合成生物学 ›› 2024, Vol. 5 ›› Issue (5): 1125-1141.DOI: 10.12211/2096-8280.2024-003
DNA存储系统中的数据写入
张宣梁, 李青婷, 王飞
- 上海交通大学化学化工学院,变革性分子前沿科学中心,致远学院,上海 200240
-
收稿日期:2024-01-02修回日期:2024-05-20出版日期:2024-10-31发布日期:2024-11-20 -
通讯作者:王飞 -
作者简介:张宣梁 (2004—),男,学士。研究方向为DNA存储与计算。 E-mail:zhangxuanliang@sjtu.edu.cn王飞 (1990—),女,博士,副教授。研究方向为DNA分子计算与信息存储。 E-mail:wangfeu@sjtu.edu.cn -
基金资助:国家重点研发计划(2022YFF1201800);国家自然科学基金(22322704)
Data writing in DNA storage systems
ZHANG Xuanliang, LI Qingting, WANG Fei
- School of Chemistry and Chemical Engineering,Frontiers Science Center for Transformative Molecules,Zhiyuan College,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2024-01-02Revised:2024-05-20Online:2024-10-31Published:2024-11-20 -
Contact:WANG Fei
摘要:
世界的数字化给人们的生活带来了极大的变化,但与此同时,史无前例的数据激增使得信息存储面临的挑战日益严峻。随着全球数据总量的指数级增长,传统存储介质将无法满足数字化带来的存储需求。使用DNA分子作为基本载体的信息存储展现出高存储密度、低维护成本和易于化学修饰等独特优势。DNA存储主要包括编码、写入、保存、检索、读取和解码六个主要步骤,其中数据的写入是实现DNA存储功能的基础。本文首先介绍DNA存储系统中体外写入数据的策略方法,主要分为将数据写入DNA序列和写入DNA结构两个部分,接着概述体内写入数据技术的发展,最后将讨论DNA存储系统中数据写入面临的写入成本高、写入速度慢等挑战,并对大规模合成高纯度DNA、改进生物酶等具有前景的应用技术进行展望。
中图分类号:
引用本文
张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141.
ZHANG Xuanliang, LI Qingting, WANG Fei. Data writing in DNA storage systems[J]. Synthetic Biology Journal, 2024, 5(5): 1125-1141.
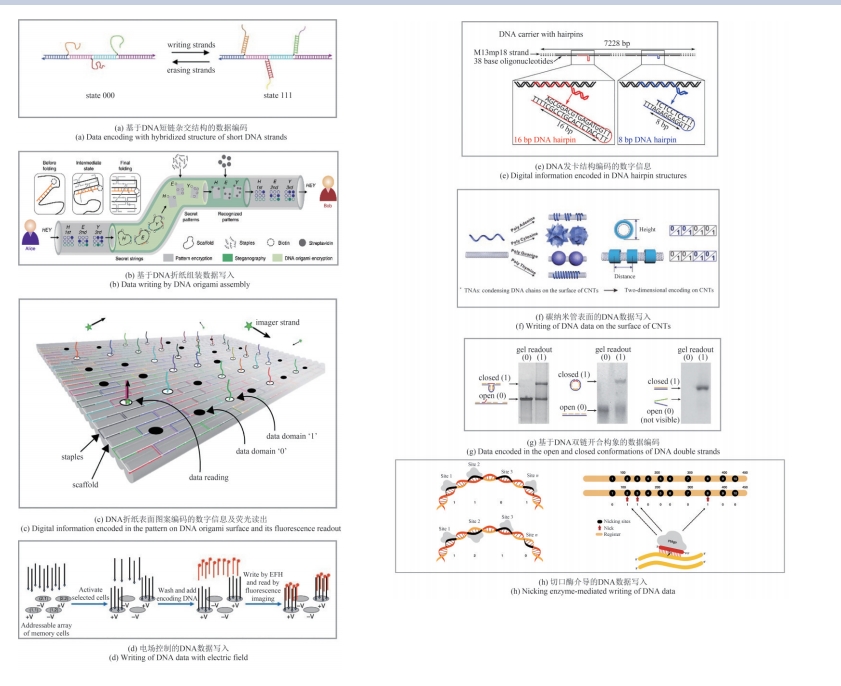
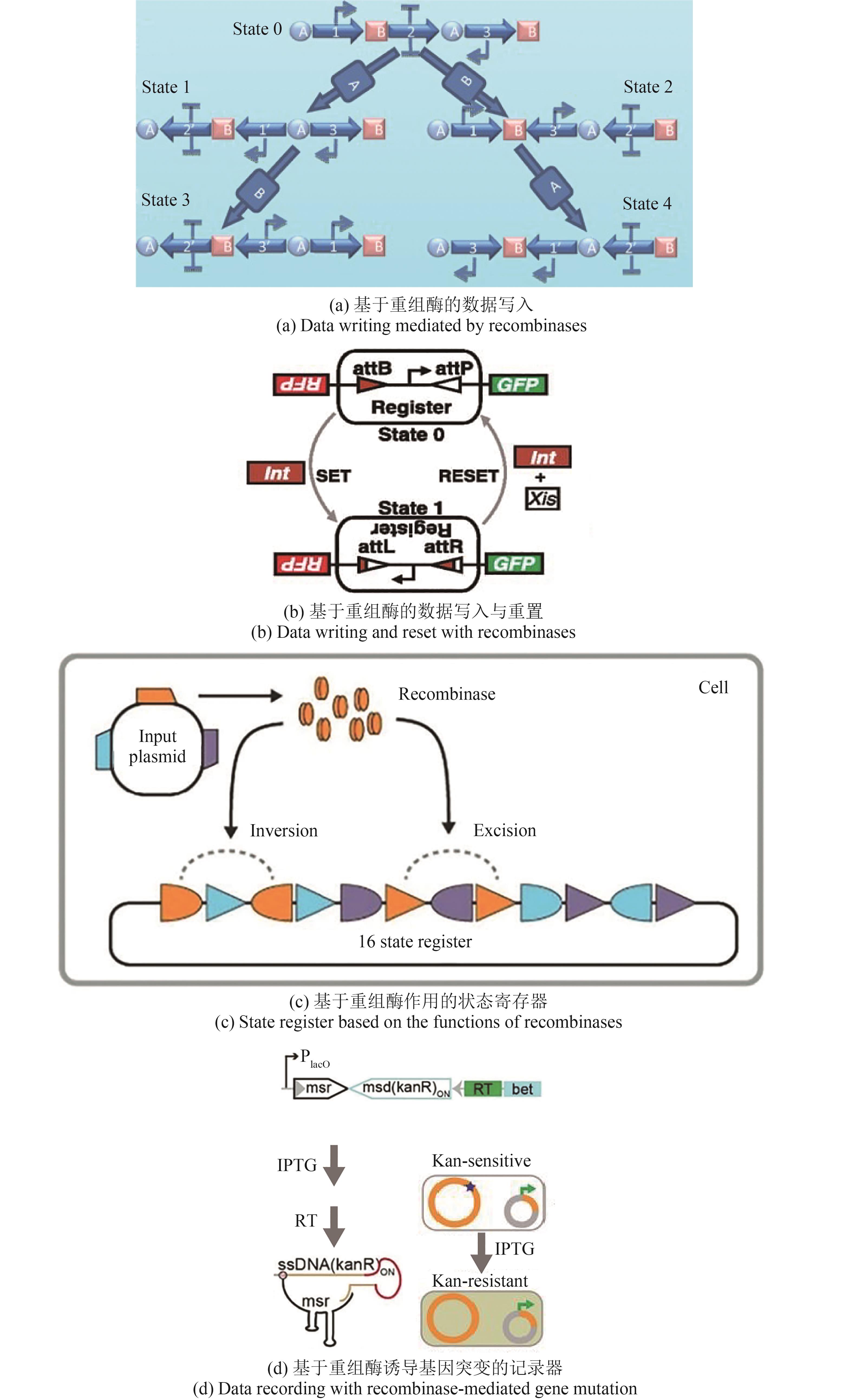
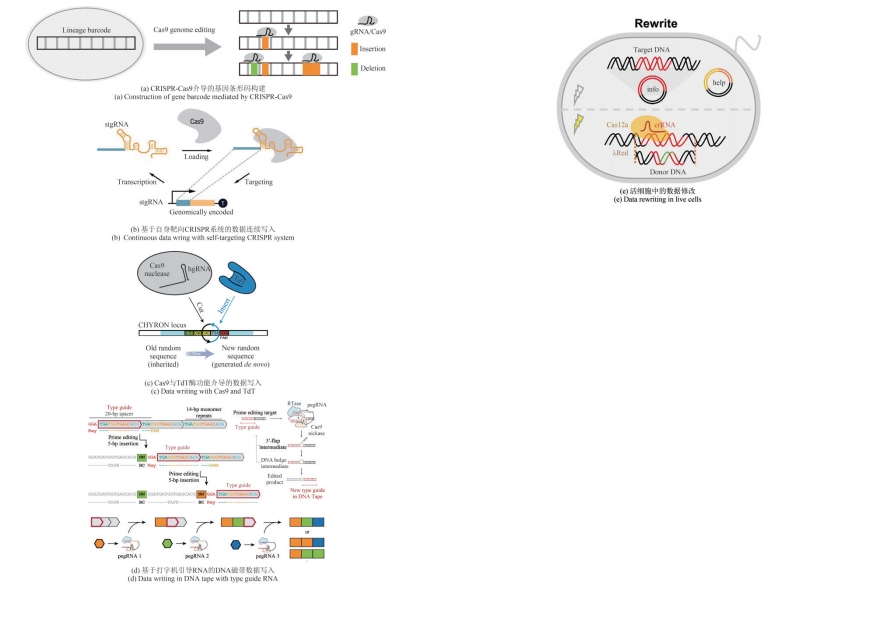
图2 DNA存储系统中基于结构的体外数据写入策略[38, 41-42, 47-50, 52]
Fig.2 In vitro data writing strategies of DNA storage systems based on the structure [38, 41-42, 47-50, 52]
| 1 | WANG S, MAO X, WANG F, et al. Data storage using DNA[J]. Advanced Materials, 2024, 36(6): e2307499. |
| 2 | DORICCHI A, PLATNICH C M, GIMPEL A, et al. Emerging approaches to DNA data storage: challenges and prospects[J]. ACS Nano, 2022, 16(11): 17552-17571. |
| 3 | HAO Y Y, LI Q, FAN C H, et al. Data storage based on DNA [J]. Small Structures, 2021, 2(2): 2000046. |
| 4 | MICHELSON A M, TODD A R. Nucleotides part ⅩⅩⅩⅡ. Synthesis of a dithymidine dinucleotide containing a 3′:5′- internucleotidic linkage[J]. Journal of the Chemical Society, 1955: 2632-2638. |
| 5 | LETSINGER R L, MAHADEVAN V. Oligonucleotide synthesis on a polymer support[J]. Journal of the American Chemical Society, 1965, 87: 3526-3527. |
| 6 | NISHIMURA S, JONES D S, KHORANA H G. Studies on polynucleotides. 48. The in vitro synthesis of a co-polypeptide containing two amino acids in alternating sequence dependent upon a DNA-like polymer containing two nucleotides in alternating sequence[J]. Journal of Molecular Biology, 1965, 13(1): 302-324. |
| 7 | TAYLOR J S, BROCKIE I R, O’DAY C L. A building block for the sequence-specific introduction of cis-syn thymine dimers into oligonucleotides. Solid-phase synthesis of TpT[c,s]pTpT[J]. Journal of the American Chemical Society, 1987, 109, 22, 6735-6742. |
| 8 | LEE H H, KALHOR R, GOELA N, et al. Terminator-free template-independent enzymatic DNA synthesis for digital information storage[J]. Nature Communications, 2019, 10(1): 2383. |
| 9 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 10 | LETSINGER R I, FINNAN J L, HEAVNER G A, et al. Phosphite coupling procedure for generating internucleotide links[J]. Journal of the American Chemical Society, 1975, 97(11): 3278-3279. |
| 11 | LETSINGER R L, LUNSFORD W B. Synthesis of thymidine oligonucleotides by phosphite triester intermediates[J]. Journal of the American Chemical Society, 1976, 98(12): 3655-3661. |
| 12 | MINHAZ UD-DEAN S M. A theoretical model for template-free synthesis of long DNA sequence[J]. Systems and Synthetic Biology, 2008, 2(3-4): 67-73. |
| 13 | MCBRIDE L J, CARUTHERS M H. An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides[J]. Tetrahedron Letters, 1983, 24(3): 245-248. |
| 14 | HOOSE A, VELLACOTT R, STORCH M, et al. DNA synthesis technologies to close the gene writing gap[J]. Nature Reviews Chemistry, 2023, 7(3): 144-161. |
| 15 | KROTZ A H, RENTEL C, GORMAN D, et al. Solution stability and degradation pathway of deoxyribonucleoside phosphoramidites in acetonitrile[J]. Nucleosides, Nucleotides & Nucleic Acids, 2004, 23(5): 767-775. |
| 16 | REN Y B, ZHANG Y, LIU Y W, et al. DNA-based concatenated encoding system for high-reliability and high-density data storage[J]. Small Methods, 2022, 6(4): e2101335. |
| 17 | CHOI Y J, RYU T H, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9(1): 6582. |
| 18 | FEI Z J, GUPTA N, LI M J, et al. Toward highly effective loading of DNA in hydrogels for high-density and long-term information storage[J]. Science Advances, 2023, 9(19): eadg9933. |
| 19 | WOODDELL C I, BURGESS R R. Use of asymmetric PCR to generate long primers and single-stranded DNA for incorporating cross-linking analogs into specific sites in a DNA probe[J]. Genome Research, 1996, 6(9): 886-892. |
| 20 | VENEZIANO R, RATANALERT S, ZHANG K M, et al. Designer nanoscale DNA assemblies programmed from the top down[J]. Science, 2016, 352(6293): 1534. |
| 21 | VENEZIANO R, SHEPHERD T R, RATANALERT S, et al. In vitro synthesis of gene-length single-stranded DNA[J]. Scientific Reports, 2018, 8(1): 6548. |
| 22 | REDDY M P, HANNA N B, FAROOQUI F. Ultrafast cleavage and deprotection of oligonucleotides synthesis and use of CAc derivatives[J]. Nucleosides & Nucleotides, 1997, 16(7/8/9): 1589-1598. |
| 23 | EFCAVITCH J W, HEINER C. Depurination as a yield decreasing mechanism in oligodeoxynucleotide synthesis[J]. Nucleosides & Nucleotides, 1985, 4(1-2): 267. |
| 24 | JENSEN M A, DAVIS R W. Template-independent enzymatic oligonucleotide synthesis (TiEOS): its history, prospects, and challenges[J]. Biochemistry, 2018, 57(12): 1821-1832. |
| 25 | GRUNBERG-MANAGO M, ORTIZ P J, OCHOA S. Enzymic synthesis of polynucleotides I. polynucleotide phosphorylase of Azotobacter vinelandii [J]. Biochimica et Biophysica Acta, 1956, 20(1): 269-284. |
| 26 | GILHAM S, SMITH M. Enzymatic synthesis of deoxyribo-oligonucleotides of defined sequence[J]. Nature: New Biology, 1972, 238(86): 233-234. |
| 27 | BOLLUM F J. Thermal conversion of nonpriming deoxyribonucleic acid to primer[J]. Journal of Biological Chemistry, 1959, 234(10): 2733-2734. |
| 28 | BOLLUM F J. Calf thymus polymerase[J]. Journal of Biological Chemistry, 1960, 235(8): 2399-2403. |
| 29 | BOLLUM F J. Oligodeoxyribonucleotide-primed reactions catalyzed by calf thymus polymerase[J]. Journal of Biological Chemistry, 1962, 237(6): 1945-1949. |
| 30 | SCHOTT H, SCHRADE H. Single-step elongation of oligodeoxynucleotides using terminal deoxynucleotidyl transferase[J]. European Journal of Biochemistry, 1984, 143(3): 613-620. |
| 31 | ENGLAND T E, UHLENBECK O C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase[J]. Biochemistry, 1978, 17(11): 2069-2076. |
| 32 | SCHMITZ C, REETZ M T. Solid-phase enzymatic synthesis of oligonucleotides[J]. Organic Letters, 1999, 1(11): 1729-1731. |
| 33 | PAN C, TABATABAEI S K, HOSSEIN TABATABAEI YAZDI S M H, et al. Rewritable two-dimensional DNA-based data storage with machine learning reconstruction[J]. Nature Communications, 2022, 13(1): 2984. |
| 34 | KAUFMANN G, KALLENBACH N R. Determination of recognition sites of T4 RNA ligase on the 3'-OH and 5'-P termini of polyribonucleotide chains[J]. Nature, 1975, 254(5499): 452-454. |
| 35 | VERARDO D, ADELIZZI B, RODRIGUEZ-PINZON D A, et al. Multiplex enzymatic synthesis of DNA with single-base resolution[J]. Science Advances, 2023, 9(27): eadi0263. |
| 36 | ZHANG C, WU R F, SUN F J, et al. Parallel molecular data storage by printing epigenetic bits on DNA[EB/OL]. bioRxiv: 2023.12.15.571646. (2023-12-15)[2023-12-16]. . |
| 37 | GONG Z Y, SONG L F, PEI G S, et al. Engineering DNA materials for sustainable data storage using a DNA movable-type system[J]. Engineering, 2023, 29: 130-136. |
| 38 | SHIN J S, PIERCE N A. Rewritable memory by controllable nanopatterning of DNA[J]. Nano Letters, 2004, 4(5): 905-909. |
| 39 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440(7082): 297-302. |
| 40 | LIN C X, JUNGMANN R, LEIFER A M, et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA[J]. Nature Chemistry, 2012, 4(10): 832-839. |
| 41 | ZHANG Y N, WANG F, CHAO J, et al. DNA origami cryptography for secure communication[J]. Nature Communications, 2019, 10(1): 5469. |
| 42 | DICKINSON G D, MORTUZA G M, CLAY W, et al. An alternative approach to nucleic acid memory[J]. Nature Communications, 2021, 12(1): 2371. |
| 43 | PARDATSCHER G, BRACHA D, DAUBE S S, et al. DNA condensation in one dimension[J]. Nature Nanotechnology, 2016, 11(12): 1076-1081. |
| 44 | WANG S P, CAI X Q, WANG L H, et al. DNA orientation-specific adhesion and patterning of living mammalian cells on self-assembled DNA monolayers[J]. Chemical Science, 2016, 7(4): 2722-2727. |
| 45 | HUANG F J, XU H G, TAN W H, et al. Multicolor and erasable DNA photolithography[J]. ACS Nano, 2014, 8(7): 6849-6855. |
| 46 | NGUYEN H H, PARK J, HWANG S, et al. On-chip fluorescence switching system for constructing a rewritable random access data storage device[J]. Scientific Reports, 2018, 8(1): 337. |
| 47 | SONG Y, KIM S, HELLER M J, et al. DNA multi-bit non-volatile memory and bit-shifting operations using addressable electrode arrays and electric field-induced hybridization[J]. Nature Communications, 2018, 9(1): 281. |
| 48 | CHEN K K, KONG J L, ZHU J B, et al. Digital data storage using DNA nanostructures and solid-state nanopores[J]. Nano Letters, 2019, 19(2): 1210-1215. |
| 49 | ZHANG Y Y, LI F, LI M, et al. Encoding carbon nanotubes with tubular nucleic acids for information storage[J]. Journal of the American Chemical Society, 2019, 141(44): 17861-17866. |
| 50 | HALVORSEN K, WONG W P. Binary DNA nanostructures for data encryption[J]. PLoS One, 2012, 7(9): e44212. |
| 51 | CHANDRASEKARAN A R, LEVCHENKO O, PATEL D S, et al. Addressable configurations of DNA nanostructures for rewritable memory[J]. Nucleic Acids Research, 2017, 45(19): 11459-11465. |
| 52 | TABATABAEI S K, WANG B Y, ATHREYA N B M, et al. DNA punch cards for storing data on native DNA sequences via enzymatic nicking[J]. Nature Communications, 2020, 11(1): 1742. |
| 53 | ZHANG J X, FANG J Z, DUAN W, et al. Predicting DNA hybridization kinetics from sequence[J]. Nature Chemistry, 2018, 10(1): 91-98. |
| 54 | ZHU J B, ERMANN N, CHEN K K, et al. Image encoding using multi-level DNA barcodes with nanopore readout[J]. Small, 2021, 17(28): e2100711. |
| 55 | GE Z L, GU H Z, LI Q, et al. Concept and development of framework nucleic acids[J]. Journal of the American Chemical Society, 2018, 140(51): 17808-17819. |
| 56 | DEY S, FAN C H, GOTHELF K V, et al. DNA origami [J]. Nature Reviews Methods Primers, 2021, 1(1): 13. |
| 57 | NGUYEN B H, TAKAHASHI C N, GUPTA G, et al. Scaling DNA data storage with nanoscale electrode wells[J]. Science Advances, 2021, 7(48): eabi6714. |
| 58 | ZHAO M Y, CHEN Y H, WANG K X, et al. DNA-directed nanofabrication of high-performance carbon nanotube field-effect transistors[J]. Science, 2020, 368(6493): 878-881. |
| 59 | GOPINATH A, THACHUK C, MITSKOVETS A, et al. Absolute and arbitrary orientation of single-molecule shapes[J]. Science, 2021, 371(6531): eabd6179. |
| 60 | DOUGLAS S M, DIETZ H, LIEDL T, et al. Self-assembly of DNA into nanoscale three-dimensional shapes[J]. Nature, 2009, 459(7245): 414-418. |
| 61 | HAM T S, LEE S K, KEASLING J D, et al. Design and construction of a double inversion recombination switch for heritable sequential genetic memory[J]. PLoS One, 2008, 3(7): e2815. |
| 62 | BONNET J, SUBSOONTORN P, ENDY D. Rewritable digital data storage in live cells via engineered control of recombination directionality[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(23): 8884-8889. |
| 63 | ROQUET N, SOLEIMANY A P, FERRIS A C, et al. Synthetic recombinase-based state machines in living cells[J]. Science, 2016, 353(6297): aad8559. |
| 64 | FARZADFARD F, LU T K. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations[J]. Science, 2014, 346(6211): 1256272. |
| 65 | HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and Archaea[J]. Science, 2010, 327(5962): 167-170. |
| 66 | SHETH R U, YIM S S, WU F L, et al. Multiplex recording of cellular events over time on CRISPR biological tape[J]. Science, 2017, 358(6369): 1457-1461. |
| 67 | SCHMIDT F, CHEREPKOVA M Y, PLATT R J. Transcriptional recording by CRISPR spacer acquisition from RNA[J]. Nature, 2018, 562(7727): 380-385. |
| 68 | McKENNA A, FINDLAY G M, GAGNON J A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing[J]. Science, 2016, 353(6298): aaf7907. |
| 69 | CHAN M M, SMITH Z D, GROSSWENDT S, et al. Molecular recording of mammalian embryogenesis[J]. Nature, 2019, 570(7759): 77-82. |
| 70 | PERLI S D, CUI C H, LU T K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells[J]. Science, 2016, 353(6304): aag0511. |
| 71 | LOVELESS T B, GROTTS J H, SCHECHTER M W, et al. Lineage tracing and analog recording in mammalian cells by single-site DNA writing[J]. Nature Chemical Biology, 2021, 17(6): 739-747. |
| 72 | REES H A, LIU D R. Base editing: precision chemistry on the genome and transcriptome of living cells[J]. Nature Reviews Genetics, 2018, 19(12): 770-788. |
| 73 | CHEN P J, LIU D R. Prime editing for precise and highly versatile genome manipulation[J]. Nature Reviews Genetics, 2023, 24(3): 161-177. |
| 74 | ZOU Z P, YE B C. Long-term rewritable report and recording of environmental stimuli in engineered bacterial populations[J]. ACS Synthetic Biology, 2020, 9(9): 2440-2449. |
| 75 | TANG W X, LIU D R. Rewritable multi-event analog recording in bacterial and mammalian cells[J]. Science, 2018, 360(6385): eaap8992. |
| 76 | CHOI J H, CHEN W, MINKINA A, et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing[J]. Nature, 2022, 608(7921): 98-107. |
| 77 | LIU Y Y, REN Y B, LI J J, et al. In vivo processing of digital information molecularly with targeted specificity and robust reliability[J]. Science Advances, 2022, 8(31): eabo7415. |
| 78 | SADREMOMTAZ A, GLASS R F, GUERRERO J E, et al. Digital data storage on DNA tape using CRISPR base editors[J]. Nature Communications, 2023, 14(1): 6472. |
| 79 | LIM C K, YEOH J W, KUNARTAMA A A, et al. A biological camera that captures and stores images directly into DNA[J]. Nature Communications, 2023, 14(1): 3921. |
| 80 | BELL N A W, KEYSER U F. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores[J]. Nature Nanotechnology, 2016, 11(7): 645-651. |
| 81 | JONES D L, LEROY P, UNOSON C, et al. Kinetics of dCas9 target search in Escherichia coli [J]. Science, 2017, 357(6358): 1420-1424. |
| 82 | DUCANI C, KAUL C, MOCHE M, et al. Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides[J]. Nature Methods, 2013, 10(7): 647-652. |
| 83 | ZHANG Q, XIA K, JIANG M, et al. Catalytic DNA-assisted mass production of arbitrary single-stranded DNA[J]. Angewandte Chemie International Edition, 2023, 62(5): e202212011. |
| 84 | PRAETORIUS F, KICK B, BEHLER K L, et al. Biotechnological mass production of DNA origami[J]. Nature, 2017, 552(7683): 84-87. |
| 85 | PRAETORIUS F, DIETZ H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes[J]. Science, 2017, 355(6331): eaam5488. |
| 86 | PACKER M S, LIU D R. Methods for the directed evolution of proteins[J]. Nature Reviews Genetics, 2015, 16(7): 379-394. |
| 87 | MORRISON M S, PODRACKY C J, LIU D V R. The developing toolkit of continuous directed evolution[J]. Nature Chemical Biology, 2020, 16(6): 610-619. |
| 88 | ESVELT K M, CARLSON J C, LIU D R. A system for the continuous directed evolution of biomolecules[J]. Nature, 2011, 472(7344): 499-503. |
| 89 | MILLER S M, WANG T N, RANDOLPH P B, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs[J]. Nature Biotechnology, 2020, 38(4): 471-481. |
| 90 | LU X Y, LI J L, LI C Y, et al. Enzymatic DNA synthesis by engineering terminal deoxynucleotidyl transferase[J]. ACS Catalysis, 2022, 12(5): 2988-2997. |
| 91 | LOVELOCK S L, CRAWSHAW R, BASLER S, et al. The road to fully programmable protein catalysis[J]. Nature, 2022, 606(7912): 49-58. |
| 92 | YOUNG B E, KUNDU N, SCZEPANSKI J T. Mirror-image oligonucleotides: history and emerging applications[J]. Chemistry, 2019, 25(34): 7981-7990. |
| 93 | FAN C Y, DENG Q, ZHU T F. Bioorthogonal information storage in L-DNA with a high-fidelity mirror-image Pfu DNA polymerase[J]. Nature Biotechnology, 2021, 39(12): 1548-1555. |
| 94 | WANG Z M, XU W L, LIU L, et al. A synthetic molecular system capable of mirror-image genetic replication and transcription[J]. Nature Chemistry, 2016, 8(7): 698-704. |
| 95 | PECH A, ACHENBACH J, JAHNZ M, et al. A thermostable D-polymerase for mirror-image PCR[J]. Nucleic Acids Research, 2017, 45(7): 3997-4005. |
| 96 | CHEN J, CHEN M Y, ZHU T F. Directed evolution and selection of biostable L-DNA aptamers with a mirror-image DNA polymerase[J]. Nature Biotechnology, 2022, 40(11): 1601-1609. |
| 97 | GEERTSEMA H J, AIMOLA G, FABRICIUS V, et al. Left-handed DNA-PAINT for improved super-resolution imaging in the nucleus[J]. Nature Biotechnology, 2021, 39(5): 551-554. |
| 98 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36(3): 242-248 |
| 99 | 黄小罗, 戴俊彪. 人工DNA合成技术:DNA数据存储的基石[J]. 合成生物学, 2021, 2(3): 335-353. |
| HUANG X L, DAI J B. DNA synthesis technology: foundation of DNA data storage[J]. Synthetic Biology Journal, 2021, 2(3): 335-353. | |
| 100 | 闫汉, 肖鹏峰, 刘全俊, 等. DNA微阵列原位化学合成[J]. 合成生物学, 2021, 2(3): 354-370. |
| YAN H, XIAO P F, LIU Q J, et al. In situ chemical synthesis of DNA microarrays[J]. Synthetic Biology Journal, 2021, 2(3): 354-370. |
| [1] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [2] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [3] | 施茜, 吴园园, 杨洋. DNA纳米技术与合成生物学[J]. 合成生物学, 2022, 3(2): 302-319. |
| [4] | 平质, 张颢龄, 陈世宏, 倪鸣, 徐讯, 朱砂, 沈玥. Chamaeleo: DNA存储碱基编解码算法的可拓展集成与系统评估平台[J]. 合成生物学, 2021, 2(3): 412-427. |
| [5] | 陈大明, 张学博, 刘晓, 马悦, 熊燕. 从全球专利分析看DNA合成与信息存储技术发展趋势[J]. 合成生物学, 2021, 2(3): 399-411. |
| [6] | 杨洋, 樊春海. 基于人工染色体的DNA信息存储前沿进展[J]. 合成生物学, 2021, 2(3): 305-308. |
| [7] | 黄小罗, 戴俊彪. 人工DNA合成技术:DNA数据存储的基石[J]. 合成生物学, 2021, 2(3): 335-353. |
| [8] | 闫汉, 肖鹏峰, 刘全俊, 陆祖宏. DNA微阵列原位化学合成[J]. 合成生物学, 2021, 2(3): 354-370. |
| [9] | 韩明哲, 陈为刚, 宋理富, 李炳志, 元英进. DNA信息存储:生命系统与信息系统的桥梁[J]. 合成生物学, 2021, 2(3): 309-322. |
| [10] | 董一名, 孙法家, 武瑞君, 钱珑. DNA数字信息存储的研究进展[J]. 合成生物学, 2021, 2(3): 323-334. |
| [11] | 彭凯, 逯晓云, 程健, 刘莹, 江会锋, 郭晓贤. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||