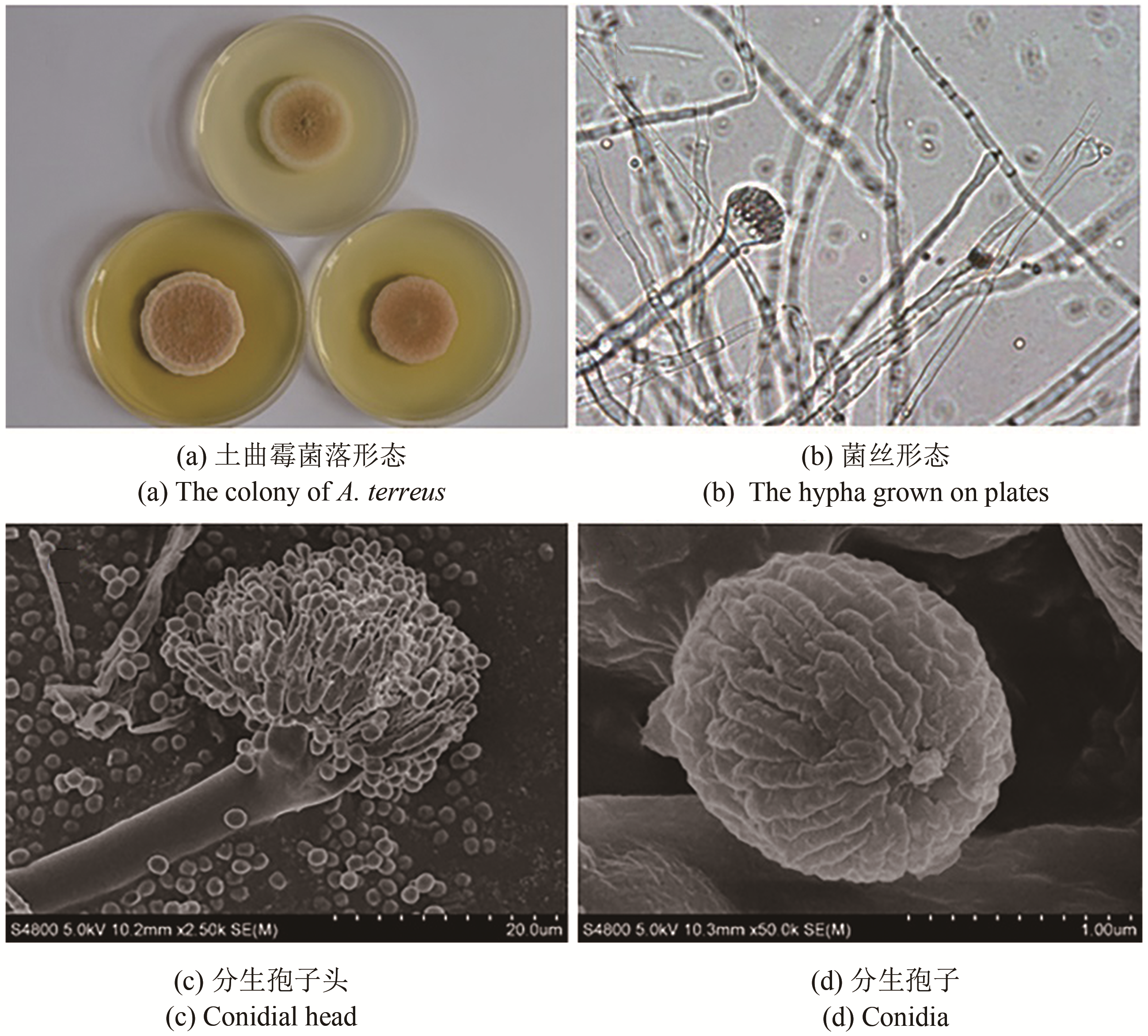
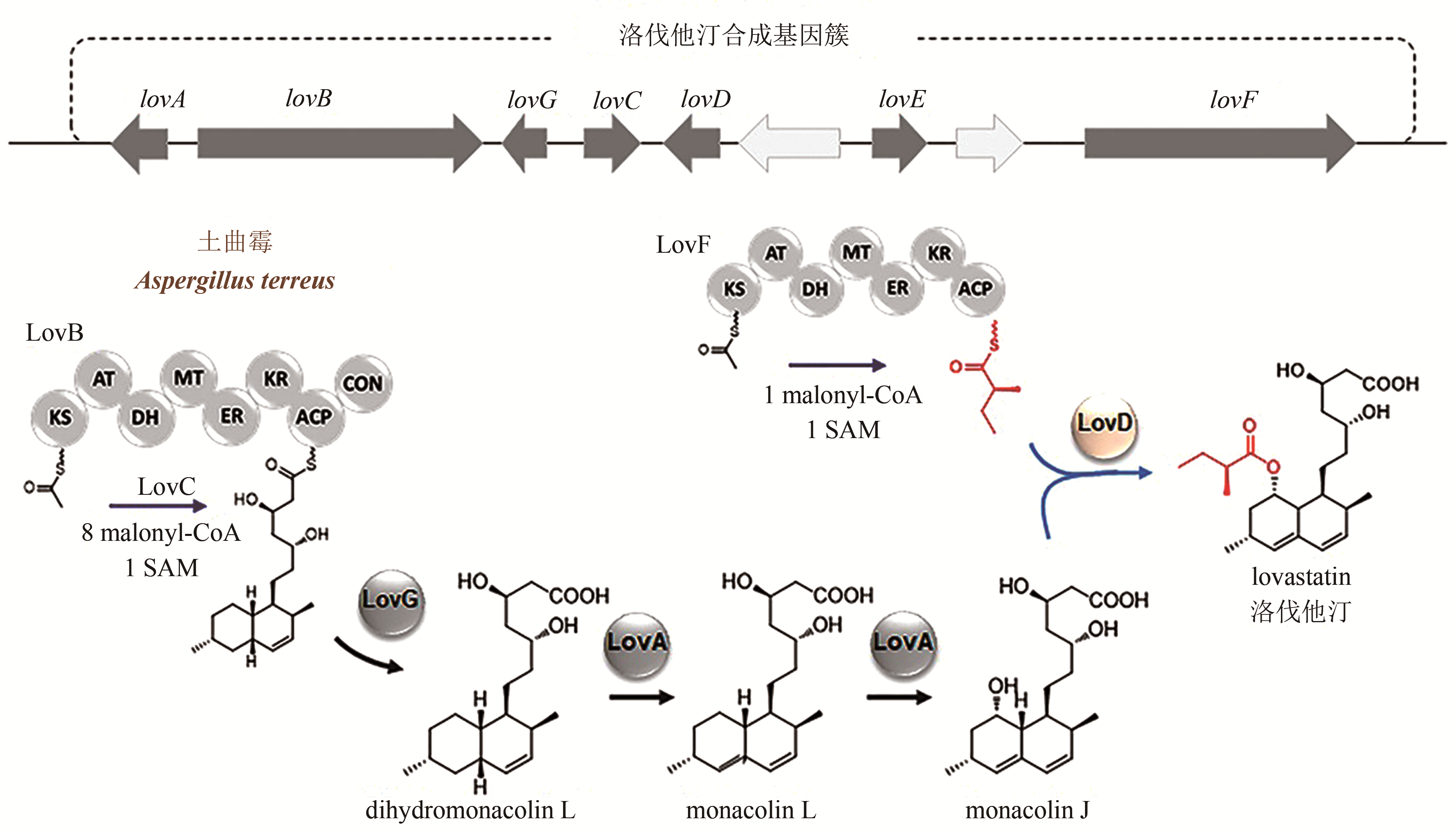
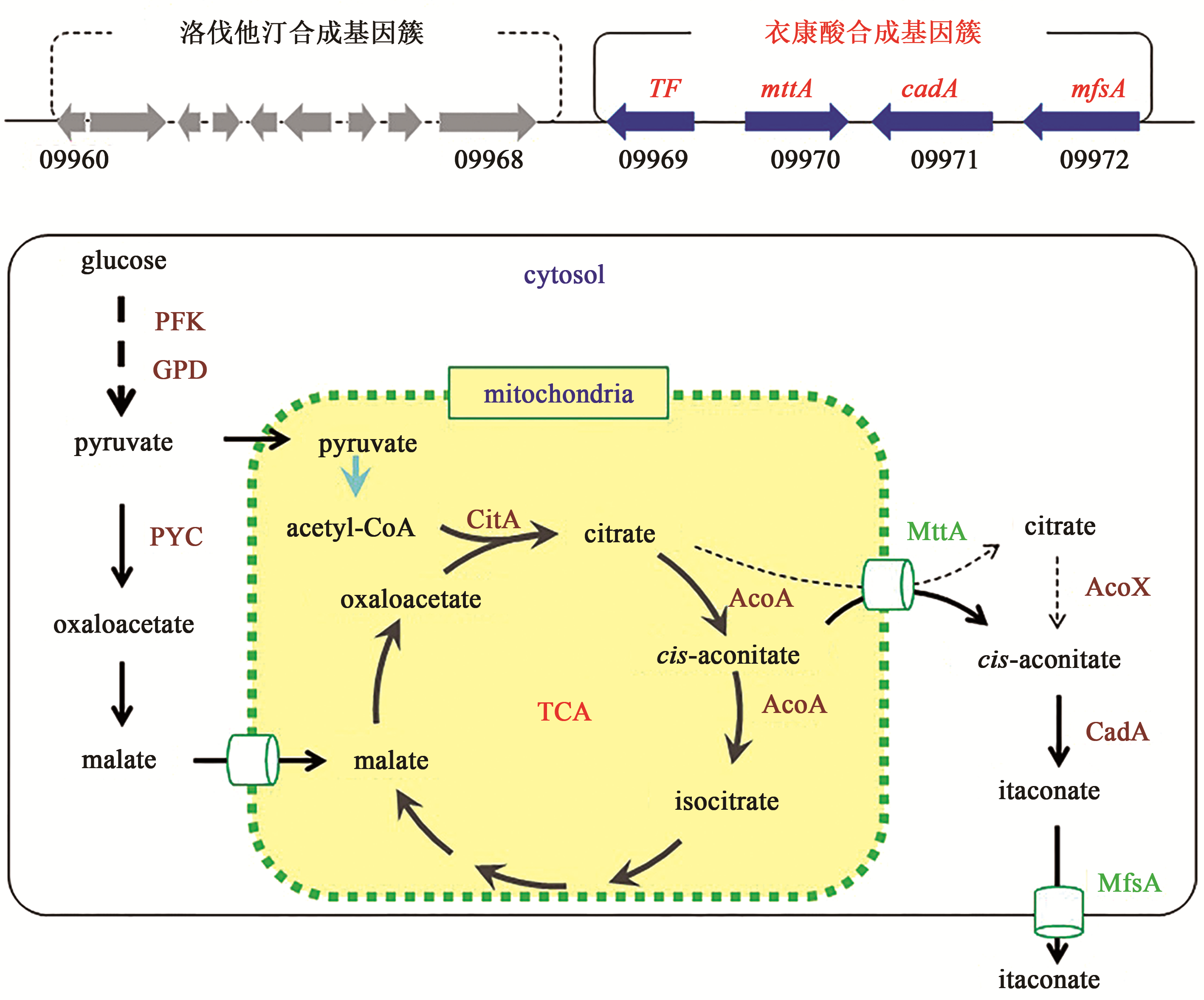
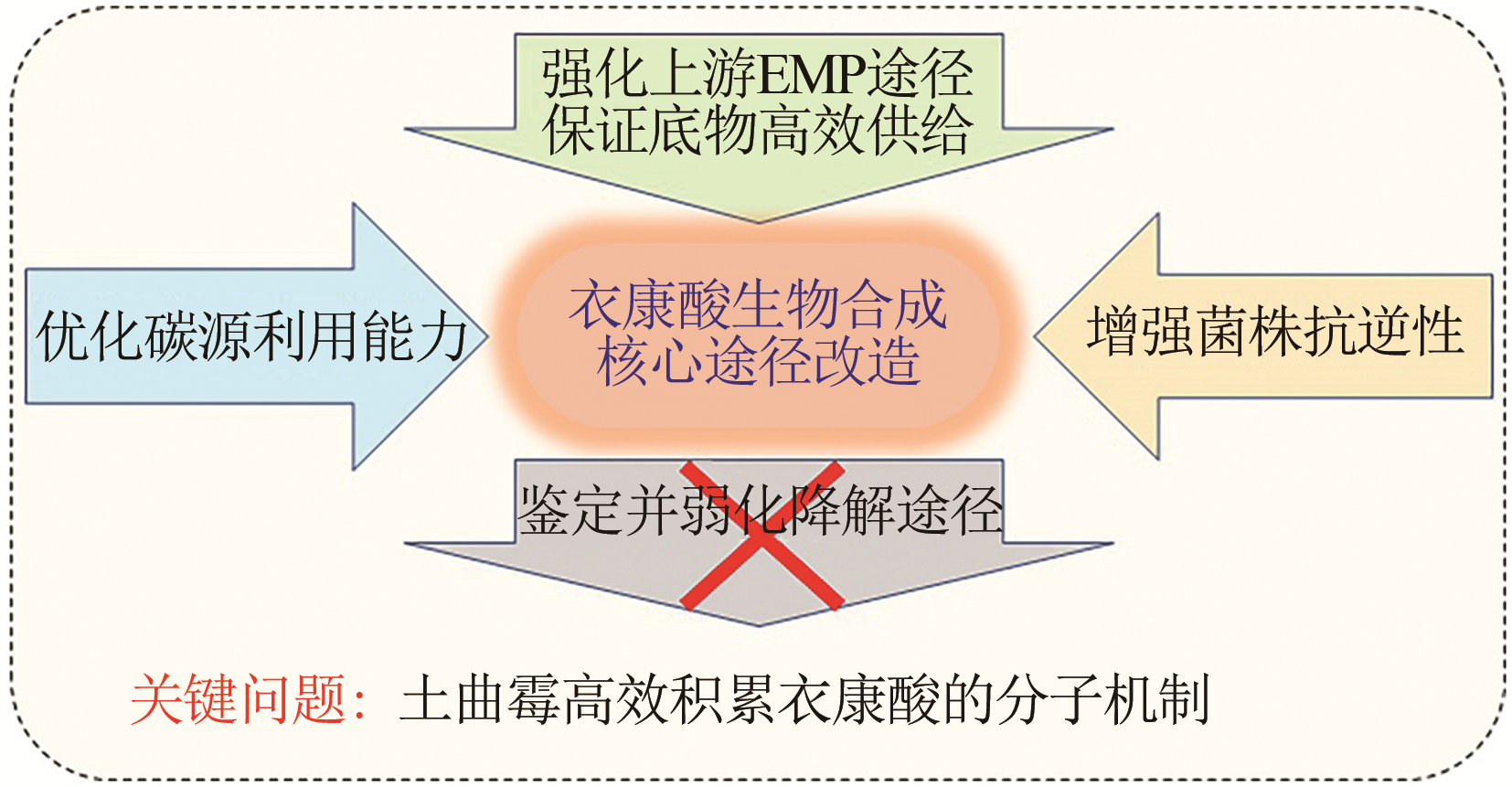
Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (2): 187-211.DOI: 10.12211/2096-8280.2020-002
• Invited Review • Previous Articles Next Articles
Progress and prospect for synthetic biology research of the industrial filamentous fungi Aspergillus terreus
HUANG Xuenian, TANG Shen, LV Xuefeng
- Shandong Provincial Key Laboratory of Synthetic Biology, Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao 266101, Shandong,China
-
Received:2020-02-26Revised:2020-03-23Online:2020-08-04Published:2020-04-30
工业丝状真菌土曲霉合成生物技术研究进展及展望
黄雪年, 唐慎, 吕雪峰
- 中国科学院青岛生物能源与过程研究所,山东省合成生物学重点实验室,中国科学院生物燃料重点实验室,山东 青岛 266101
-
作者简介:黄雪年(1986—),男,博士,副研究员,主要从事丝状真菌天然产物生物合成研究。E-mail:huangxn@qibebt.ac.cn
吕雪峰(1974—),男,博士,研究员,主要从事光合蓝细菌、丝状真菌等微生物的代谢工程与合成生物学研究。E-mail:lvxf@qibebt.ac.cn -
基金资助:中国科学院科技服务网络计划(STS计划)(KFS-STS-QYZX-118);山东省泰山学者人才工程;山东省自然科学基金(ZR2019BC014)
CLC Number:
Cite this article
HUANG Xuenian, TANG Shen, LV Xuefeng. Progress and prospect for synthetic biology research of the industrial filamentous fungi Aspergillus terreus[J]. Synthetic Biology Journal, 2020, 1(2): 187-211.
黄雪年, 唐慎, 吕雪峰. 工业丝状真菌土曲霉合成生物技术研究进展及展望[J]. 合成生物学, 2020, 1(2): 187-211.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-002
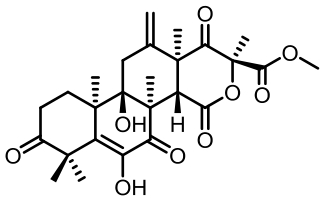
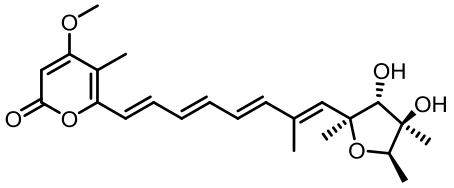
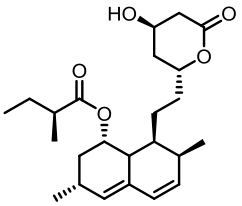
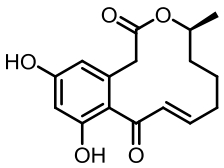
| 序号 | 核心基因编号 | 类型 | 代表性产物 | 参考文献 |
|---|---|---|---|---|
| 1 | ATEG_00145 (terA) | nrPKS |
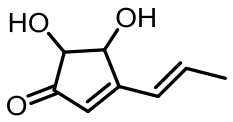
terrein | [ |
| 2 | ATEG_03432 | nrPKS |
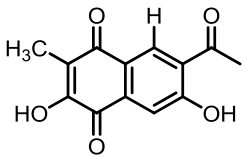
6-acetyl-2,7-dihydroxy-3-methylnaphthalene-1,4-dione | [ |
| 3 | ATEG_06275 (atX) | hrPKS |
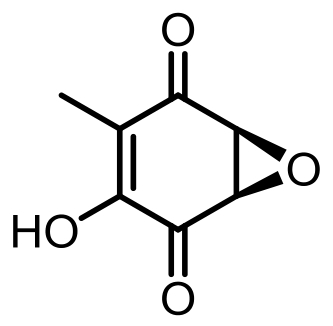
terreic acid | [ |
| 4 | ATEG_08451 (gedC) | nrPKS |
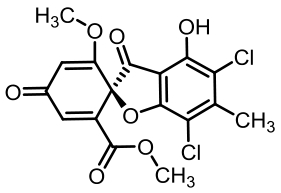
geodin | [ |
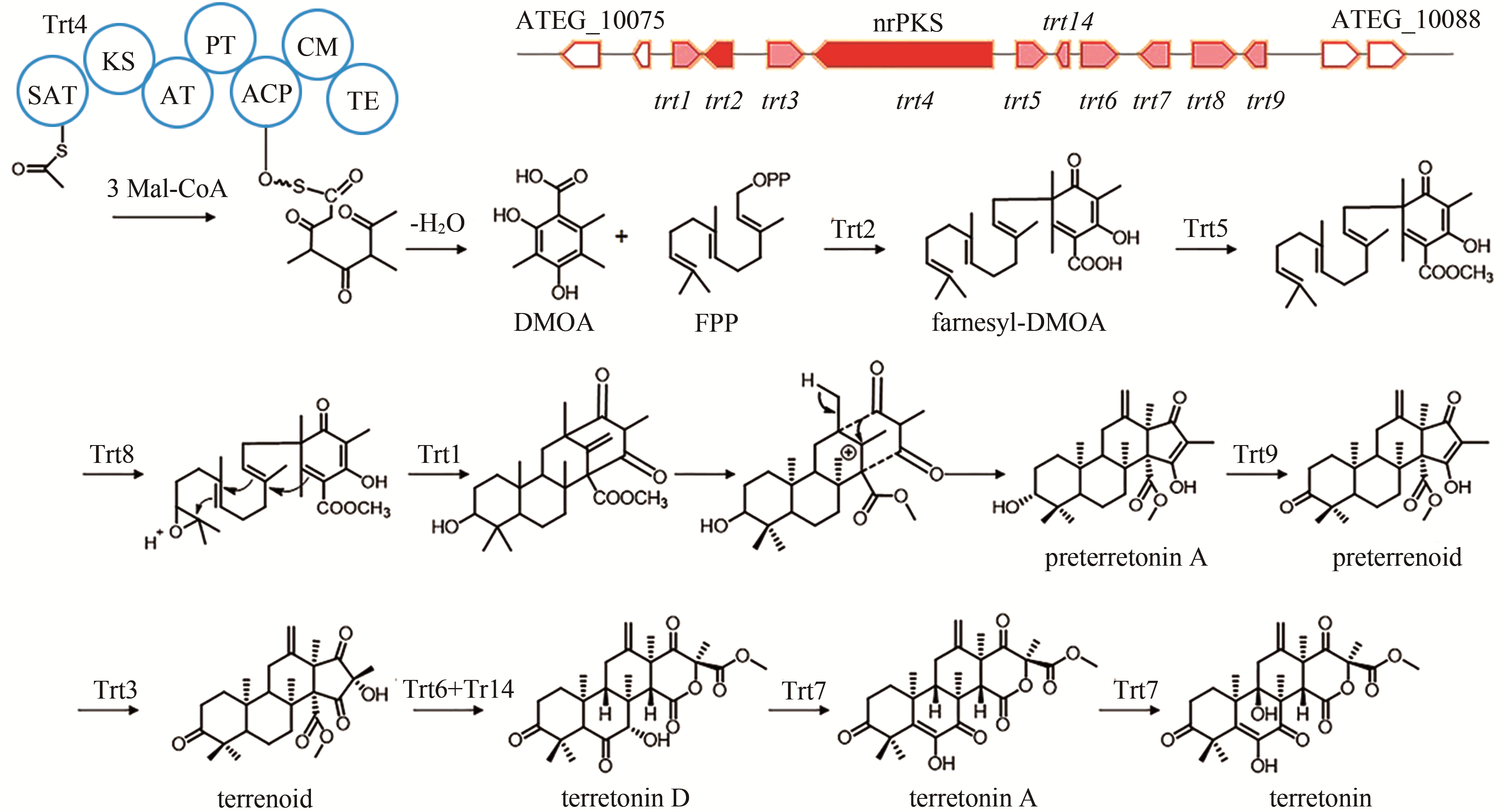
| 5 | ATEG_10080 (trt4) | nrPKS |
terretonin | [ |
| 6 | ATEG_09617 (ctvA) | hrPKS |
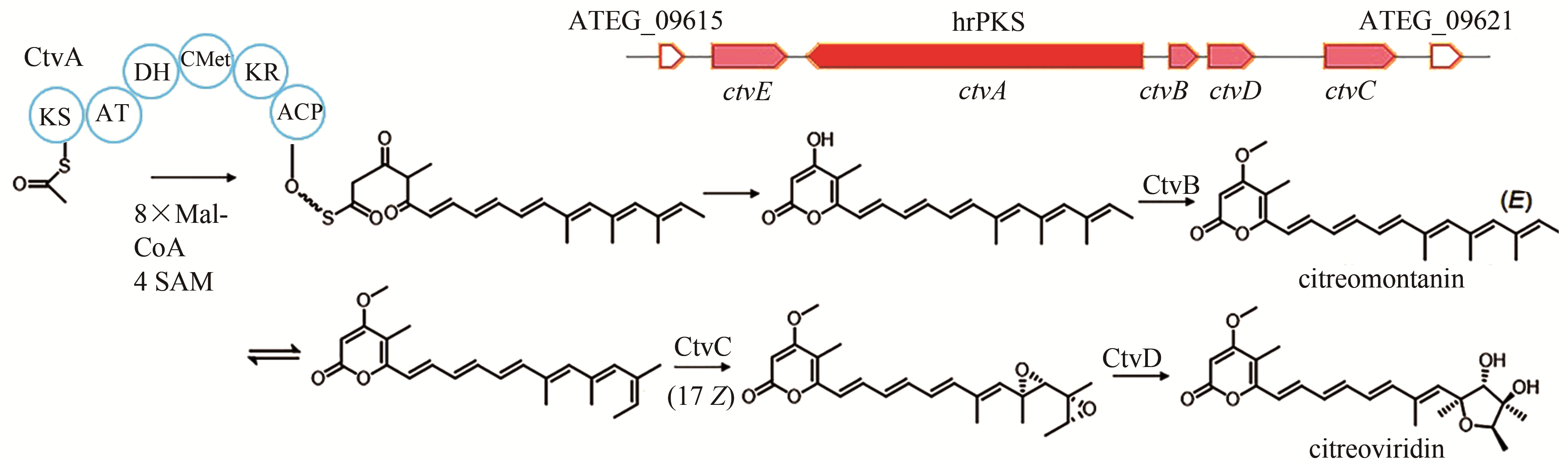
citreoviridin | [ |
| 7 | ATEG_09961 (lovB) | hrPKS |
lovastatin | [ |
| ATEG_09968 (lovF) | hrPKS | |||
| 8 | Atcurs1 | hrPKS |
10,11-dehydrocurvularin | [ |
| Atcurs2 | nrPKS | |||
| 9 | ATEG_03629 | nrPKS |
azasperpyranone A | [ |
| ATEG_03630 | NRPS-like | |||
| ATEG_07659 | hrPKS | |||
| ATEG_07661 | nrPKS | |||
| 10 | ATEG_03470 (ataP) | NRPS |
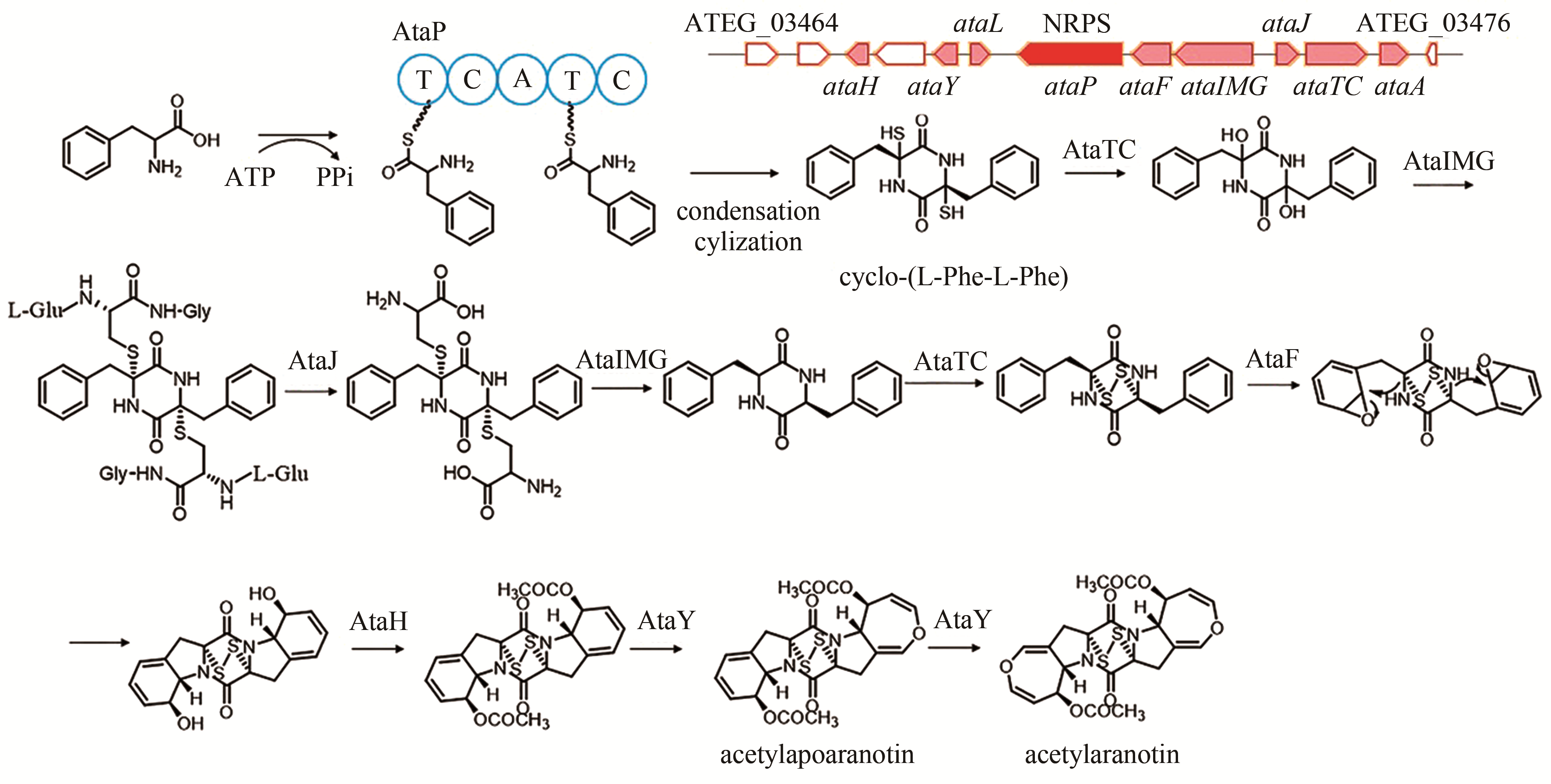
acetylaranotin | [ |
| 11 | ATEG_10305 (anaPS) | NRPS |
asterrelenin | [ |
| 12 | ATEG_09064 (apmB) | NRPS |
asperphenamate | [ |
| ATEG_09068 (apmA) | NRPS | |||
| 13 | ATEG_00700 (atqA) | NRPS-like |
asterriquinones CT5 | [ |
| 14 | ATEG_02004 (apvA) | NRPS-like |
aspulvinones B1 | [ |
| 15 | ATEG_02815 (btyA) | NRPS-like |
butyrolactones I | [ |
| 16 | ATEG_03563 (atmelA) | NRPS-like |
aspulvinone E (asp-melanin前体) | [ |
| 17 | ATEG_08899 (pgnA) | NRPS-like |
phenguignardic acid | [ |
| 18 | ATEG_00325 | PKS-NRPS |
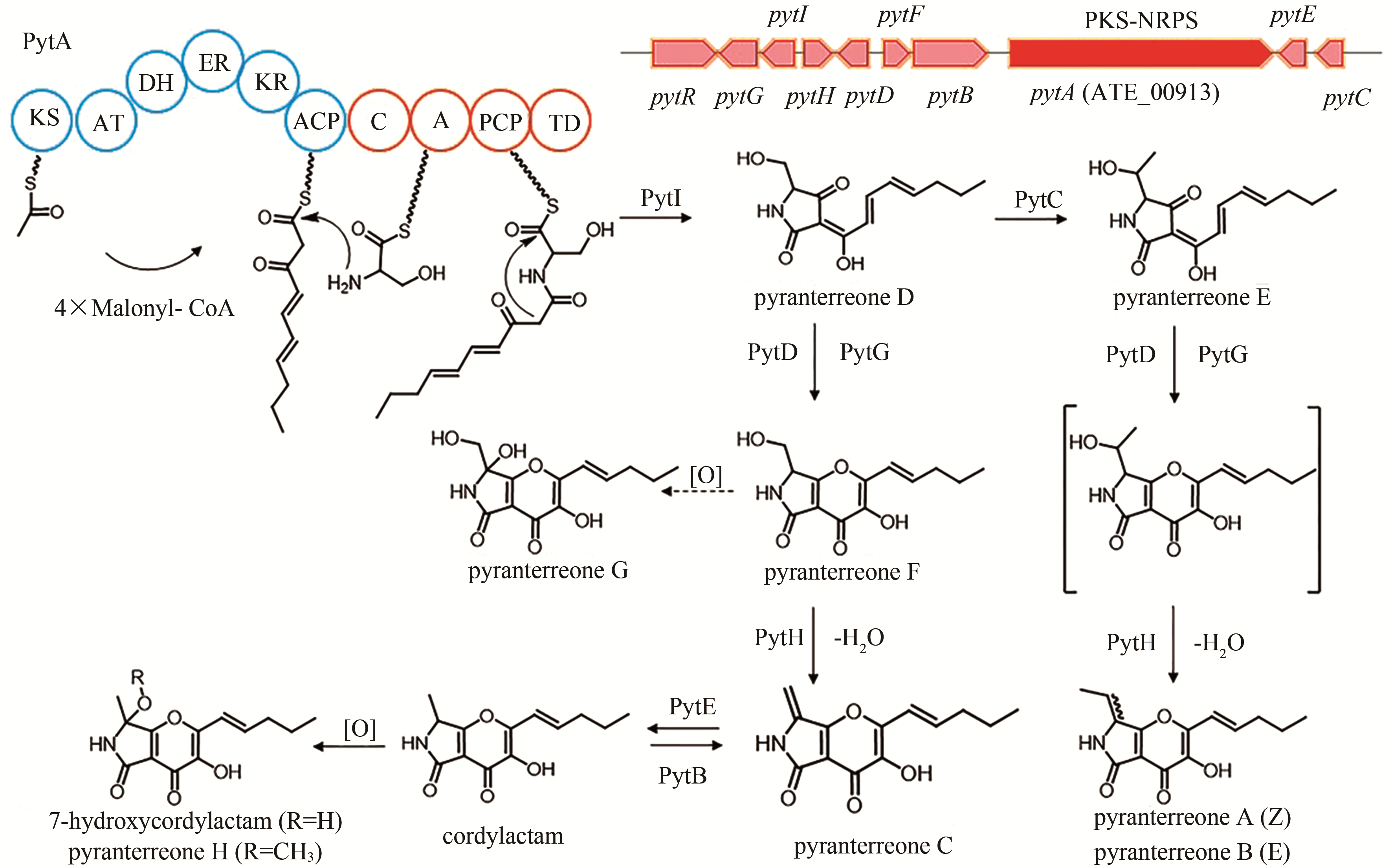
isoflavipucine | [ |
| 19 | ATEG_00913 (pytA) | PKS-NRPS |
pyranterreone C | [ |
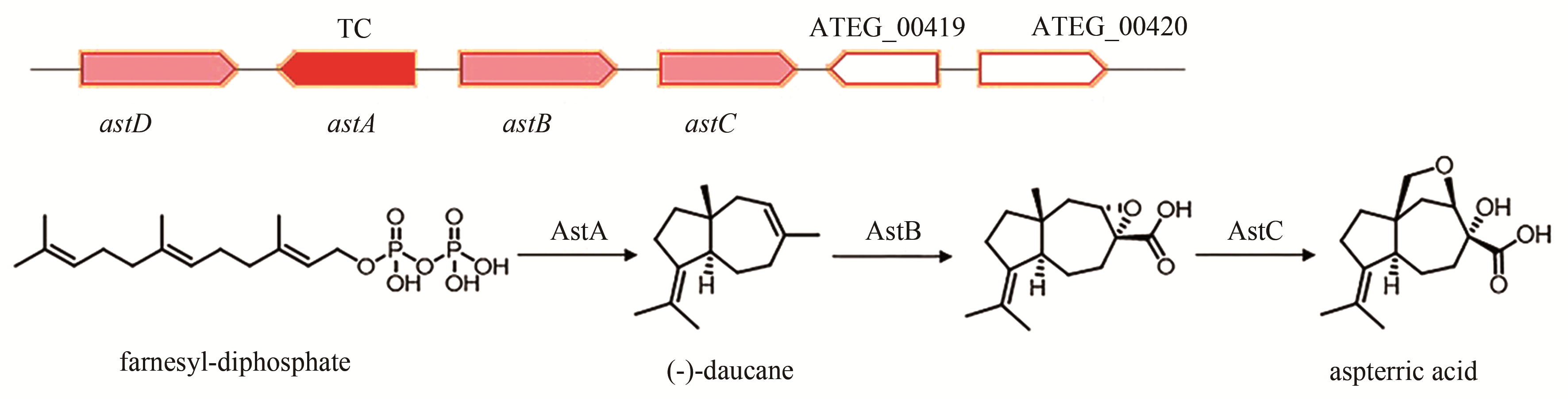
| 20 | ATEG_04416 (astA) | TC |
aspterric acid | [ |
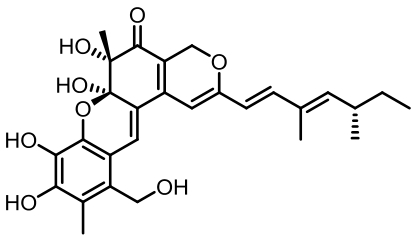
Tab. 1 Biosynthetic gene clusters and products identified in A. terreus
| 序号 | 核心基因编号 | 类型 | 代表性产物 | 参考文献 |
|---|---|---|---|---|
| 1 | ATEG_00145 (terA) | nrPKS |
terrein | [ |
| 2 | ATEG_03432 | nrPKS |
6-acetyl-2,7-dihydroxy-3-methylnaphthalene-1,4-dione | [ |
| 3 | ATEG_06275 (atX) | hrPKS |
terreic acid | [ |
| 4 | ATEG_08451 (gedC) | nrPKS |
geodin | [ |
| 5 | ATEG_10080 (trt4) | nrPKS |
terretonin | [ |
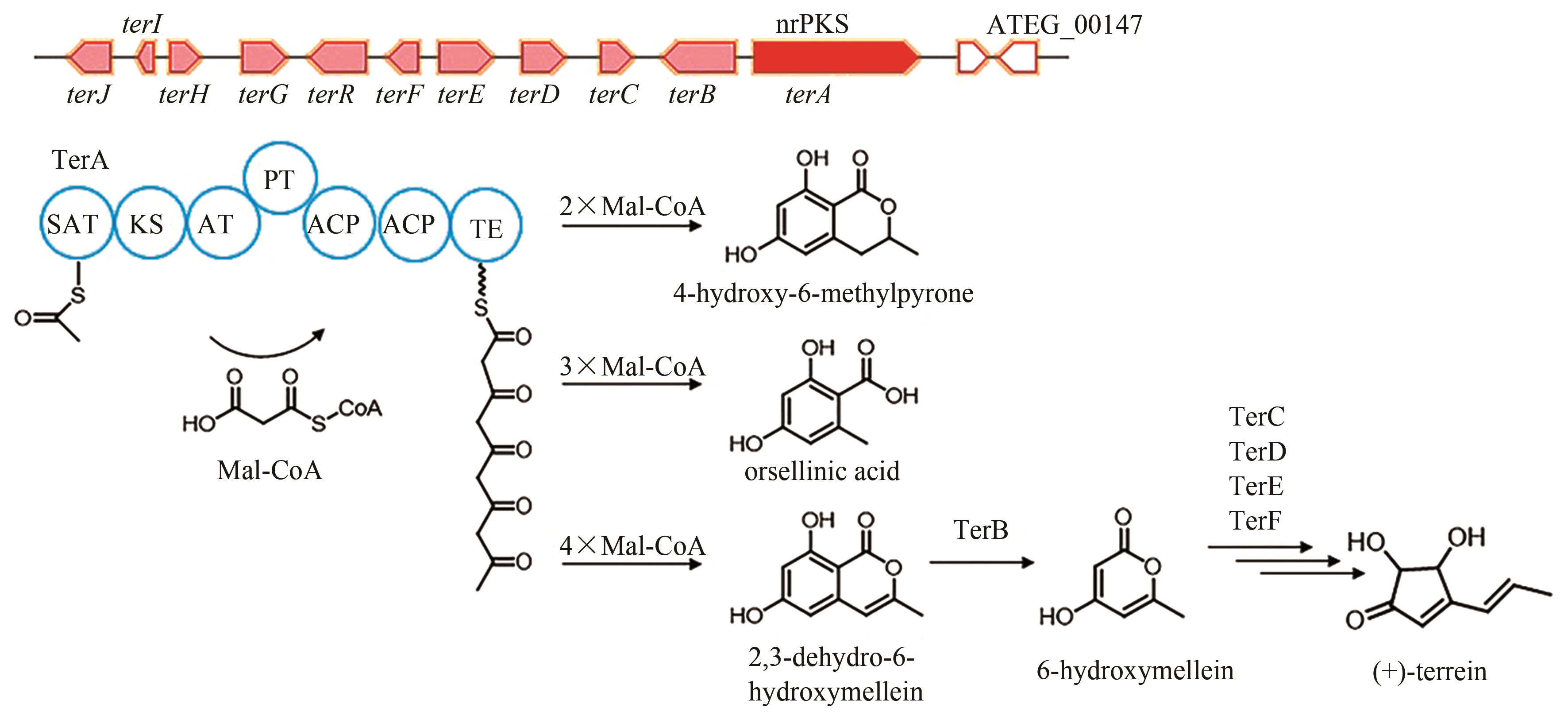
| 6 | ATEG_09617 (ctvA) | hrPKS |
citreoviridin | [ |
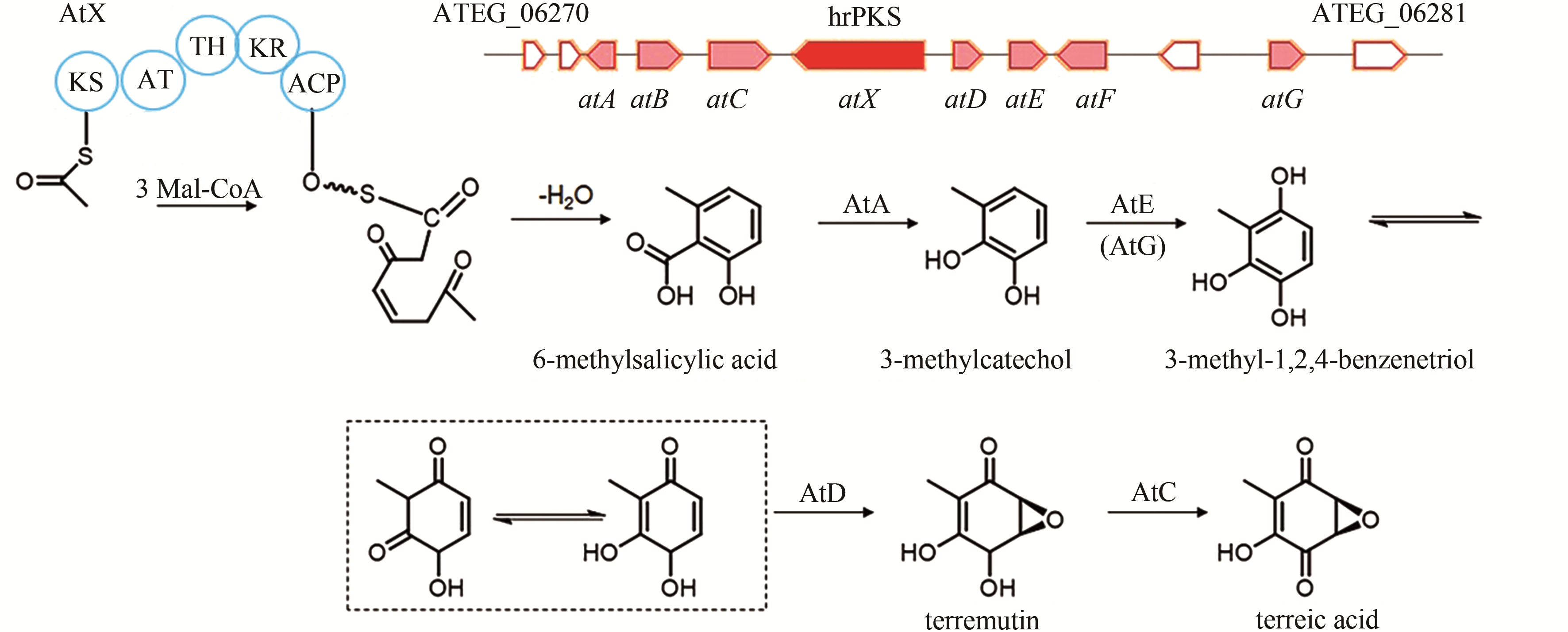
| 7 | ATEG_09961 (lovB) | hrPKS |
lovastatin | [ |
| ATEG_09968 (lovF) | hrPKS | |||
| 8 | Atcurs1 | hrPKS |
10,11-dehydrocurvularin | [ |
| Atcurs2 | nrPKS | |||
| 9 | ATEG_03629 | nrPKS |
azasperpyranone A | [ |
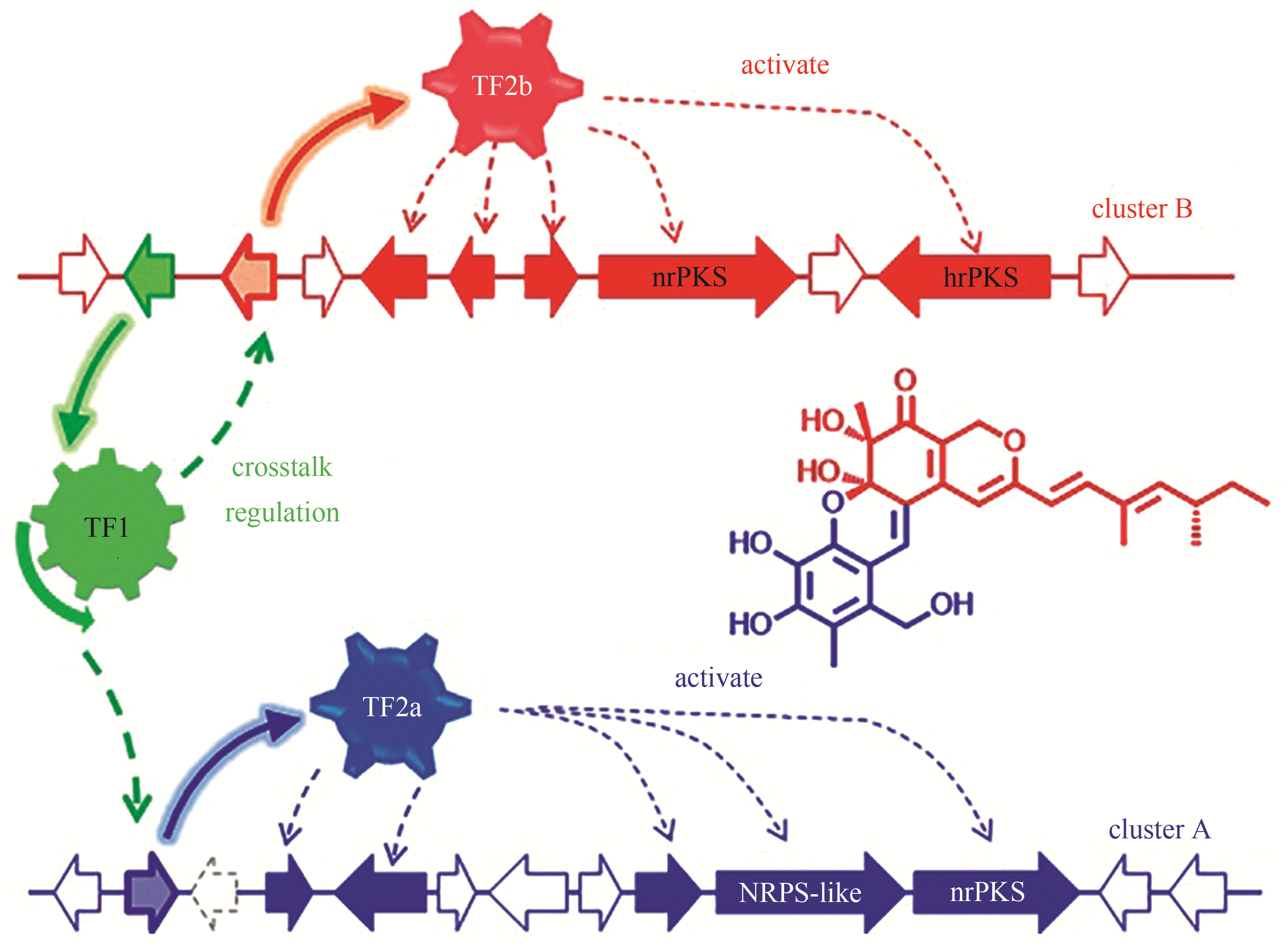
| ATEG_03630 | NRPS-like | |||
| ATEG_07659 | hrPKS | |||
| ATEG_07661 | nrPKS | |||
| 10 | ATEG_03470 (ataP) | NRPS |
acetylaranotin | [ |
| 11 | ATEG_10305 (anaPS) | NRPS |
asterrelenin | [ |
| 12 | ATEG_09064 (apmB) | NRPS |
asperphenamate | [ |
| ATEG_09068 (apmA) | NRPS | |||
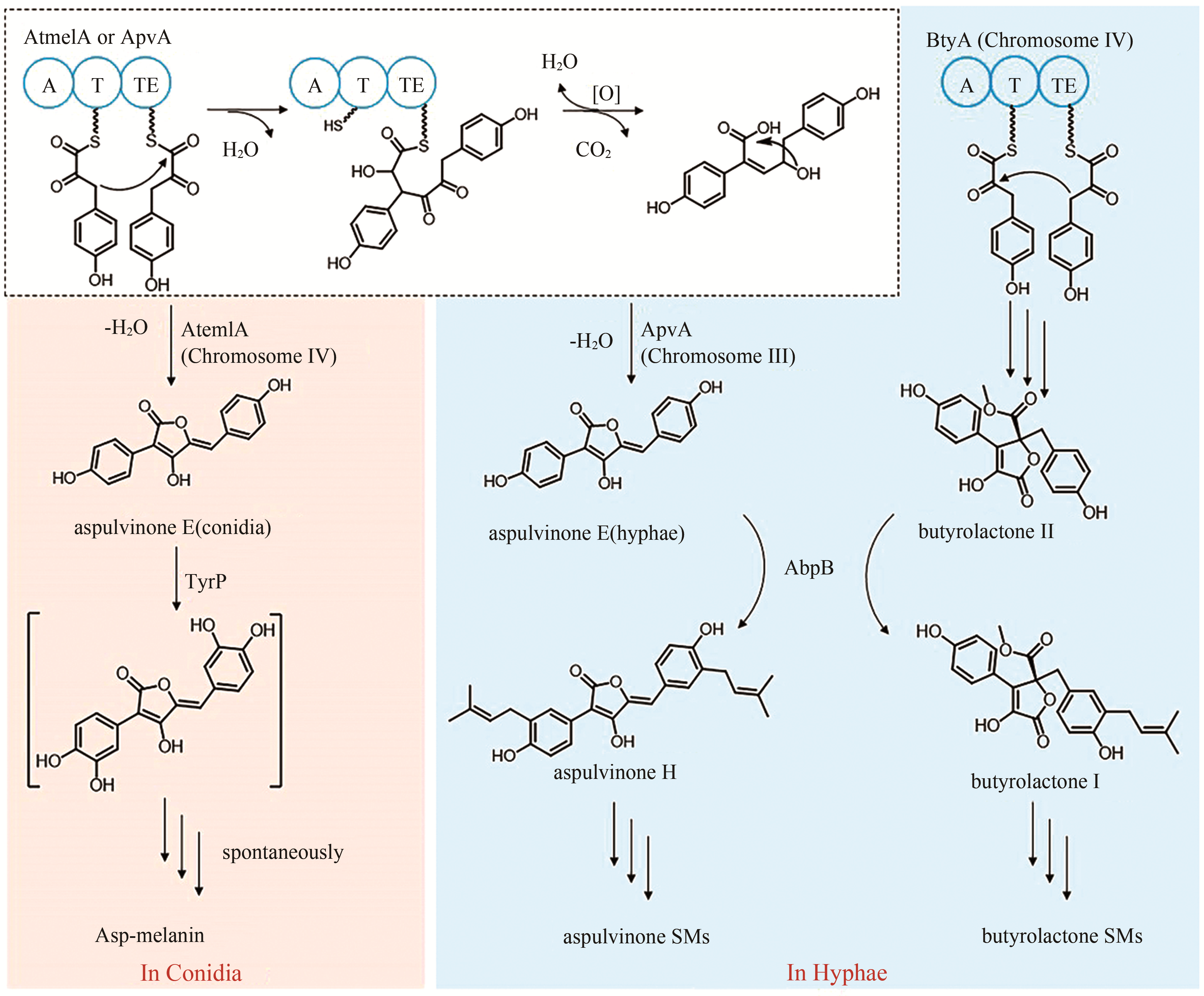
| 13 | ATEG_00700 (atqA) | NRPS-like |
asterriquinones CT5 | [ |
| 14 | ATEG_02004 (apvA) | NRPS-like |
aspulvinones B1 | [ |
| 15 | ATEG_02815 (btyA) | NRPS-like |
butyrolactones I | [ |
| 16 | ATEG_03563 (atmelA) | NRPS-like |
aspulvinone E (asp-melanin前体) | [ |
| 17 | ATEG_08899 (pgnA) | NRPS-like |
phenguignardic acid | [ |
| 18 | ATEG_00325 | PKS-NRPS |
isoflavipucine | [ |
| 19 | ATEG_00913 (pytA) | PKS-NRPS |
pyranterreone C | [ |
| 20 | ATEG_04416 (astA) | TC |
aspterric acid | [ |
| 70 | AMAIKE S, KELLER N P. Distinct roles for vea and laea in development and pathogenesis of Aspergillus flavus [J]. Eukaryotic Cell, 2009, 8(7): 1051-1060. |
| 71 | BRAKHAGE A A. Regulation of fungal secondary metabolism[J]. Nature Reviews Microbiology, 2013, 11(1): 21-32. |
| 72 | KENNEDY J, TURNER G. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans [J]. Molecular and General Genetics, 1996, 253(11): 189-197. |
| 73 | MAIYA S, GRUNDMANN A, LI S M, et al. The fumitremorgin gene cluster of Aspergillus fumigatus: Identification of a gene encoding brevianamide F synthetase[J]. Chembiochem, 2006, 7(7): 1062-1069. |
| 74 | YIN W B, KELLER N P. Transcriptional regulatory elements in fungal secondary metabolism[J]. Journal of Microbiology, 2011, 49(3): 329-339. |
| 75 | CHIANG Y M, SZEWCZYK E, DAVIDSON A D, et al. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans [J]. Journal of the American Chemical Society, 2009, 131(8): 2965-2970. |
| 76 | TANG S, ZHANG W, LI Z M, et al. Discovery and characterization of a PKS-NRPS hybrid in Aspergillus terreus by genome mining [J]. Journal of Natural Products, 2020. DOI: 10.1021/acs.jnatprod.9b01140 . |
| 77 | LYU H N, LIU H W, KELLER N P, et al. Harnessing diverse transcriptional regulators for natural product discovery in fungi.[J]. Natural Product Reports, 2020. 37(1): 6-16. |
| 78 | LAZARUS C M, WILLIAMS K, BAILEY A M. Reconstructing fungal natural product biosynthetic pathways[J]. Natural Product Reports, 2014, 31(10): 1339-1347. |
| 79 | PFEIFER B A, ADMIRAAL S J, GRAMAJO H, et al. Biosynthesis of complex polyketides in a metabolically engineered strain of E-coli [J]. Science, 2001, 291(5509): 1790-1792. |
| 80 | TAN G Y, LIU T G. Rational synthetic pathway refactoring of natural products biosynthesis in actinobacteria[J]. Metabolic Engineering, 2016, 39: 228-236. |
| 81 | CHIANG Y M, OAKLEY C E, AHUJA M, et al. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans [J]. Journal of the American Chemical Society, 2013, 135(20): 7720-7731. |
| 82 | HILL R A, CARTER R H, STAUNTON J. Biosynthesis of terrein, a metabolite of Aspergillus terreus Thom[J]. Journal of the Chemical Society, Chemicial Communications, 1975, 10: 381-385. |
| 83 | ZAEHLE C, GRESSLER M, SHELEST E, et al. Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity[J]. Chemistry & Biology, 2014, 21(6): 719-731. |
| 84 | GUO C J, SUN W W, BRUNO K S, et al. Molecular genetic characterization of terreic acid pathway in Aspergillus terreus [J]. Organic Letters, 2014, 16(20): 5250-5253. |
| 85 | KONG C X, HUANG H Z, XUE Y, et al. Heterologous pathway assembly reveals molecular steps of fungal terreic acid biosynthesis[J]. Scientific Reports, 2018, 8(1): 2116. |
| 86 | AWAKAWA T, YOKOTA K, FUNA N, et al. Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase[J]. Chemistry & Biology, 2009, 16(6): 613-623. |
| 87 | NIELSEN M T, NIELSEN J B, ANYAOGU D C, et al. Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR based approach[J]. PLoS One, 2013, 8(8): E72871. |
| 88 | GUO C J, KNOX B P, CHIANG Y M, et al. Molecular genetic characterization of a cluster in A. terreus for biosynthesis of the meroterpenoid terretonin[J]. Organic Letters, 2012, 14(22): 5684-5687. |
| 89 | MATSUDA Y, IWABUCHI T, WAKIMOTO T, et al. Uncovering the unusual D-ring construction in terretonin biosynthesis by collaboration of a multifunctional cytochrome P450 and a unique isomerase[J]. Journal of the American Chemical Society, 2015, 137(9): 3393-3401. |
| 90 | LIN T S, CHIANG Y M, WANG C C. Biosynthetic pathway of the reduced polyketide product citreoviridin in Aspergillus terreus var. aureus revealed by heterologous expression in Aspergillus nidulans [J]. Organic Letters, 2016, 18(6): 1366-1369. |
| 91 | XU Y Q, ESPINOSA-ARTILES P, SCHUBERT V, et al. Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat shock response-modulating anticancer fungal polyketide from Aspergillus terreus [J]. Applied and Environmental Microbiology, 2013, 79(6): 2038-2047. |
| 92 | HUANG X N, ZHANG W, TANG S, et al. Collaborative biosynthesis of a class of bioactive azaphilones by two separate gene clusters containing four PKS/NRPSs with transcriptional crosstalk in fungi[J]. Angewandte Chemie. International Edition, 2020. DOI: 10.1002/anie.201915514 . |
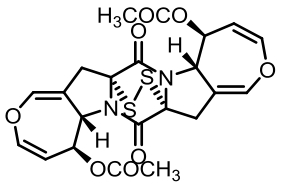
| 93 | GUO C J, YEH H H, CHIANG Y M, et al. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis[J]. Journal of the American Chemical Society, 2013, 135(19): 7205-7213. |
| 1 | ARABATZIS M, VELEGRAKI A. Sexual reproduction in the opportunistic human pathogen Aspergillus terreus [J]. Mycologia, 2013, 105(1): 71-79. |
| 2 | GUO C J, KNOX B P, SANCHEZ J F, et al. Application of an efficient gene targeting system linking secondary metabolites to their biosynthetic genes in Aspergillus terreus [J]. Organic Letters, 2013, 15(14): 3562-3565. |
| 3 | KUBODERA T, YAMASHITA N, NISHIMURA A. Transformation of Aspergillus sp and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae [J]. Bioscience Biotechnology and Biochemistry, 2002, 66(2): 404-406. |
| 4 | PRABHA V L, PUNEKAR N S. Genetic transformation in Aspergilli: tools of the trade[J]. Indian Journal of Biochemistry & Biophysics, 2004, 41(5): 205-215. |
| 5 | TEVZ G, BENCINA M, LEGISA M. Enhancing itaconic acid production by Aspergillus terreus [J]. Applied Microbiology and Biotechnology, 2010, 87(5): 1657-1664. |
| 6 | WANKA F, CAIRNS T, BOECKER S, et al. Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus [J]. Fungal Genetics and Biology, 2016, 89: 72-83. |
| 7 | SUN W W, GUO C J, WANG C C. Characterization of the product of a nonribosomal peptide synthetase-like (NRPS-like) gene using the doxycycline dependent Tet-on system in Aspergillus terreus [J]. Fungal Genetics and Biology, 2016, 89: 84-88. |
| 8 | VILLANUEVA A, MACCABE A P, BUESA J, et al. Apparent mRNA instability in Aspergillus nidulans and Aspergillus terreus of a heterologous cDNA encoding the major capsid antigen of Rotavirus [J]. Revista Iberoamericana de Micología, 1999, 16(3): 130-135. |
| 9 | HUANG X N, LU X F, LI J J. Cloning, characterization and application of a glyceraldehyde-3-phosphate dehydrogenase promoter from Aspergillus terreus [J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(3): 585-592. |
| 10 | HUANG X N, CHEN M, LU X F, et al. Direct production of itaconic acid from liquefied corn starch by genetically engineered Aspergillus terreus [J]. Microbial Cell Factories, 2014, 13: 108. |
| 11 | NINOMIYA Y, SUZUKI K, ISHII C, et al. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining[J]. PNAS, 2004, 101(33): 12248-12253. |
| 12 | HUANG X N, CHEN M, LI J J, al et, Establishing an efficient gene-targeting system in an itaconic-acid producing Aspergillus terreus strain [J]. Biotechnology Letters, 2016, 38(9): 1603-1610. |
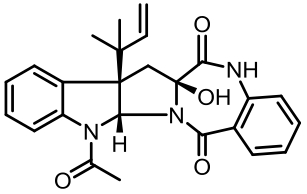
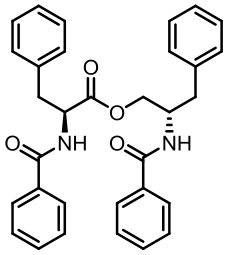
| 94 | LI W, FAN A L, WANG L, et al. Asperphenamate biosynthesis reveals a novel two-module NRPS system to synthesize amino acid esters in fungi[J]. Chemical Science, 2018, 9(9): 2589-2594. |
| 95 | GUO C J, SUN W W, BRUNO K S, et al. Spatial regulation of a common precursor from two distinct genes generates metabolite diversity[J]. Chemical Science, 2015, 6(10): 5913-5921. |
| 96 | GRESSLER M, ZAEHLE C, SCHERLACH K, et al. Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus [J]. Chemistry & Biology, 2011, 18(2): 198-209. |
| 97 | YAN Y, LIU Q K, ZANG X, et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action[J]. Nature, 2018, 559(7714): 415-418. |
| 98 | JONGH W A D, NIELSEN J. Enhanced citrate production through gene insertion in Aspergillus niger [J]. Metabolic Engineering, 2008, 10(2): 87-96. |
| 99 | HEINE D, MEYER F, HERTWECK C, et al. Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals[J]. eLife, 2015, 4: E07861. |
| 100 | BORUTA T, BIZUKOJC M. Culture-based and sequence-based insights into biosynthesis of secondary metabolites by Aspergillus terreus ATCC 20542[J]. Journal of Biotechnology, 2014, 175: 53-62. |
| 101 | PATRON N J, WALLER R F, COZIJNSEN A J, et al. Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes[J]. BMC Evolutionary Biology, 2007, 7(1): 174-188. |
| 102 | GEIB E, GRESSLER M, VIEDIERNIKOVA I, et al. A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress[J]. Cell Chemical Biology, 2016, 23(5): 587-597. |
| 103 | MCINTYRE C R, SCOTT F E, SIMPSON T J, et al. Application of stable isotope labelling methodology to the biosynthesis of the mycotoxin, terretonin, by Aspergillus terreus: incorporation of 13C-labelled acetates and methionine, 2H- and 13C, 18O-labelled ethyl 3,5-dimethylorsellinate and oxygen-18 gas[J]. Tetrahedron, 1989, 45(8): 2307-2321. |
| 104 | WANG M, BEISSNER M, ZHAO H M. Aryl-aldehyde formation in fungal polyketides: discovery and characterization of a distinct biosynthetic mechanism[J]. Chemistry & Biology, 2014, 21(2): 257-263. |
| 105 | 吕雪峰, 黄雪年, 齐飞飞, 等. 一种积累大黄素的基因工程菌株及其构建方法和应用: CN201910650149.7 [P]. 2019-07-18. |
| 13 | MIZUTANI O, MASAKI K, GOMI K, et al. Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue[J]. Applied and Environmental Microbiology, 2012, 78(12): 4126-4133. |
| 14 | SONG R J, ZHAI Q, SUN L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective[J]. Applied Microbiology and Biotechnology, 2019, 103(17): 6919-6932. |
| 15 | 李红花, 刘钢. CRISPR/Cas9 在丝状真菌基因组编辑中的应用[J]. 遗传, 2017, 39(5): 355-367. |
| LI H X, LIU G. The application of CRISPR/Cas9 in genome editing of filamentous fungi[J]. Hereditas (Beijing), 2017, 39(5): 355-367. | |
| 16 | YAMAMOTO S, OOSHIMA Y, NAKATA M, et al. Efficient gene-targeting in rat embryonic stem cells by CRISPR/Cas and generation of human kynurenine aminotransferase II (KAT II) knock-in rat[J]. Transgenic Research, 2015, 24(6): 991-1001. |
| 17 | LIU R, CHEN L, JIANG Y, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discovery, 2015. 1: 15007. |
| 18 | KATAYAMA T, TANAKA Y, OKABE T, et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae [J]. Biotechnology Letters, 2016, 38(4): 637-642. |
| 19 | KUIVANEN J, WANG Y M J, RICHARD P. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9[J]. Microbial Cell Factories, 2016, 15(1): 210. |
| 20 | ZHENG X M, ZHENG P, ZHANG K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger [J]. ACS Synthetic Biology, 2019, 8(7): 1568-1574. |
| 21 | SCHUSTER M, SCHWEIZER G, REISSMANN S, et al. Genome editing in Ustilago maydis using the CRISPR-Cas system[J]. Fungal Genetics & Biology, 2016, 89: 3-9. |
| 22 | LIU Q, GAO R R, LI J E, et al. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering[J]. Biotechnology for Biofuels, 2017, 10: 1. |
| 23 | ENDO A. The discovery and development of HMG-CoA reductase inhibitors[J]. Journal of Lipid Research, 1992, 33(11): 1569-1582. |
| 105 | LV X F, HUANG X N, QI F F, et al. Genetically engineered strain that accumulates emodin, and its construction method and application: CN201910650149.7[P]. 2019-07-18. |
| 106 | 吕雪峰, 黄雪年, 耿策, 等. 一种乌头酸的土曲霉菌株及其构建方法与应用: CN201910649851.1 [P]. 2019-07-18. |
| LV X F, HUANG X N, GENG C, et al. An aconitic acid producing Aspergillus terreus strain and its construction method and application: CN201910649851.1 [P]. 2019-07-18. | |
| 24 | KENNEDY J, AUCLAIR K, KENDREW S G, et al. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis[J]. Science, 1999, 284(5418): 1368-1372. |
| 25 | MA S M, LI J W, CHOI J W, et al. Complete reconstitution of a highly reducing iterative polyketide synthase[J]. Science, 2009, 326(5952): 589-592. |
| 26 | XU W., CHOOI Y H, CHOI J W, et al. LovG: the thioesterase required for dihydromonacolin L release and lovastatin nonaketide synthase turnover in lovastatin biosynthesis[J]. Angewandte Chemie International Edition, 2013, 52(25): 6472-6475. |
| 27 | BARRIUSO J, NGUYEN D T, LI J W, et al. Double oxidation of the cyclic nonaketide dihydromonacolin L to monacolin J by a single cytochrome P450 monooxygenase, LovA[J]. Journal of the American Chemical Society, 2011, 133(21): 8078-8081. |
| 28 | XIE X K, MEEHAN M J, XU W, et al. Acyltransferase mediated polyketide release from a fungal megasynthase[J]. Journal of the American Chemical Society, 2009, 131(24): 8388-8389. |
| 29 | BOK J W, KELLER N P. LaeA, a regulator of secondary metabolism in Aspergillus spp[J]. Eukaryotic Cell, 2004, 3(2): 527-535. |
| 30 | HASAN H, RAHIM M H ABD, CAMPBELL L, et al. Overexpression of acetyl-CoA carboxylase in Aspergillus terreus to increase lovastatin production[J]. New Biotechnology, 2018, 44: 64-71. |
| 31 | ITOH H, MIURA A, MATSUI M, et al. Knockout of the SREBP system increases production of the polyketide FR901512 in filamentous fungal sp. No. 14919 and lovastatin in Aspergillus terreus ATCC20542[J]. Applied Microbiology and Biotechnology, 2018, 102(3): 1393-1405. |
| 32 | XIE X K, WATANABE K, WOJCICKI W A, et al. Biosynthesis of lovastatin analogs with a broadly specific acyltransferase[J]. Cell Chemical Biology, 2006, 13(11): 1161-1169. |
| 33 | JIMENEZ-OSES G, OSUNA S, GAO X, et al. The role of distant mutations and allosteric regulation on LovD active site dynamics[J]. Nature Chemical Biology, 2014, 10(6): 431-436. |
| 34 | XIE X K, TANG Y. Efficient synthesis of simvastatin by use of Whole-Cell biocatalysis[J]. Applied & Environmental Microbiology, 2007, 73(7): 2054-2060 |
| 35 | XIE X K, PASHKOV I, GAO X, et al. Rational improvement of simvastatin synthase solubility in Escherichia coli leads to higher whole-cell biocatalytic activity[J]. Biotechnology & Bioengineering, 2010, 102(1): 20-28. |
| 36 | KOMAGATA D, YAMASHITA H, ENDO A. Microbial conversion of compactin (ML-236B) to ML-236A[J]. Journal of Antibiotics, 1986, 39(11): 1574-1577. |
| 37 | SCHIMMEL T G, BORNEMAN W S, CONDER M J. Purification and characterization of a lovastatin esterase from Clonostachys compactiuscula [J]. Applied and Environmental Microbiology, 1997, 63(4): 1307-11. |
| 38 | CHEN L C, LAI Y K, WU S C, et al. Production by Clonostachys compactiuscula of a lovastatin esterase that converts lovastatin to monacolin J[J]. Enzyme and Microbial Technology, 2006, 39(5): 1051-1059. |
| 39 | MORGAN B, BURK M, LEVIN M, et al. Methods for making simvastatin and intermediates: WO2005040107[P]. 2005-05-06. |
| 40 | HUANG X N, LIANG Y J, YANG Y, et al. Single-step production of the simvastatin precursor monacolin J by engineering of an industrial strain of Aspergillus terreus [J]. Metabolic Engineering, 2017, 42: 109-114. |
| 41 | LIANG Y J, LU X F. Structural insights into the catalytic mechanism of lovastatin hydrolase[J]. Journal of Biological Chemistry, 2020, 295(4): 1047-1055. |
| 42 | LIANG B, HUANG X N, TENG Y, et al. Enhanced single-step bioproduction of the simvastatin precursor monacolin J in an industrial strain of Aspergillus terreus by employing the evolved lovastatin hydrolase[J]. Biotechnology Journal, 2018, 13(6): E1800094. |
| 43 | HUANG X N, TANG S, ZHENG L H, et al. Construction of an efficient and robust Aspergillus terreus cell factory for monacolin J production[J]. ACS Synthetic Biology, 2019, 8(4): 818-825. |
| 44 | KLEMENT T, BUCHS J. Itaconic acid—a biotechnological process in change[J]. Bioresource Technology, 2013, 135: 422-31. |
| 45 | WERPY T A, HOLLADAY J E, WHIT J F. Top value added chemicals from biomass: I. Results of screening for potential candidates from sugars and synthesis gas[R]. Richland: Department of Energy, Pacific Northwest National Laboratory, 2004: 3-20. |
| 46 | KANAMASA S, DWIARTI L, OKABE M, et al. Cloning and functional characterization of the cis-aconitic acid decarboxylase (CAD) gene from Aspergillus terreus [J]. Applied Microbiology and Biotechnology, 2008, 80(2): 223-229. |
| 47 | LI A, LUIJK N V, BEEK M T, et al. A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus [J]. Fungal Genetics and Biology, 2011, 48(6): 602-611. |
| 48 | LAI L S, HUNG C S, LO C C. Effects of lactose and glucose on production of itaconic acid and lovastatin by Aspergillus terreus ATCC 20542[J]. Journal of Bioscience and Bioengineering, 2007, 104(1): 9-13. |
| 49 | STEIGER M G, BLUMHOFF M L, MATTANOVICH D, et al. Biochemistry of microbial itaconic acid production[J]. Frontiers in Microbiology, 2013, 4(1): 23. |
| 50 | CAPUDER M, SOLAR T, BENCINA M, et al. Highly active, citrate inhibition resistant form of Aspergillus niger 6-phosphofructo-1-kinase encoded by a modified pfkA gene[J]. Journal of Biotechnology, 2009, 144(1): 51-57. |
| 51 | HUANG X N, LU X F, LI Y M, et al. Improving itaconic acid production through genetic engineering of an industrial Aspergillus terreus strain[J]. Microbial Cell Factories, 2014, 13(1): 119. |
| 52 | LIN Y H, LI Y F, HUANG M C, et al. Intracellular expression of Vitreoscilla hemoglobin in Aspergillus terreus to alleviate the effect of a short break in aeration during culture[J]. Biotechnology Letters, 2004, 26(13): 1067-1072. |
| 53 | LI Q, BAI Z H, O'DONNELL A, et al. Oxidative stress in fungal fermentation processes: the roles of alternative respiration[J]. Biotechnology Letters, 2011, 33(3): 457-467. |
| 54 | CHEN M, HUANG X, ZHONG C W, et al. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus [J]. Applied Microbiology and Biotechnology, 2016, 100(17): 7541-7548. |
| 55 | BLUMHOFF M L, STEIGER M G, MATTANOVICH D, et al. Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger [J]. Metabolic Engineering, 2013, 19: 26-32. |
| 56 | KUENZ A, KRULL S. Biotechnological production of itaconic acid-things you have to know[J]. Applied Microbiology and Biotechnology, 2018, 102(9): 3901-3914. |
| 57 | BRAKHAGE A A, SCHROECKH V. Fungal secondary metabolites-strategies to activate silent gene clusters[J]. Fungal Genetics and Biology : FG & B, 2011, 48(1): 15-22. |
| 58 | CHIANG Y M, CHANG S L, OAKLEY B R, et al. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms[J]. Current Opinion in Chemical Biology, 2011, 15(1): 137-143. |
| 59 | ROMSDAHL J, WANG C C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012—2018)[J]. Medicinal Chemistry Communications, 2019, 10(6): 840-866. |
| 60 | RUTLEDGE P J, CHALLIS G L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters[J]. Nature Reviews Microbiology, 2015, 13(8): 509-523. |
| 61 | BODE H B, BETHE B, HFS R, et al. Big effects from small changes: possible ways to explore nature's chemical diversity[J]. ChemBioChem, 2002, 3(7): 619-627. |
| 62 | CICHEWICZ R H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners[J]. Natural Product Reports, 2010, 27(1): 11-22. |
| 63 | SHWAB E K, BOK J W, TRIBUS M, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus [J]. Eukaryotic Cell, 2007, 6(9): 1656-1664. |
| 64 | BOK J W, CHIANG Y M, SZEWCZYK E, et al. Chromatin-level regulation of biosynthetic gene clusters[J]. Nature Chemical Biology, 2009, 5(7): 462-464. |
| 65 | SZEWCZYK E, CHIANG Y M, OAKLEY C E, et al. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans [J]. Applied and Environmental Microbiology, 2008, 74(24): 7607-7612. |
| 66 | HENKE M T, SOUKUP A A, GOERING A W, et al. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans [J]. ACS Chemical Biology, 2016, 11(8): 2117-2123. |
| 67 | PERRIN R M, FEDOROVA N D, BOK J W, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA[J]. PLoS Pathogens, 2007, 3(4): E50. |
| 68 | SUGUI J A, PARDO J, CHANG Y C, et al. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus [J]. Eukaryotic Cell, 2007, 6(9): 1552-1561. |
| 69 | BAYRAM O, KRAPPMANN S, NI M, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism[J]. Science, 2008, 320(5882): 1504-1506. |
| [1] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [2] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||