG E M, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review[J]. Biotechnology Advances, 2015, 33(8): 1582-1614. |
| 11 | MATHUR S, HOSKINS C. Drug development: lessons from nature[J]. Biomedical Reports, 2017, 6(6): 612-614. |
| 12 | NEWMAN D J. Natural products and drug discovery[J]. National Science Review, 2022, 9(11): nwac206. |
| 13 | ATANASOV A G, ZOTCHEV S B, DIRSCH V M, et al. Natural products in drug discovery: advances and opportunities[J]. Nature Reviews Drug Discovery, 2021, 20(3): 200-216. |
| 14 | ZHU F, QIN C, TAO L, et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(31): 12943-12948. |
| 15 | LIU X Y, QIN Y. Industrial total synthesis of natural medicines[J]. Natural Product Reports, 2023, 40(11): 1694-1700. |
| 16 | 张发光, 曲戈, 孙周通, 等. 从化学合成到生物合成——天然产物全合成新趋势[J]. 合成生物学, 2021, 2(5): 674-696. |
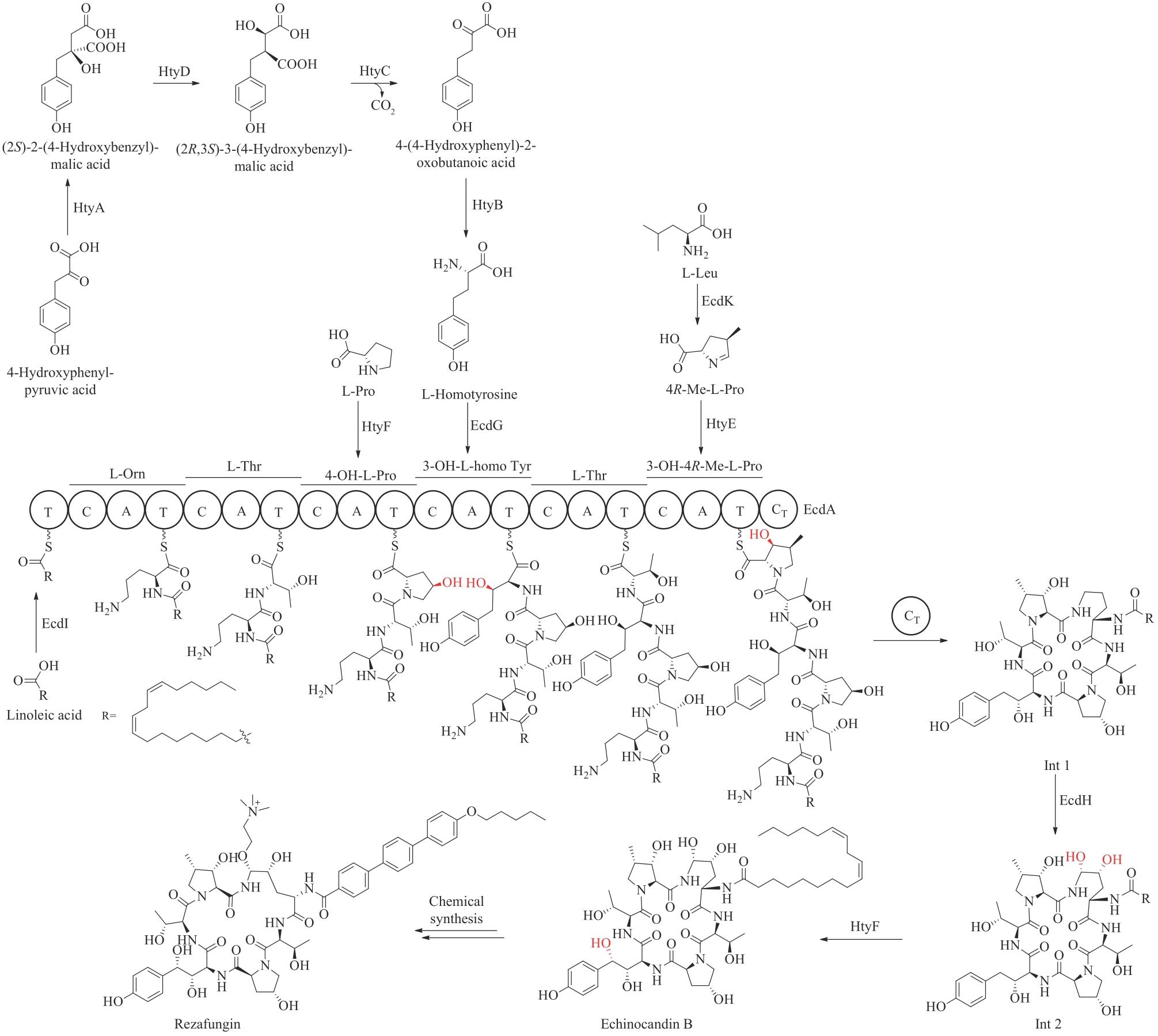
| ZHANG F G, QU G, SUN Z T, et al. From chemical synthesis to biosynthesis: trends toward total synthesis of natural products[J]. Synthetic Biology Journal, 2021, 2(5): 674-696. | |
| 17 | 李晓军, 张万斌, 高栓虎. 复杂天然产物全合成: 化学合成与生物合成结合的策略[J]. 有机化学, 2018, 38(9): 2185-2198. |
| LI X J, ZHANG W B, GAO S H. Total synthesis of complex natural products: combination of chemical synthesis and biosynthesis strategies[J]. Chinese Journal of Organic Chemistry, 2018, 38(9): 2185-2198. | |
| 18 | 贺俊斌, 孟松, 潘海学, 等. 多酶催化串联策略在复杂天然产物合成中的应用[J]. 合成生物学, 2020, 1(2): 226-246. |
| HE J B, MENG S, PAN H X, et al. Applications of the multienzyme-catalyzed tandem strategy in the synthesis of complex natural products[J]. Synthetic Biology Journal, 2020, 1(2): 226-246. | |
| 19 | HARVEY A L, EDRADA-EBEL R, QUINN R J. The re-emergence of natural products for drug discovery in the genomics era[J]. Nature Reviews Drug Discovery, 2015, 14(2): 111-129. |
| 20 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs from 1981 to 2014[J]. Journal of Natural Products, 2016, 79(3): 629-661. |
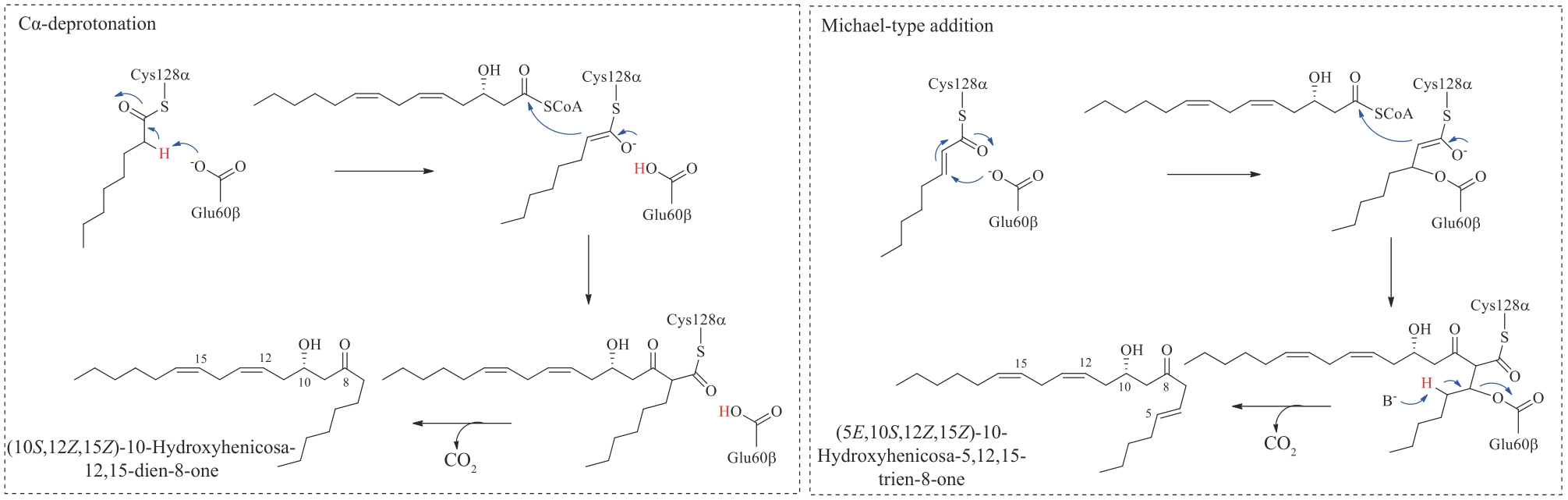
| 21 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83(3): 770-803. |
| 22 | THOMFORD N E, SENTHEBANE D A, ROWE A, et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery[J]. International Journal of Molecular Sciences, 2018, 19(6): 1578. |
| 23 | KHALAF K, TORNESE P, COCCO A, et al. Tauroursodeoxycholic acid: a potential therapeutic tool in neurodegenerative diseases[J]. Translational Neurodegeneration, 2022, 11(1): 33. |
| 24 | JOYCE S A, BRACHMANN A O, GLAZER I, et al. Bacterial biosynthesis of a multipotent stilbene[J]. Angewandte Chemie International Edition, 2008, 47(10): 1942-1945. |
| 25 | STANCZYK F Z, ARCHER D F. Biosynthesis of estetrol in human pregnancy: potential pathways[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2023, 232: 106359. |
| 26 | TAHIR M N, SHAHBAZI F, RONDEAU-GAGNÉ S, et al. The biosynthesis of the cannabinoids[J]. Journal of Cannabis Research, 2021, 3(1): 7. |
| 27 | RIORDAN J F. Angiotensin Ⅱ: biosynthesis, molecular recognition, and signal transduction[J]. Cellular and Molecular Neurobiology, 1995, 15(6): 637-651. |
| 28 | AUCHUS R J. Overview of dehydroepiandrosterone biosynthesis[J]. Seminars in Reproductive Medicine, 2004, 22(4): 281-288. |
| 29 | MASSCHELEIN J, JENNER M, CHALLIS G L. Antibiotics from Gram-negative bacteria: a comprehensive overview and selected biosynthetic highlights[J]. Natural Product Reports, 2017, 34(7): 712-783. |
| 30 | ŠARENAC T M, MIKOV M. Bile acid synthesis: from nature to the chemical modification and synthesis and their applications as drugs and nutrients[J]. Frontiers in Pharmacology, 2018, 9: 939. |
| 31 | GARCIA-EFFRON G. Rezafungin-mechanisms of action, susceptibility and resistance: similarities and differences with the other echinocandins[J]. Journal of Fungi, 2020, 6(4): 262. |
| 32 | APGAR J M, WILKENING R R, PARKER D L JR, et al. Ibrexafungerp: an orally active β-1,3-glucan synthesis inhibitor[J]. Bioorganic & Medicinal Chemistry Letters, 2021, 32: 127661. |
| 33 | WANG Y. Voclosporin (Lupkynis), a macrocyclic peptide inhibitor of calcineurin for the treatment of lupus nephritis[M/OL]// LI J J. Current drug synthesis. 1st ed. New York: Wiley, 2022: 323-338 [2023-11-01]. . |
| 34 | PETERSON H, KIRCIK L, ARMSTRONG A W. Clascoterone cream 1%: mechanism of action, efficacy, and safety of a novel, first-in-class topical antiandrogen therapy for acne[J]. Journal of Drugs in Dermatology, 2023, 22(6): SF350992s7-SF350992s14. |
| 35 | PRESSER A, FEICHTINGER A, BUZZI S. A simplified and scalable synthesis of artesunate[J]. Monatshefte Fur Chemie, 2017, 148(1): 63-68. |
| 36 | MARTÍNEZ-MONTEAGUDO S I, ENTESHARI M, METZGER L. Lactitol: production, properties, and applications[J]. Trends in Food Science & Technology, 2019, 83: 181-191. |
| 37 | WATKINS R R, FILE T M. Lefamulin: a novel semisynthetic pleuromutilin antibiotic for community-acquired bacterial pneumonia[J]. Clinical Infectious Diseases, 2020, 71(10): 2757-2762. |
| 38 | XU L H, MA J, SHI L F, et al. Design, synthesis and characterizations of prodrugs of brexanolone[J]. Bioorganic & Medicinal Chemistry Letters, 2023, 90: 129344. |
| 39 | SENSI P. History of the development of rifampin[J]. Clinical Infectious Diseases, 1983, 5(): S402-S406. |
| 40 | DURÃES F, SOUSA E. Omadacycline: a newly approved antibacterial from the class of tetracyclines[J]. Pharmaceuticals, 2019, 12(2): 63. |
| 41 | CLARK J A, BURGESS D S. Plazomicin: a new aminoglycoside in the fight against antimicrobial resistance[J]. Therapeutic Advances in Infectious Disease, 2020, 7: 2049936120952604. |
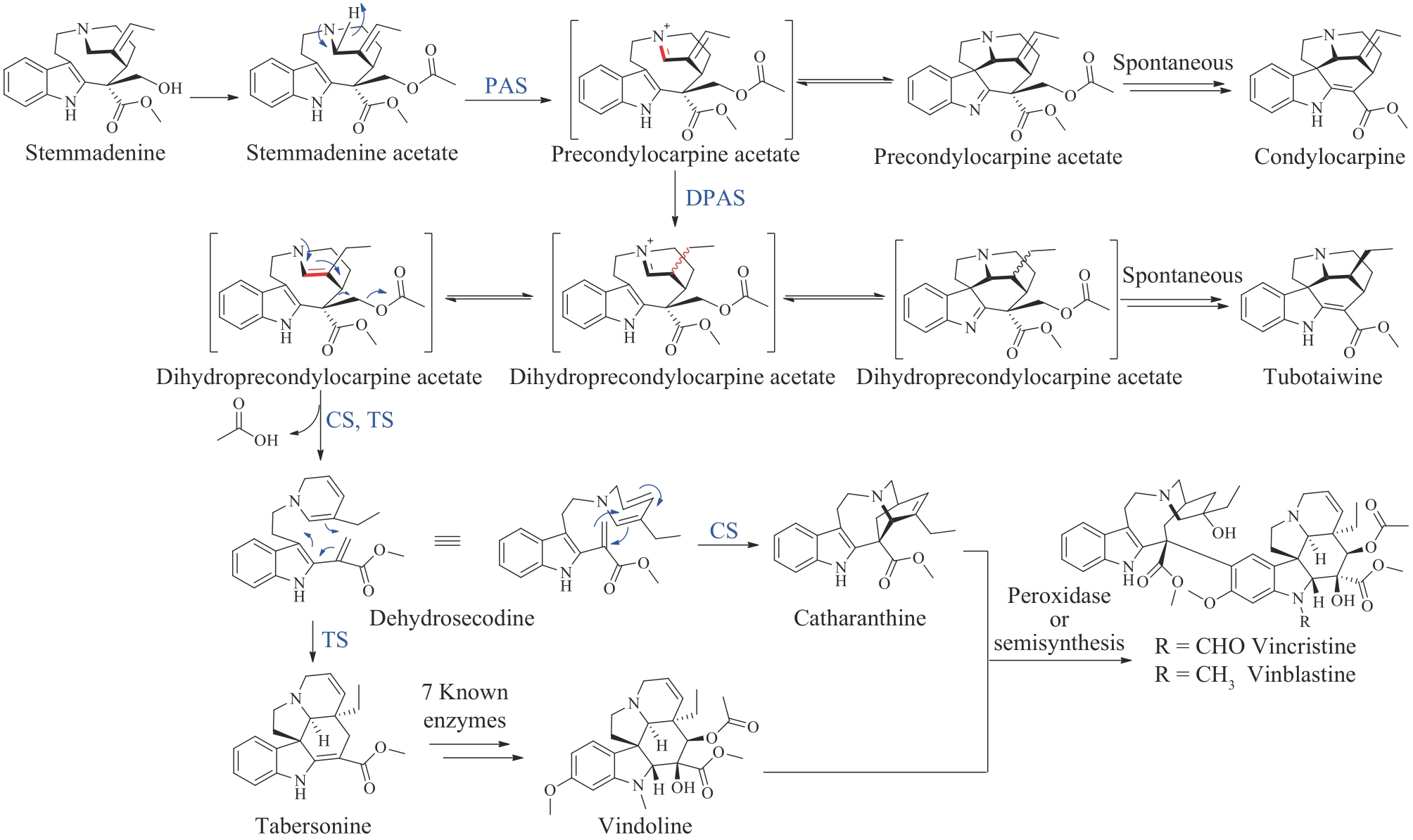
| 42 | COBB R, BOECKH A. Moxidectin: a review of chemistry, pharmacokinetics and use in horses[J]. Parasites & Vectors, 2009, 2(S2): S5. |
| 43 | RONN M, ZHU Z J, HOGAN P C, et al. Process R&D of eravacycline: the first fully synthetic fluorocycline in clinical development[J]. Organic Process Research & Development, 2013, 17(5): 838-845. |
| 44 | BUNICK C G, KERI J, TANAKA S K, et al. Antibacterial mechanisms and efficacy of sarecycline in animal models of infection and inflammation[J]. Antibiotics, 2021, 10(4): 439. |
| 45 | STONE R M, MANLEY P W, LARSON R A, et al. Midostaurin: its odyssey from discovery to approval for treating acute myeloid leukemia and advanced systemic mastocytosis[J]. Blood Advances, 2018, 2(4): 444-453. |
| 46 | NAGUNURI G C L, KATANGOOR V, SURIGILLA V, et al. Efficient process for obeticholic acid: synthesis, structural assignment, and control strategy for diastereoisomeric impurities[J]. Organic Process Research & Development, 2022, 26(12): 3265-3275. |
| 47 | BERRIO ESCOBAR J, PASTRANA RESTREPO M H, GALEANO JARAMILLO E, et al. Synthesis and cytotoxic activity of per-acetylated and halogenated derivatives of nucleosides in breast cancer cells[J]. Ars Pharmaceutica, 2017, 58(4): 145-154. |
| 48 | HUGHES D L. Patent review of manufacturing routes to fifth-generation cephalosporin drugs. Part 1, Ceftolozane[J]. Organic Process Research & Development, 2017, 21(3): 430-443. |
| 49 | SARAVOLATZ L D, STEIN G E. Oritavancin: a long-half-life lipoglycopeptide[J]. Clinical Infectious Diseases, 2015, 61(4): 627-632. |
| 50 | ZHANG D Z, ZHANG F, LIU W. A KAS-Ⅲ heterodimer in lipstatin biosynthesis nondecarboxylatively condenses C8 and C14 fatty acyl-CoA substrates by a variable mechanism during the establishment of a C22 aliphatic skeleton[J]. Journal of the American Chemical Society, 2019, 141(9): 3993-4001. |
| 51 | SILVERBERG L J, DILLON J L, VEMISHETTI P, et al. Efficient synthesis of the anticancer drug etoposide 4′- phosphate: use of benzylic ether-protecting groups on the carbohydrate segment1 [J]. Organic Process Research & Development, 2000, 4(1): 34-42. |
| 52 | ISHIKAWA H, COLBY D A, SETO S, et al. Total synthesis of vinblastine, vincristine, related natural products, and key structural analogues[J]. Journal of the American Chemical Society, 2009, 131(13): 4904-4916. |
| 53 | NETT R S, LAU W, SATTELY E S. Discovery and engineering of colchicine alkaloid biosynthesis[J]. Nature, 2020, 584(7819): 148-153. |
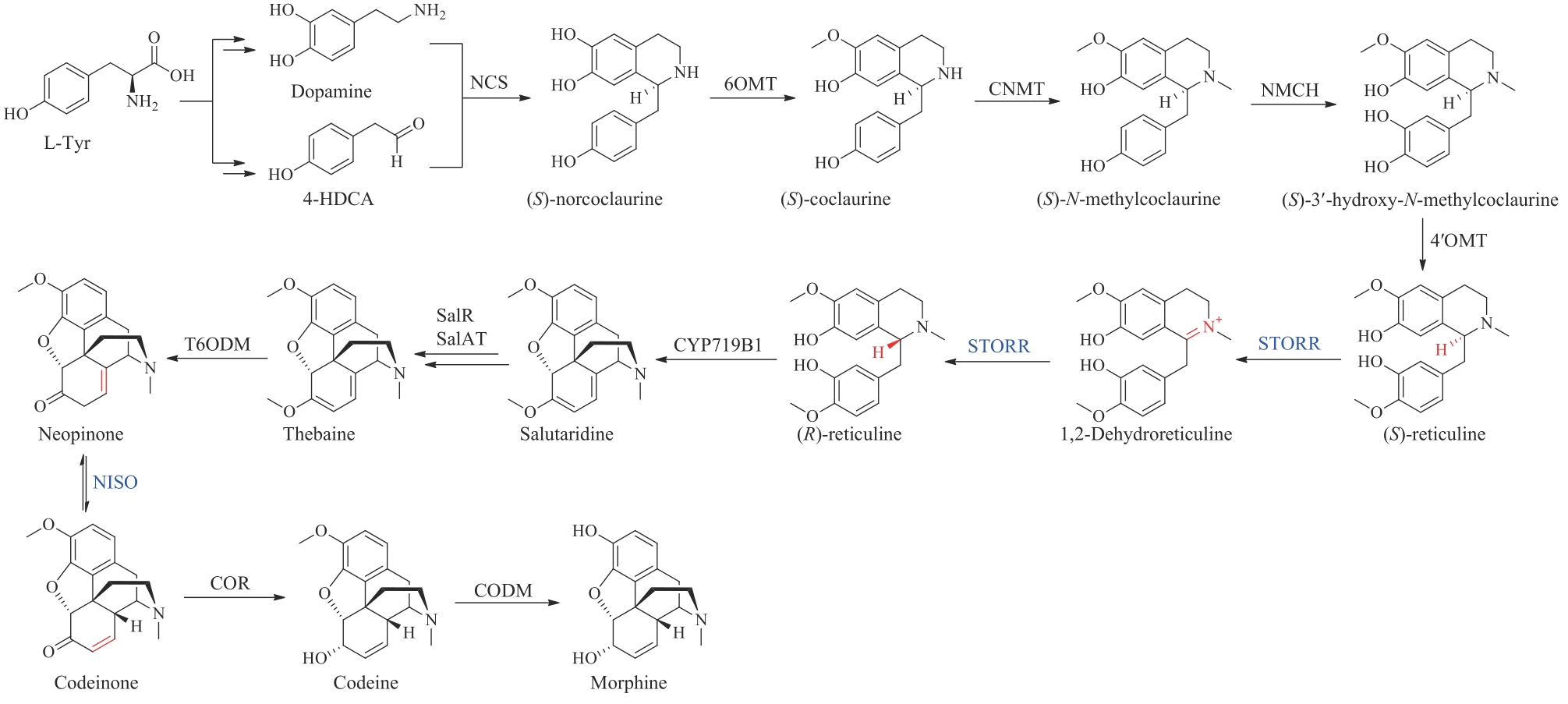
| 54 | DASTMALCHI M, CHEN X, HAGEL J M, et al. Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy[J]. Nature Chemical Biology, 2019, 15(4): 384-390. |
| 55 | NOCQUET P A, OPATZ T. Total synthesis of (±)-scopolamine: challenges of the tropane ring[J]. European Journal of Organic Chemistry, 2016, 2016(6): 1156-1164. |
| 56 | WARREN M J, RAUX E, SCHUBERT H L, et al. The biosynthesis of adenosylcobalamin (vitamin B12)[J]. Natural Product Reports, 2002, 19(4): 390-412. |
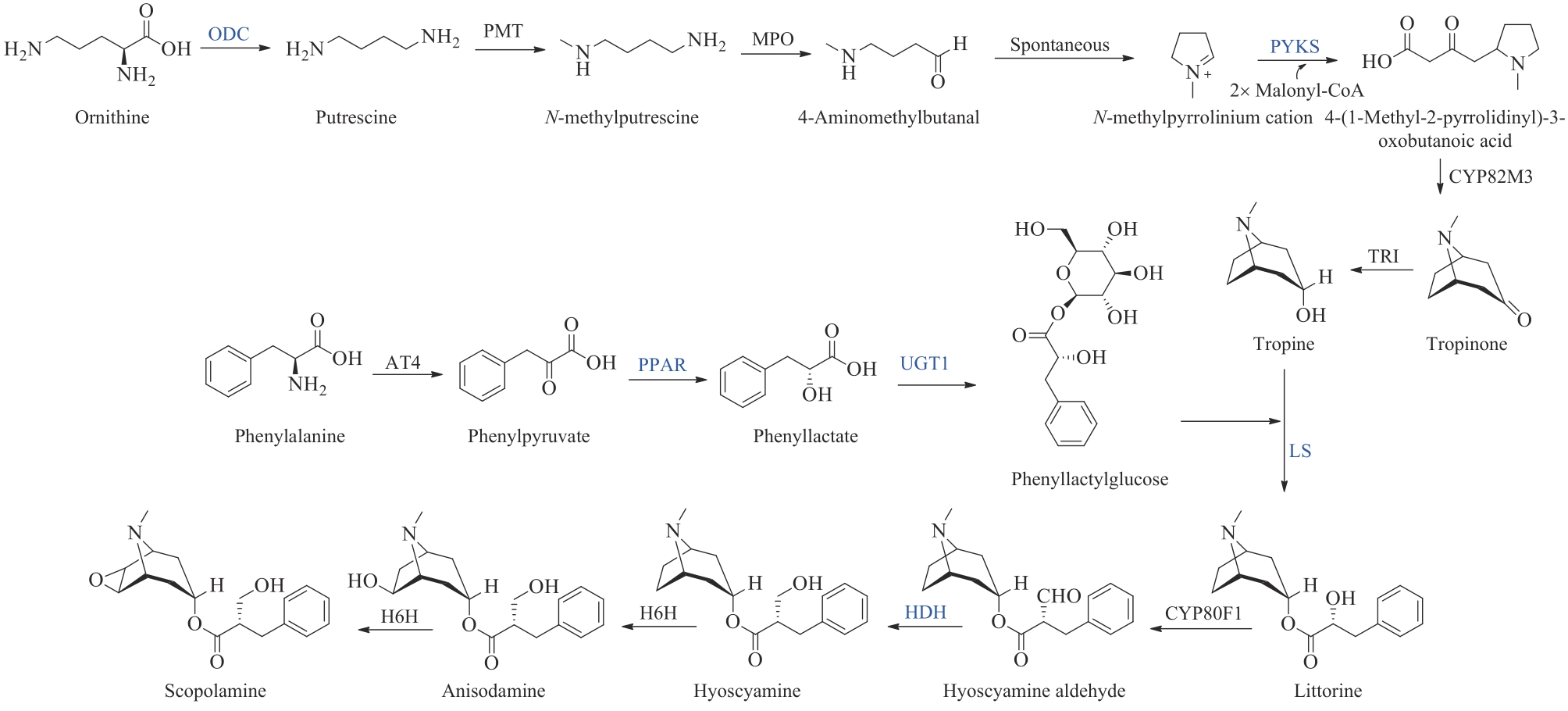
| 57 | HOWAT S, PARK B, OH I S, et al. Paclitaxel: biosynthesis, production and future prospects[J]. New Biotechnology, 2014, 31(3): 242-245. |
| 58 | STAELS B, FONSECA V A. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration[J]. Diabetes Care, 2009, 32(S2): S237-S245. |
| 59 | GUZIOR D V, QUINN R A. Review: microbial transformations of human bile acids[J]. Microbiome, 2021, 9(1): 140. |
| 60 | HONDA A, SALEN G, SHEFER S, et al. Regulation of 25- and 27-hydroxylation side chain cleavage pathways for cholic acid biosynthesis in humans, rabbits, and mice: assay of enzyme activities by high-resolution gas chromatography;-mass spectrometry[J]. Journal of Lipid Research, 2000, 41(3): 442-451. |
| 61 | KRIAA A, MARIAULE V, JABLAOUI A, et al. Bile acids: key players in inflammatory bowel diseases?[J]. Cells, 2022, 11(5): 901. |
| 62 | MARION S, DESHARNAIS L, STUDER N, et al. Biogeography of microbial bile acid transformations along the murine gut[J]. Journal of Lipid Research, 2020, 61(11): 1450-1463. |
| 63 | VANG S, LONGLEY K, STEER C J, et al. The unexpected uses of urso- and tauroursodeoxycholic acid in the treatment of non-liver diseases[J]. Global Advances in Health and Medicine, 2014, 3(3): 58-69. |
| 64 | AHN T K, KIM K T, JOSHI H P, et al. Therapeutic potential of tauroursodeoxycholic acid for the treatment of osteoporosis[J]. International Journal of Molecular Sciences, 2020, 21(12): 4274. |
| 65 | KUSACZUK M. Tauroursodeoxycholate-bile acid with chaperoning activity: molecular and cellular effects and therapeutic perspectives[J]. Cells, 2019, 8(12): 1471. |
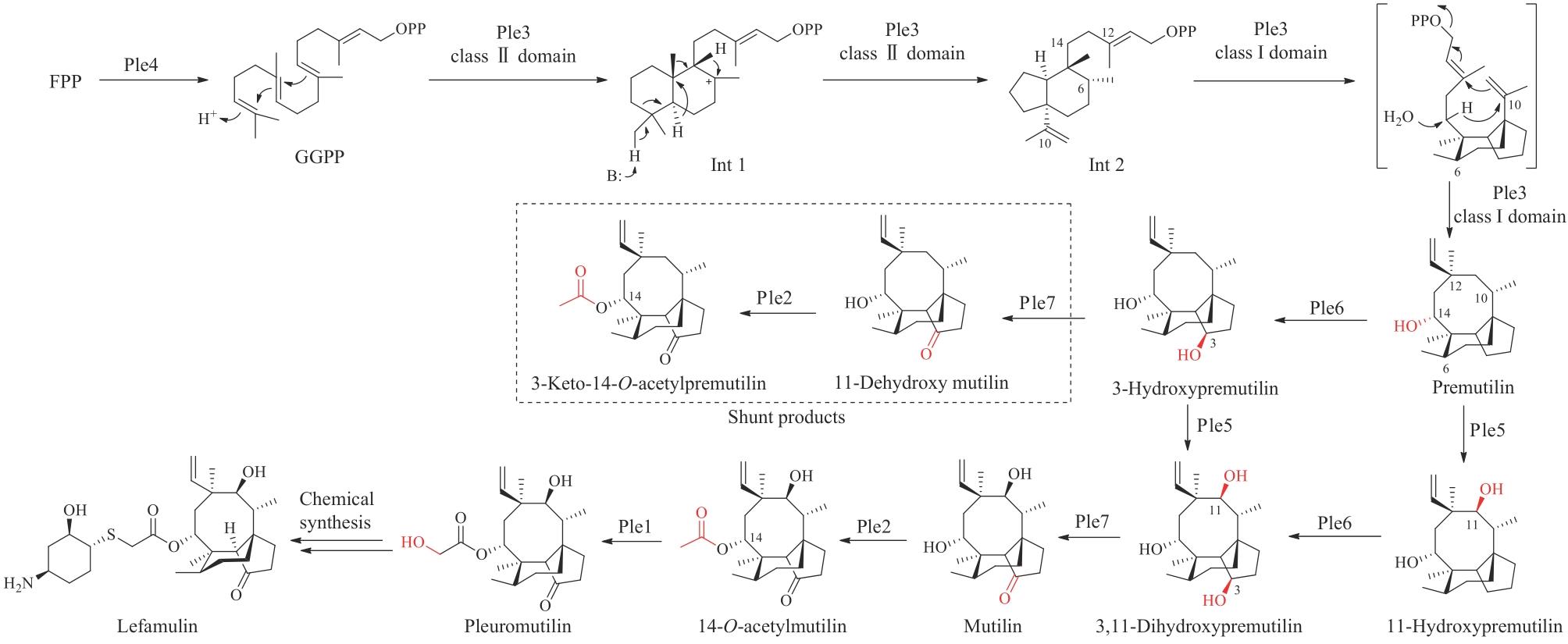
| 66 | HUIJGHEBAERT S M, HOFMANN A F. Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures[J]. Journal of Lipid Research, 1988, 27(7): 742-752. |
| 67 | FERRANDI E E, BERTOLESI G M, POLENTINI F, et al. In search of sustainable chemical processes: cloning, recombinant expression, and functional characterization of the 7α- and 7β-hydroxysteroid dehydrogenases from Clostridium absonum [J]. Applied Microbiology and Biotechnology, 2012, 95(5): 1221-1233. |
| 68 | SHONSEY E M, WHEELER J, JOHNSON M, et al. Synthesis of bile acid coenzyme A thioesters in the amino acid conjugation of bile acids[M/OL]// Methods in enzymology. Amsterdam: Elsevier, 2005, 400: 360-373 [2023-11-01]. . |
| 69 | SHONSEY E M, SFAKIANOS M, JOHNSON M, et al. Bile acid coenzyme A: amino acid N-acyltransferase in the amino acid conjugation of bile acids[M/OL]// Methods in enzymology. Amsterdam: Elsevier, 2005, 400: 374-394 [2023-11-01]. . |
| 70 | JI Q Z, TAN J, ZHU L C, et al. Preparing tauroursodeoxycholic acid (TUDCA) using a double-enzyme-coupled system[J]. Biochemical Engineering Journal, 2016, 105: 1-9. |
| 71 | SONG C, WANG B C, TAN J, et al. Discovery of tauroursodeoxycholic acid biotransformation enzymes from the gut microbiome of black bears using metagenomics[J]. Scientific Reports, 2017, 7: 45495. |
| 72 | XU Y P, YANG L, ZHAO S J, et al. Large-scale production of tauroursodeoxycholic acid products through fermentation optimization of engineered Escherichia coli cell factory[J]. Microbial Cell Factories, 2019, 18(1): 34. |
| 73 | JIN L N, YANG L, ZHAO S J, et al. A green strategy to produce potential substitute resource for bear bile using engineered Saccharomyces cerevisiae [J]. Bioresources and Bioprocessing, 2022, 9(1): 32. |
| 74 | SONG P, ZHANG X, FENG W, et al. Biological synthesis of ursodeoxycholic acid[J]. Frontiers in Microbiology, 2023, 14: 1140662. |
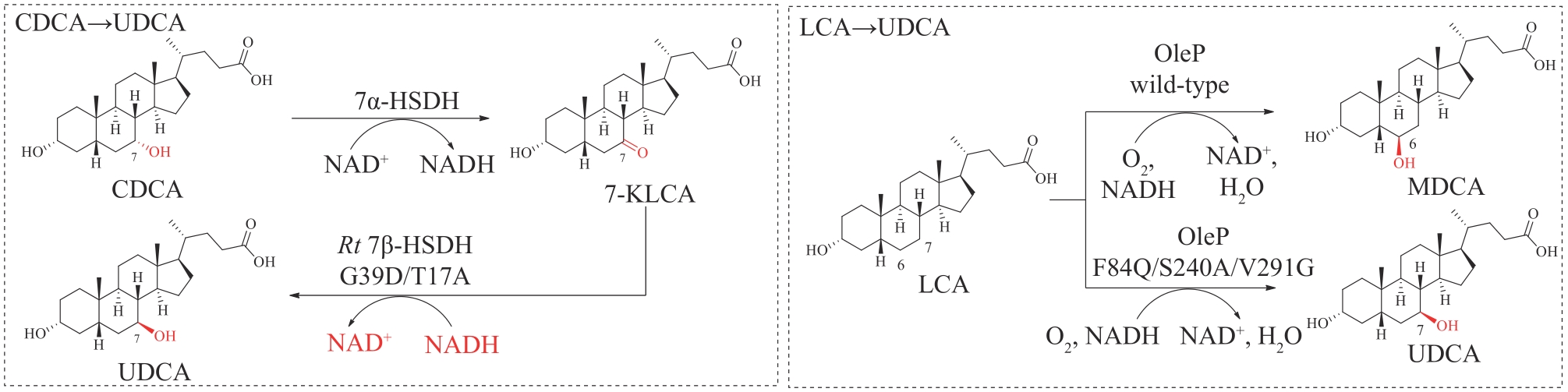
| 75 | ZHENG M M, CHEN K C, WANG R F, et al. Engineering 7β-hydroxysteroid dehydrogenase for enhanced ursodeoxycholic acid production by multiobjective directed evolution[J]. Journal of Agricultural and Food Chemistry, 2017, 65(6): 1178-1185. |
| 76 | ZHENG M M, CHEN F F, LI H, et al. Continuous production of ursodeoxycholic acid by using two cascade reactors with co-immobilized enzymes[J]. ChemBioChem, 2018, 19(4): 347-353. |
| 77 | YOU Z N, CHEN Q, SHI S C, et al. Switching cofactor dependence of 7β-hydroxysteroid dehydrogenase for cost-effective production of ursodeoxycholic acid[J]. ACS Catalysis, 2019, 9(1): 466-473. |
| 78 | GROBE S, BADENHORST C P S, BAYER T, et al. Engineering regioselectivity of a P450 monooxygenase enables the synthesis of ursodeoxycholic acid via 7β-hydroxylation of lithocholic acid[J]. Angewandte Chemie International Edition, 2021, 60(2): 753-757. |
| 79 | COELINGH BENNINK H J T, HOLINKA C F, DICZFALUSY E. Estetrol review: profile and potential clinical applications[J]. Climacteric, 2008, 11(): 47-58. |
| 80 | GABAI G, MONGILLO P, GIARETTA E, et al. Do dehydroepiandrosterone (DHEA) and its sulfate (DHEAS) play a role in the stress response in domestic animals?[J]. Frontiers in Veterinary Science, 2020, 7: 588835. |
| 81 | MILLER K K M, CAI J, RIPP S L, et al. Stereo- and regioselectivity account for the diversity of dehydroepiandrosterone (DHEA) metabolites produced by liver microsomal cytochromes P450[J]. Drug Metabolism and Disposition, 2004, 32(3): 305-313. |
| 82 | HOLINKA C F, DICZFALUSY E, COELINGH BENNINK H J T. Estetrol: a unique steroid in human pregnancy[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2008, 110(1-2): 138-143. |
| 83 | CANTINEAU R, KREMERS P, DE GRAEVE J, et al. 15- and 16-hydroxylations of androgens and estrogens in the human fetal liver: a critical step in estetrol biosynthesis[J]. Journal of Steroid Biochemistry, 1985, 22(2): 195-201. |
| 84 | FURUE M, HASHIMOTO-HACHIYA A, TSUJI G. Aryl hydrocarbon receptor in atopic dermatitis and psoriasis[J]. International Journal of Molecular Sciences, 2019, 20(21): 5424. |
| 85 | BISSONNETTE R, VASIST L S, BULLMAN J N, et al. Systemic pharmacokinetics, safety, and preliminary efficacy of topical AhR agonist tapinarof: results of a phase 1 study[J]. Clinical Pharmacology in Drug Development, 2018, 7(5): 524-531. |
| 86 | BISSONNETTE R, STEIN GOLD L, RUBENSTEIN D S, et al. Tapinarof in the treatment of psoriasis: a review of the unique mechanism of action of a novel therapeutic aryl hydrocarbon receptor-modulating agent[J]. Journal of the American Academy of Dermatology, 2021, 84(4): 1059-1067. |
| 87 | ELEFTHERIANOS I, BOUNDY S, JOYCE S A, et al. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(7): 2419-2424. |
| 88 | LI J, CHEN G, WU H, et al. Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens [J]. Applied and Environmental Microbiology, 1995, 61(12): 4329-4333. |
| 89 | HU K J, LI J X, WEBSTER J M. Quantitative analysis of a bacteria-derived antibiotic in nematode-infected insects using HPLC-UV and TLC-UV methods[J]. Journal of Chromatography B: Biomedical Sciences and Applications, 1997, 703(1-2): 177-183. |
| 90 | FUCHS S W, BOZHÜYÜK K A, KRESOVIC D, et al. Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occuring ketosynthase[J]. Angewandte Chemie International Edition, 2013, 52(15): 4108-4112. |
| 91 | MORI T, AWAKAWA T, SHIMOMURA K, et al. Structural insight into the enzymatic formation of bacterial stilbene[J]. Cell Chemical Biology, 2016, 23(12): 1468-1479. |
| 92 | KAVAKLI S, GRAMMBITTER G L C, BODE H B. Biosynthesis of the multifunctional isopropylstilbene in Photorhabdus laumondii involves cross-talk between specialized and primary metabolism[J]. Tetrahedron, 2022, 128: 133116. |
| 93 | PARK H B, GODDARD T N, OH J, et al. Bacterial autoimmune drug metabolism transforms an immunomodulator into structurally and functionally divergent antibiotics[J]. Angewandte Chemie International Edition, 2020, 59(20): 7871-7880. |
| 94 | LAPRAIRIE R B, BAGHER A M, KELLY M E M, et al. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor[J]. British Journal of Pharmacology, 2015, 172(20): 4790-4805. |
| 95 | PERTWEE R G. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Δ9-tetrahydrocannabinol, cannabidiol and Δ9-tetrahydrocannabivarin[J]. British Journal of Pharmacology, 2008, 153(2): 199-215. |
| 96 | ELSOHLY M A, SLADE D. Chemical constituents of marijuana: the complex mixture of natural cannabinoids[J]. Life Sciences, 2005, 78(5): 539-548. |
| 97 | LIVINGSTON S J, QUILICHINI T D, BOOTH J K, et al. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation[J]. The Plant Journal, 2020, 101(1): 37-56. |
| 98 | TAURA F, TANAKA S, TAGUCHI C, et al. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway[J]. FEBS Letters, 2009, 583(12): 2061-2066. |
| 99 | GAGNE S J, STOUT J M, LIU E W, et al. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(31): 12811-12816. |
| 100 | TAN Z G, CLOMBURG J M, GONZALEZ R. Synthetic pathway for the production of olivetolic acid in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(8): 1886-1896. |
| 101 | KEARSEY L J, PRANDI N, KARUPPIAH V, et al. Structure of the Cannabis sativa olivetol-producing enzyme reveals cyclization plasticity in type Ⅲ polyketide synthases[J]. The FEBS Journal, 2020, 287(8): 1511-1524. |
| 102 | YANG X M, MATSUI T, KODAMA T, et al. Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa [J]. The FEBS Journal, 2016, 283(6): 1088-1106. |
| 103 | TAURA F, SIRIKANTARAMAS S, SHOYAMA Y, et al. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa [J]. FEBS Letters, 2007, 581(16): 2929-2934. |
| 104 | FELLERMEIER M, ZENK M H. Prenylation of olivetolate by a hemp transferase yields cannabigerolic acid, the precursor of tetrahydrocannabinol[J]. FEBS Letters, 1998, 427(2): 283-285. |
| 105 | TAURA F, TANAYA R, SIRIKANTARAMAS S. Recent advances in cannabinoid biochemistry and biotechnology[J]. ScienceAsia, 2019, 45(5): 399. |
| 106 | PERROTIN-BRUNEL H, BUIJS W, VAN SPRONSEN J, et al. Decarboxylation of Δ9-tetrahydrocannabinol: kinetics and molecular modeling[J]. Journal of Molecular Structure, 2011, 987(1-3): 67-73. |
| 107 | WANG M, WANG Y H, AVULA B, et al. Decarboxylation study of acidic cannabinoids: a novel approach using ultra-high-performance supercritical fluid chromatography/photodiode array-mass spectrometry[J]. Cannabis and Cannabinoid Research, 2016, 1(1): 262-271. |
| 108 | LUO X Z, REITER M A, D’ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567(7746): 123-126. |
| 109 | VALLIERE M A, KORMAN T P, ARBING M A, et al. A bio-inspired cell-free system for cannabinoid production from inexpensive inputs[J]. Nature Chemical Biology, 2020, 16(12): 1427-1433. |
| 110 | TORGERSON J S, HAUPTMAN J, BOLDRIN M N, et al. XENical in the prevention of diabetes in obese subjects (XENDOS) study: a randomized study of orlistat as an adjunct to lifestyle changes for the prevention of type 2 diabetes in obese patients[J]. Diabetes Care, 2004, 27(1): 155-161. |
| 111 | KUMAR P, DUBEY K K. Current trends and future prospects of lipstatin: a lipase inhibitor and pro-drug for obesity[J]. RSC Advances, 2015, 5(106): 86954-86966. |
| 112 | KRIDEL S J, AXELROD F, ROZENKRANTZ N, et al. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity[J]. Cancer Research, 2004, 64(6): 2070-2075. |
| 113 | WEIBEL E K, HADVARY P, HOCHULI E, et al. Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. Ⅰ. Producing organism, fermentation, isolation and biological activity[J]. The Journal of Antibiotics, 1987, 40(8): 1081-1085. |
| 114 | EISENREICH W, KUPFER E, WEBER W, et al. Tracer studies with crude U-13C-lipid mixtures[J]. Journal of Biological Chemistry, 1997, 272(2): 867-874. |
| 115 | EISENREICH W, KUPFER E, STOHLER P, et al. Biosynthetic origin of a branched chain analogue of the lipase inhibitor, lipstatin[J]. Journal of Medicinal Chemistry, 2003, 46(19): 4209-4212. |
| 116 | GOESE M, EISENREICH W, KUPFER E, et al. Biosynthetic origin of hydrogen atoms in the lipase inhibitor lipstatin[J]. The Journal of Biological Chemistry, 2000, 275(28): 21192-21196. |
| 117 | GOESE M, EISENREICH W, KUPFER E, et al. Biosynthesis of lipstatin. Incorporation of multiply deuterium-labeled (5Z, 8Z)-tetradeca-5,8-dienoic acid and octanoic acid[J]. The Journal of Organic Chemistry, 2001, 66(13): 4673-4678. |
| 118 | SCHUHR C A, EISENREICH W, GOESE M, et al. Biosynthetic precursors of the lipase inhibitor lipstatin[J]. The Journal of Organic Chemistry, 2002, 67(7): 2257-2262. |
| 119 | BAI T L, ZHANG D Z, LIN S J, et al. Operon for biosynthesis of lipstatin, the beta-lactone inhibitor of human pancreatic lipase[J]. Applied and Environmental Microbiology, 2014, 80(24): 7473-7483. |
| 120 | KRASNER C N, MCMEEKIN D S, CHAN S, et al. A phase Ⅱ study of trabectedin single agent in patients with recurrent ovarian cancer previously treated with platinum-based regimens[J]. British Journal of Cancer, 2007, 97(12): 1618-1624. |
| 121 | TAVECCHIO M, NATOLI C, UBEZIO P, et al. Dynamics of cell cycle phase perturbations by trabectedin (ET-743) in nucleotide excision repair (NER)-deficient and NER-proficient cells, unravelled by a novel mathematical simulation approach[J]. Cell Proliferation, 2007, 40(6): 885-904. |
| 122 | WANG J L, WANG P F, ZENG Z, et al. Trabectedin in cancers: mechanisms and clinical applications[J]. Current Pharmaceutical Design, 2022, 28(24): 1949-1965. |
| 123 | PETEK B J, LOGGERS E T, POLLACK S M, et al. Trabectedin in soft tissue sarcomas[J]. Marine Drugs, 2015, 13(2): 974-983. |
| 124 | D’INCALCI M, BADRI N, GALMARINI C M, et al. Trabectedin, a drug acting on both cancer cells and the tumour microenvironment[J]. British Journal of Cancer, 2014, 111(4): 646-650. |
| 125 | SCHOFIELD M M, JAIN S, PORAT D, et al. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743[J]. Environmental Microbiology, 2015, 17(10): 3964-3975. |
| 126 | RATH C M, JANTO B, EARL J, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743[J]. ACS Chemical Biology, 2011, 6(11): 1244-1256. |
| 127 | IRSCHIK H, TROWITZSCH-KIENAST W, GERTH K, et al. Saframycin Mx1, a new natural saframycin isolated from a myxobacterium[J]. The Journal of Antibiotics, 1988, 41(8): 993-998. |
| 128 | MIKAMI Y, TAKAHASHI K, YAZAWA K, et al. Biosynthetic studies on saframycin A, a quinone antitumor antibiotic produced by Streptomyces lavendulae [J]. The Journal of Biological Chemistry, 1985, 260(1): 344-348. |
| 129 | IKEDA Y, MATSUKI H, OGAWA T, et al. Safracins, new antitumor antibiotics. Ⅱ. Physicochemical properties and chemical structures[J]. The Journal of Antibiotics, 1983, 36(10): 1284-1289. |
| 130 | KLUEPFEL D, BAKER H A, PIATTONI G, et al. Naphthyridinomycin, a new broad-spectrum antibiotic[J]. The Journal of Antibiotics, 1975, 28(7): 497-502. |
| 131 | TOMITA F, TAKAHASHI K, TAMAOKI T. Quinocarcin, a novel antitumor antibiotic. 3. Mode of action[J]. The Journal of Antibiotics, 1984, 37(10): 1268-1272. |
| 132 | KOKETSU K, WATANABE K, SUDA H, et al. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms[J]. Nature Chemical Biology, 2010, 6(6): 408-410. |
| 133 | PENG C, PU J Y, SONG L Q, et al. Hijacking a hydroxyethyl unit from a central metabolic ketose into a nonribosomal peptide assembly line[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(22): 8540-8545. |
| 134 | FU C Y, TANG M C, PENG C, et al. Biosynthesis of 3-hydroxy-5-methyl-O-methyltyrosine in the saframycin/safracin biosynthetic pathway[J]. Journal of Microbiology and Biotechnology, 2009, 19(5): 439-446. |
| 135 | TANG M C, FU C Y, TANG G L. Characterization of SfmD as a heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis[J]. Journal of Biological Chemistry, 2012, 287(7): 5112-5121. |
| 136 | DRAKE E J, GULICK A M. Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis[J]. ACS Chemical Biology, 2011, 6(11): 1277-1286. |
| 137 | LE V H, INAI M, WILLIAMS R M, et al. Ecteinascidins. A review of the chemistry, biology and clinical utility of potent tetrahydroisoquinoline antitumor antibiotics[J]. Natural Product Reports, 2015, 32(2): 328-347. |
| 138 | LAMOTH F. Novel therapeutic approaches to invasive candidiasis: considerations for the clinician[J]. Infection and Drug Resistance, 2023, 16: 1087-1097. |
| 139 | MIESEL L, LIN K Y, ONG V. Rezafungin treatment in mouse models of invasive candidiasis and aspergillosis: insights on the PK/PD pharmacometrics of rezafungin efficacy[J]. Pharmacology Research & Perspectives, 2019, 7(6): e00546. |
| 140 | BENZ F, KNÜSEL F, NÜESCH J, et al. Stoffwechselprodukte von Mikroorganismen 143. Mitteilung. echinocandin B, ein neuartiges polypeptid-antibioticum aus Aspergillus nidulans var. echinulatus: isolierung und bausteine[J]. Helvetica Chimica Acta, 1974, 57(8): 2459-2477. |
| 141 | SCHWARTZ R E, GIACOBBE R A, BLAND J A, et al. L-671,329, a new antifungal agent. Ⅰ. Fermentation and isolation[J]. The Journal of Antibiotics, 1989, 42(2): 163-167. |
| 142 | FROMTLING R A, ABRUZZO G K. L-671,329, a new antifungal agent. Ⅲ. In vitro activity, toxicity and efficacy in comparison to aculeacin[J]. The Journal of Antibiotics, 1989, 42(2): 174-178. |
| 143 | WICHMANN C F, LIESCH J M, SCHWARTZ R E. L-671,329, a new antifungal agent. Ⅱ. Structure determination[J]. The Journal of Antibiotics, 1989, 42(2): 168-173. |
| 144 | KELLER-JUSLÉN C, KUHN M, LOOSLI H R, et al. Struktur des cyclopeptid-antibiotikums sl 7810 (=echinocandinb)[J]. Tetrahedron Letters, 1976, 17(46): 4147-4150. |
| 145 | MIZUNO K, YAGI A, SATOI S, et al. Studies on aculeacin. Ⅰ. Isolation and characterization of aculeacin A[J]. The Journal of Antibiotics, 1977, 30(4): 297-302. |
| 146 | SATOI S, YAGI A, ASANO K, et al. Studies on aculeacin. Ⅱ. Isolation and characterization of aculeacins B, C, D, E, F and G[J]. The Journal of Antibiotics, 1977, 30(4): 303-307. |
| 147 | NORRIS T, VANALSTEN J, HUBBS S, et al. Commercialization and late-stage development of a semisynthetic antifungal API: anidulafungin/D-fructose (Eraxis)[J]. Organic Process Research & Development, 2008, 12(3): 447-455. |
| 148 | BALKOVEC J M, HUGHES D L, MASUREKAR P S, et al. Discovery and development of first in class antifungal caspofungin (CANCIDAS®) — a case study[J]. Natural Product Reports, 2014, 31(1): 15-34. |
| 149 | FUJIE A. Discovery of micafungin (FK463): a novel antifungal drug derived from a natural product lead[J]. Pure and Applied Chemistry, 2007, 79(4): 603-614. |
| 150 | CACHO R A, JIANG W, CHOOI Y H, et al. Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440[J]. Journal of the American Chemical Society, 2012, 134(40): 16781-16790. |
| 151 | HÜTTEL W, YOUSSAR L, GRÜNING B A, et al. Echinocandin B biosynthesis: a biosynthetic cluster from Aspergillus nidulans NRRL 8112 and reassembly of the subclusters Ecd and Hty from Aspergillus pachycristatus NRRL 11440 reveals a single coherent gene cluster[J]. BMC Genomics, 2016, 17: 570. |
| 152 | JIANG W, CACHO R A, CHIOU G, et al. EcdGHK are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold[J]. Journal of the American Chemical Society, 2013, 135(11): 4457-4466. |
| 153 | MATTAY J, HOUWAART S, HÜTTEL W. Cryptic production of trans-3-hydroxyproline in echinocandin B biosynthesis[J]. Applied and Environmental Microbiology, 2018, 84(7): e02370-17. |
| 154 | CHEN L, YUE Q, ZHANG X Y, et al. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis [J]. BMC Genomics, 2013, 14: 339. |
| 155 | WEI T Y, ZHENG Y, WAN M Y, et al. Analysis of FR901379 biosynthetic genes in Coleophoma empetri by clustered regularly interspaced short palindromic repeats/Cas9-based genomic manipulation[J]. ACS Chemical Biology, 2022, 17(8): 2130-2141. |
| 156 | FORTINEZ C M, BLOUDOFF K, HARRIGAN C, et al. Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module[J]. Nature Communications, 2022, 13(1): 548. |
| 157 | LI Y, LAN N, XU L J, et al. Biosynthesis of pneumocandin lipopeptides and perspectives for its production and related echinocandins[J]. Applied Microbiology and Biotechnology, 2018, 102(23): 9881-9891. |
| 158 | YUE Q, CHEN L, ZHANG X L, et al. Evolution of chemical diversity in echinocandin lipopeptide antifungal metabolites[J]. Eukaryotic Cell, 2015, 14(7): 698-718. |
| 159 | HÜTTEL W. Structural diversity in echinocandin biosynthesis: the impact of oxidation steps and approaches toward an evolutionary explanation[J]. Zeitschrift für Naturforschung C, 2017, 72(1/2): 1-20. |
| 160 | LAN N, PERLATTI B, KVITEK D J, et al. Acrophiarin (antibiotic S31794/F-1) from Penicillium arenicola shares biosynthetic features with both Aspergillus- and Leotiomycete-type echinocandins[J]. Environmental Microbiology, 2020, 22(6): 2292-2311. |
| 161 | HÜTTEL W. Echinocandins: structural diversity, biosynthesis, and development of antimycotics[J]. Applied Microbiology and Biotechnology, 2021, 105(1): 55-66. |
| 162 | JIANG K L, LUO P, WANG X X, et al. Insight into advances for the biosynthetic progress of fermented echinocandins of antifungals[J]. Microbial Biotechnology, 2023, 17(1): e14359. |
| 163 | MEN P, WANG M, LI J D, et al. Establishing an efficient genetic manipulation system for sulfated echinocandin producing fungus Coleophoma empetri [J]. Frontiers in Microbiology, 2021, 12: 734780. |
| 164 | MEN P, GENG C, ZHANG X, et al. Biosynthesis mechanism, genome mining and artificial construction of echinocandin O-sulfonation[J]. Metabolic Engineering, 2022, 74: 160-167. |
| 165 | ZHOU Z, ZWELLING L A, GANAPATHI R, et al. Enhanced etoposide sensitivity following adenovirus-mediated human topoisomerase Ⅱ α gene transfer is independent of topoisomerase Ⅱ β [J]. British Journal of Cancer, 2001, 85(5): 747-751. |
| 166 | CANEL C, MORAES R M, DAYAN F E, et al. Podophyllotoxin[J]. Phytochemistry, 2000, 54(2): 115-120. |
| 167 | DINKOVA-KOSTOVA A T, GANG D R, DAVIN L B, et al. (+)-pinoresinol/(+)-lariciresinol reductase from Forsythia intermedia [J]. Journal of Biological Chemistry, 1996, 271(46): 29473-29482. |
| 168 | DAVIN L B, WANG H B, CROWELL A L, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center[J]. Science, 1997, 275(5298): 362-367. |
| 169 | XIA Z Q, COSTA M A, PELISSIER H C, et al. Secoisolariciresinol dehydrogenase purification, cloning, and functional expression. Implications for human health protection[J]. The Journal of Biological Chemistry, 2001, 276(16): 12614-12623. |
| 170 | MARQUES J V, KIM K W, LEE C, et al. Next generation sequencing in predicting gene function in podophyllotoxin biosynthesis[J]. The Journal of Biological Chemistry, 2013, 288(1): 466-479. |
| 171 | LAU W, SATTELY E S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone[J]. Science, 2015, 349(6253): 1224-1228. |
| 172 | SCHULTZ B J, KIM S Y, LAU W, et al. Total biosynthesis for milligram-scale production of etoposide intermediates in a plant chassis[J]. Journal of the American Chemical Society, 2019, 141(49): 19231-19235. |
| 173 | SALERNI B L, BATES D J, ALBERSHARDT T C, et al. Vinblastine induces acute, cell cycle phase-independent apoptosis in some leukemias and lymphomas and can induce acute apoptosis in others when Mcl-1 is suppressed[J]. Molecular Cancer Therapeutics, 2010, 9(4): 791-802. |
| 174 | DHAMODHARAN R, JORDAN M A, THROWER D, et al. Vinblastine suppresses dynamics of individual microtubules in living interphase cells[J]. Molecular Biology of the Cell, 1995, 6(9): 1215-1229. |
| 175 | HARRISON T S, LYSENG-WILLIAMSON K A. Vincristine sulfate liposome injection: a guide to its use in refractory or relapsed acute lymphoblastic leukemia[J]. BioDrugs, 2013, 27(1): 69-74. |
| 176 | O’CONNOR S E, MARESH J J. Chemistry and biology of monoterpene indole alkaloid biosynthesis[J]. Natural Product Reports, 2006, 23(4): 532-547. |
| 177 | DUGÉ DE BERNONVILLE T, CLASTRE M, BESSEAU S, et al. Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine[J]. Phytochemistry, 2015, 113: 9-23. |
| 178 | CAPUTI L, FRANKE J, FARROW S C, et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle [J]. Science, 2018, 360(6394):1235-1239. |
| 179 | ZHANG J, HANSEN L G, GUDICH O, et al. A microbial supply chain for production of the anti-cancer drug vinblastine[J]. Nature, 2022, 609(7926): 341-347. |
| 180 | GAO J C, ZUO Y M, XIAO F, et al. Biosynthesis of catharanthine in engineered Pichia pastoris [J]. Nature Synthesis, 2023, 2: 231-242. |
| 181 | CAMPBELL K B, CICCI T A, VORA A K, et al. Beyond gout: colchicine use in the cardiovascular patient[J]. Hospital Pharmacy, 2015, 50(10): 859-867. |
| 182 | LEUNG Y Y, YAO HUI L L, KRAUS V B. Colchicine — update on mechanisms of action and therapeutic uses[J]. Seminars in Arthritis and Rheumatism, 2015, 45(3): 341-350. |
| 183 | DALBETH N, LAUTERIO T J, WOLFE H R. Mechanism of action of colchicine in the treatment of gout[J]. Clinical Therapeutics, 2014, 36(10): 1465-1479. |
| 184 | BHATTACHARYYA B, PANDA D, GUPTA S, et al. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin[J]. Medicinal Research Reviews, 2008, 28(1): 155-183. |
| 185 | SLOBODNICK A, SHAH B, PILLINGER M H, et al. Colchicine: old and new[J]. The American Journal of Medicine, 2015, 128(5): 461-470. |
| 186 | CHEN B, LIU X, HU Y J, et al. Enantioselective total synthesis of (-)-colchicine, (+)-demecolcinone and metacolchicine: determination of the absolute configurations of the latter two alkaloids[J]. Chemical Science, 2017, 8(7): 4961-4966. |
| 187 | LIANG X, LI L, WEI K, et al. Gram-scale, seven-step total synthesis of (-)-colchicine[J]. Organic Letters, 2021, 23(7): 2731-2735. |
| 188 | HERBERT R B. The biosynthesis of plant alkaloids and nitrogenous microbial metabolites[J]. Natural Product Reports, 2003, 20(5): 494-508. |
| 189 | LARSSON S, RØNSTED N. Reviewing Colchicaceae alkaloids — perspectives of evolution on medicinal chemistry[J]. Current Topics in Medicinal Chemistry, 2014, 14(2): 274-289. |
| 190 | WICKS C, HUDLICKY T, RINNER U. Morphine alkaloids: history, biology, and synthesis[M/OL]// The alkaloids: chemistry and biology. Amsterdam: Elsevier, 2021, 86: 145-342 [2023-12-01]. . |
| 191 | BHANDARI M, BHANDARI A, BHANDARI A. Recent updates on codeine[J]. Pharmaceutical Methods, 2011, 2(1): 3-8. |
| 192 | WINZER T, KERN M, KING A J, et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein[J]. Science, 2015, 349(6245): 309-312. |
| 193 | LAKSTYGAL A M, KOLESNIKOVA T O, KHATSKO S L, et al. DARK classics in chemical neuroscience: atropine, scopolamine, and other anticholinergic deliriant hallucinogens[J]. ACS Chemical Neuroscience, 2019, 10(5): 2144-2159. |
| 194 | KOHNEN-JOHANNSEN K L, KAYSER O. Tropane alkaloids: chemistry, pharmacology, biosynthesis and production[J]. Molecules, 2019, 24(4): 796. |
| 195 | HUANG J P, FANG C L, MA X Y, et al. Tropane alkaloids biosynthesis involves an unusual type Ⅲ polyketide synthase and non-enzymatic condensation[J]. Nature Communications, 2019, 10(1): 4036. |
| 196 | QIU F, ZENG J L, WANG J, et al. Functional genomics analysis reveals two novel genes required for littorine biosynthesis[J]. The New Phytologist, 2020, 225(5): 1906-1914. |
| 197 | QIU F, YANG C X, YUAN L N, et al. A phenylpyruvic acid reductase is required for biosynthesis of tropane alkaloids[J]. Organic Letters, 2018, 20(24): 7807-7810. |
| 198 | ZHAO T F, LI S Q, WANG J, et al. Engineering tropane alkaloid production based on metabolic characterization of ornithine decarboxylase in Atropa belladonna [J]. ACS Synthetic Biology, 2020, 9(2): 437-448. |
| 199 | QIU F, YAN Y J, ZENG J L, et al. Biochemical and metabolic insights into hyoscyamine dehydrogenase[J]. ACS Catalysis, 2021, 11(5): 2912-2924. |
| 200 | SRINIVASAN P, SMOLKE C D. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids[J]. Nature Communications, 2019, 10(1): 3634. |
| 201 | SRINIVASAN P, SMOLKE C D. Biosynthesis of medicinal tropane alkaloids in yeast[J]. Nature, 2020, 585(7826): 614-619. |
| 202 | ADHIKARY S, DUGGAL M K, NAGENDRAN S, et al. Lefamulin: a new hope in the field of community-acquired bacterial pneumonia[J]. Current Pharmacology Reports, 2022, 8(6): 418-426. |
| 203 | PAUKNER S, RIEDL R. Pleuromutilins: potent drugs for resistant bugs-mode of action and resistance[J]. Cold Spring Harbor Perspectives in Medicine, 2017, 7(1): a027110. |
| 204 | GOETHE O, HEUER A, MA X S, et al. Antibacterial properties and clinical potential of pleuromutilins[J]. Natural Product Reports, 2019, 36(1): 220-247. |
| 205 | ARIGONI D. Some studies in the biosynthesis of terpenes and related compounds[J]. Pure and Applied Chemistry, 1968, 17(3/4): 331-348. |
| 206 | BIRCH A J, HOLZAPFEL C W, RICKARDS R W. The structure and some aspects of the biosynthesis of pleuromutilin[J]. Tetrahedron, 1966, 22: 359-387. |
| 207 | LEMKE C, WHITHAM O, PETERS R J. Magnesium-specific ring expansion/contraction catalysed by the class Ⅱ diterpene cyclase from pleuromutilin biosynthesis[J]. Organic & Biomolecular Chemistry, 2020, 18(29): 5586-5588. |
| 208 | TSUKAGOSHI T, TOKIWANO T, OIKAWA H. Studies on the later stage of the biosynthesis of pleuromutilin[J]. Bioscience, Biotechnology, and Biochemistry, 2007, 71(12): 3116-3121. |
| 209 | BAILEY A M, ALBERTI F, KILARU S, et al. Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production[J]. Scientific Reports, 2016, 6: 25202. |
| 210 | ALBERTI F, KHAIRUDIN K, VENEGAS E R, et al. Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives[J]. Nature Communications, 2017, 8(1): 1831. |
| 211 | YAMANE M, MINAMI A, LIU C W, et al. Biosynthetic machinery of diterpene pleuromutilin isolated from Basidiomycete fungi[J]. ChemBioChem, 2017, 18(23): 2317-2322. |
| 212 | ALBERTI F, KHAIRUDIN K, DAVIES J A, et al. Biosynthesis of pleuromutilin congeners using an Aspergillus oryzae expression platform[J]. Chemical Science, 2023, 14(14): 3826-3833. |
| 213 | SCHAFHAUSER T, WIBBERG D, BINDER A, et al. Genome assembly and genetic traits of the pleuromutilin-producer Clitopilus passeckerianus DSM1602[J]. Journal of Fungi, 2022, 8(8): 862. |
| 214 | GUO C, DAI H Q, ZHANG M T, et al. Molecular networking assisted discovery and combinatorial biosynthesis of new antimicrobial pleuromutilins[J]. European Journal of Medicinal Chemistry, 2022, 243: 114713. |
| 215 | BASSO N, TERRAGNO N A. History about the discovery of the renin-angiotensin system[J]. Hypertension, 2001, 38(6): 1246-1249. |
| 216 | LU H, CASSIS L A, KOOI C W V, et al. Structure and functions of angiotensinogen[J]. Hypertension Research, 2016, 39(7): 492-500. |
| 217 | STREATFEILD-JAMES R M A, WILLIAMSON D, PIKE R N, et al. Angiotensinogen cleavage by renin: importance of a structurally constrained N-terminus[J]. FEBS Letters, 1998, 436(2): 267-270. |
| 218 | FANG H, LI D, KANG J, et al. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12 [J]. Nature Communications, 2018, 9(1): 4917. |
| 219 | KANG Q, FANG H, XIANG M J, et al. A synthetic cell-free 36-enzyme reaction system for vitamin B12 production[J]. Nature Communications, 2023, 14(1): 5177. |
| 220 | HU Y J, GU C C, WANG X F, et al. Asymmetric total synthesis of taxol[J]. Journal of the American Chemical Society, 2021, 143(42): 17862-17870. |
| 221 | BIGGS B W, LIM C G, SAGLIANI K, et al. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(12): 3209-3214. |
| 222 | LI J H, MUTANDA I, WANG K B, et al. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana [J]. Nature Communications, 2019, 10(1): 4850. |
| 223 | XIONG X Y, GOU J B, LIAO Q G, et al. The Taxus genome provides insights into paclitaxel biosynthesis[J]. Nature Plants, 2021, 7(8): 1026-1036. |
| 224 | XU B F, LEI L, ZHU X C, et al. Identification and characterization of L-lysine decarboxylase from Huperzia serrata and its role in the metabolic pathway of lycopodium alkaloid[J]. Phytochemistry, 2017, 136: 23-30. |
| 225 | WANG J, ZHANG Z K, JIANG F F, et al. Deciphering the biosynthetic mechanism of pelletierine in Lycopodium alkaloid biosynthesis[J]. Organic Letters, 2020, 22(21): 8725-8729. |
| 226 | NETT R S, DHO Y, TSAI C, et al. Plant carbonic anhydrase-like enzymes in neuroactive alkaloid biosynthesis[J]. Nature, 2023, 624(7990): 182-191. |
| 227 | FORMAN V, LUO D, GEU-FLORES F, et al. A gene cluster in Ginkgo biloba encodes unique multifunctional cytochrome P450s that initiate ginkgolide biosynthesis[J]. Nature Communications, 2022, 13(1): 5143. |
| 228 | ADRIO J, CUEVAS C, MANZANARES I, et al. Total synthesis and biological evaluation of tamandarin B analogues[J]. The Journal of Organic Chemistry, 2007, 72(14): 5129-5138. |
| 229 | WHITE K M, ROSALES R, YILDIZ S, et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A[J]. Science, 2021, 371(6532): 926-931. |
| 230 | BARANOVA A A, ALFEROVA V A, KORSHUN V A, et al. Modern trends in natural antibiotic discovery[J]. Life, 2023, 13(5): 1073. |
| 231 | IMAI Y, MEYER K J, IINISHI A, et al. A new antibiotic selectively kills Gram-negative pathogens[J]. Nature, 2019, 576(7787): 459-464. |
| 232 | SHUKLA R, LAVORE F, MAITY S, et al. Teixobactin kills bacteria by a two-pronged attack on the cell envelope[J]. Nature, 2022, 608(7922): 390-396. |
| 233 | WANG Z Q, KOIRALA B, HERNANDEZ Y, et al. A naturally inspired antibiotic to target multidrug-resistant pathogens[J]. Nature, 2022, 601(7894): 606-611. |
| 234 | YUAN Y J, CHENG S, BIAN G K, et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi[J]. Nature Catalysis, 2022, 5(4): 277-287. |
| 235 | ZHANG M M, WONG F T, WANG Y J, et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters[J]. Nature Chemical Biology, 2017, 13(6): 607-609. |
| 236 | MULLOWNEY M W, DUNCAN K R, ELSAYED S S, et al. Artificial intelligence for natural product drug discovery[J]. Nature Reviews Drug Discovery, 2023, 22(11): 895-916. |
| [1] | 仲泉周, 单依怡, 裴清云, 金艳芸, 王艺涵, 孟璐远, 王歆韵, 张雨鑫, 刘坤媛, 王慧中, 冯尚国. 生物合成法生产α-熊果苷的研究进展[J]. 合成生物学, 2025, 6(1): 118-135. |
| [2] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [3] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [4] | 李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517. |
| [5] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [6] | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| [7] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| [8] | 刘子健, 穆柏杨, 段志强, 王璇, 陆晓杰. 与核酸兼容的化学反应开发进展[J]. 合成生物学, 2024, 5(5): 1102-1124. |
| [9] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| [10] | 谢向前, 郭雯, 王欢, 李进. 含氨基乙烯半胱氨酸核糖体肽的生物合成与化学合成[J]. 合成生物学, 2024, 5(5): 981-996. |
| [11] | 汤志军, 胡友财, 刘文. 酶促4+2和2+2环加成反应:区域与立体选择性的理解与应用[J]. 合成生物学, 2024, 5(3): 401-407. |
| [12] | 张俊, 金诗雪, 云倩, 瞿旭东. 聚酮化合物非天然延伸单元的生物合成与结构改造应用[J]. 合成生物学, 2024, 5(3): 561-570. |
| [13] | 陈锡玮, 张华然, 邹懿. 真菌源非核糖体肽类药物生物合成及代谢工程[J]. 合成生物学, 2024, 5(3): 571-592. |
| [14] | 虞旭昶, 吴辉, 李雷. 文库构建与基因簇靶向筛选驱动的微生物天然产物高效发现[J]. 合成生物学, 2024, 5(3): 492-506. |
| [15] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||