Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (2): 141-157.DOI: 10.12211/2096-8280.2020-021
• Invited Review • Previous Articles Next Articles
Applications of synthetic biology in the treatment and prevention of infectious diseases
PU Lu, HUANG Yajia, YANG Shuai, JIN Fan
- Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen Institute of Synthetic Biology, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen 518055, Guangdong,China
-
Received:2020-03-10Revised:2020-04-22Online:2020-08-04Published:2020-04-30 -
Contact:JIN Fan
合成生物学在感染性疾病防治中的应用
蒲璐, 黄亚佳, 杨帅, 金帆
- 中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点研究室,广东 深圳 518055
-
通讯作者:金帆 -
作者简介:蒲璐(1992—),女,硕士,研究助理,研究方向为合成生物学。E-mail:lu.pu@siat.ac.cn
金帆(1978—),男,博士,研究员,研究方向为合成生物学。E-mail:fan.jin@siat.ac.cn -
基金资助:国家自然科学基金(21774117)
CLC Number:
Cite this article
PU Lu, HUANG Yajia, YANG Shuai, JIN Fan. Applications of synthetic biology in the treatment and prevention of infectious diseases[J]. Synthetic Biology Journal, 2020, 1(2): 141-157.
蒲璐, 黄亚佳, 杨帅, 金帆. 合成生物学在感染性疾病防治中的应用[J]. 合成生物学, 2020, 1(2): 141-157.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-021
| 1 | MARNER W D. Practical application of synthetic biology principles [J]. Biotechnology Journal: Healthcare Nutrition Technology, 2009, 4(10): 1406-1419. |
| 2 | LARTIGUE C, VASHEE S, ALGIRE M A, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast [J]. Science, 2009, 325(5948): 1693-1696. |
| 3 | QUAN J, SAAEM I, TANG N, et al. Parallel on-chip gene synthesis and application to optimization of protein expression [J]. Nature Biotechnology, 2011, 29(5): 449. |
| 4 | KOSURI S, EROSHENKO N, LEPROUST E M, et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips [J]. Nature Biotechnology, 2010, 28(12): 1295. |
| 5 | NANDAGOPAL N, ELOWITZ M B. Synthetic biology: integrated gene circuits [J]. Science, 2011, 333(6047): 1244-1248. |
| 6 | TIGGES M, FUSSENEGGER M. Recent advances in mammalian synthetic biology-design of synthetic transgene control networks [J]. Current Opinion in Biotechnology, 2009, 20(4): 449-460. |
| 7 | WEBER W, FUSSENEGGER M. Synthetic gene networks in mammalian cells [J]. Current Opinion in Biotechnology, 2010, 21(5): 690-696. |
| 8 | WIN M N, LIANG J C, SMOLKE C D. Frameworks for programming biological function through RNA parts and devices [J]. Chemistry & Biology, 2009, 16(3): 298-310. |
| 9 | BENENSON Y. RNA-based computation in live cells [J]. Current Opinion in Biotechnology, 2009, 20(4): 471-478. |
| 10 | CANTON B, LABNO A, ENDY D. Refinement and standardization of synthetic biological parts and devices [J]. Nature Biotechnology, 2008, 26(7): 787-793. |
| 11 | HOOSHANGI S, THIBERGE S, WEISS R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade [J]. PNAS, 2005, 102(10): 3581-3586. |
| 12 | KRAMER B P, FISCHER M, FUSSENEGGER M. Semi-synthetic mammalian gene regulatory networks [J]. Metabolic Engineering, 2005, 7(4): 241-250. |
| 13 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 14 | KRAMER B P, VIRETTA A U, DAOUD-EL BABA M, et al. An engineered epigenetic transgene switch in mammalian cells [J]. Nature Biotechnology, 2004, 22(7): 867-870. |
| 15 | DANINO T, MONDRAGóN-PALOMINO O, TSIMRING L, et al. A synchronized quorum of genetic clocks [J]. Nature, 2010, 463(7279): 326-330. |
| 16 | RUDER W C, LU T, COLLINS J J. Synthetic biology moving into the clinic [J]. Science, 2011, 333(6047): 1248-1252. |
| 17 | MAY M. Synthetic biology's clinical applications [J]. Science, 2015, 349(6255): 1564-1566. |
| 18 | ANDERSON R M, ANDERSON B, MAY R M. Infectious diseases of humans: dynamics and control [M]. Oxford: Oxford University Press, 1992. |
| 19 | KEELING M J, ROHANI P. Modeling infectious diseases in humans and animals [M]. Princeton: Princeton University Press, 2011. |
| 20 | KOLLEF M H, SHERMAN G, WARD S, et al. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients [J]. Chest, 1999, 115(2): 462-474. |
| 21 | PARDEE K, GREEN A A, TAKAHASHI M K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components [J]. Cell, 2016, 165(5): 1255-1266. |
| 22 | ABUDAYYEH O O, GOOTENBERG J S, KONERMANN S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector [J]. Science, 2016, 353(6299): AAf5573. |
| 23 | GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2 [J]. Science, 2017, 356(6336): 438-442. |
| 24 | GOOTENBERG J S, ABUDAYYEH O O, KELLNER M J, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6 [J]. Science, 2018, 360(6387): 439-444. |
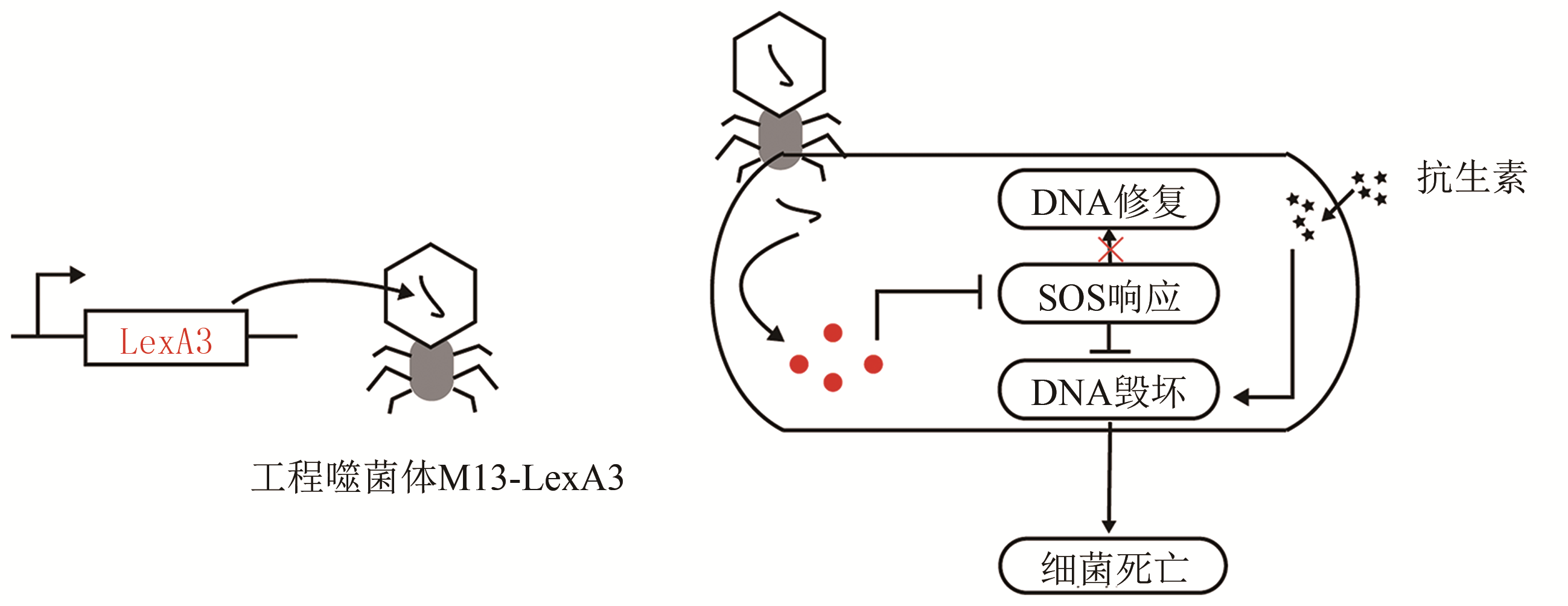
| 25 | ENGLISH M A, SOENKSEN L R, GAYET R V, et al. Programmable CRISPR-responsive smart materials [J]. Science, 2019, 365(6455): 780-785. |
| 26 | FREIJE C A, MYHRVOLD C, BOEHM C K, et al. Programmable inhibition and detection of RNA viruses using Cas13 [J]. Molecular Cell, 2019, 76(5): 826-837, E11. |
| 27 | CHEN J S, MA E, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity [J]. Science, 2018, 360(6387): 436-439. |
| 28 | LI S Y, CHENG Q X, LIU J K, et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA [J]. Cell Research, 2018, 28(4): 491-493. |
| 29 | LI S Y, CHENG Q X, WANG J M, et al. CRISPR-Cas12a-assisted nucleic acid detection [J]. Cell Discovery, 2018, 4(1): 20. |
| 30 | MALEK A, RAAD I. Catheter- and device-related infections in critically Ill cancer patients [M]. Berlin: Springer, 2020: 1401-1417. |
| 31 | HøIBY N, CIOFU O, JOHANSEN H K, et al. The clinical impact of bacterial biofilms [J]. International Journal of Oral Science, 2011, 3(2): 55-65. |
| 32 | FLEMMING H C, WINGENDER J. The biofilm matrix [J]. Nature Reviews Microbiology, 2010, 8(9): 623-633. |
| 33 | DONLAN R M. Biofilm formation: a clinically relevant microbiological process [J]. Clinical Infectious Diseases, 2001, 33(8): 1387-1392. |
| 34 | STEWART P S, COSTERTON J W. Antibiotic resistance of bacteria in biofilms [J]. The Lancet, 2001, 358(9276): 135-138. |
| 35 | CHUA S L, LIU Y, YAM J K H, et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles [J]. Nature Communications, 2014, 5(1): 4462. |
| 36 | LU T K, COLLINS J J. Dispersing biofilms with engineered enzymatic bacteriophage [J]. PNAS, 2007, 104(27): 11197-11202. |
| 37 | YEHL K, LEMIRE S, YANG A C, et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis [J]. Cell, 2019, 179(2): 459-469, E9. |
| 38 | FU W, FORSTER T, MAYER O, et al. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system [J]. Antimicrobial Agents and Chemotherapy, 2010, 54(1): 397-404. |
| 39 | ALVES D, GAUDION A, BEAN J, et al. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation [J]. Appl. Environ. Microbiol., 2014, 80(21): 6694-6703. |
| 40 | GUTIÉRREZ D, VANDENHEUVEL D, MARTíNEZ B, et al. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono-and dual-species staphylococcal biofilms [J]. Appl. Environ. Microbiol., 2015, 81(10): 3336-3348. |
| 41 | LEHMAN S M, DONLAN R M. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model [J]. Antimicrobial Agents and Chemotherapy, 2015, 59(2): 1127-1137. |
| 42 | ALVES D R, PEREZ‐ESTEBAN P, KOT W, et al. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions [J]. Microbial Biotechnology, 2016, 9(1): 61-74. |
| 43 | RYAN E M, ALKAWAREEK M Y, DONNELLY R F, et al. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro [J]. FEMS Immunology & Medical Microbiology, 2012, 65(2): 395-398. |
| 44 | RAHMAN M, KIM S, KIM S M, et al. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin [J]. Biofouling, 2011, 27(10): 1087-1093. |
| 45 | BEDI M S, VERMA V, CHHIBBER S. Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055 [J]. World Journal of Microbiology and Biotechnology, 2009, 25(7): 1145. |
| 46 | KOHANSKI M A, DWYER D J, HAYETE B, et al. A common mechanism of cellular death induced by bactericidal antibiotics [J]. Cell, 2007, 130(5): 797-810. |
| 47 | LU T K, COLLINS J J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy [J]. PNAS, 2009, 106(12): 4629-4634. |
| 48 | 吴楠楠, 朱同玉. 噬菌体在实体器官移植中的应用 [J]. 器官移植, 2019(4): 410-415. |
| WU N N, ZHU T Y. Application of phage in solid organ transplantation [J]. Organ Transplantation, 2019(4): 410-415. | |
| 49 | RIGLAR D T, SILVER P A. Engineering bacteria for diagnostic and therapeutic applications [J]. Nature Reviews Microbiology, 2018, 16(4): 214-225. |
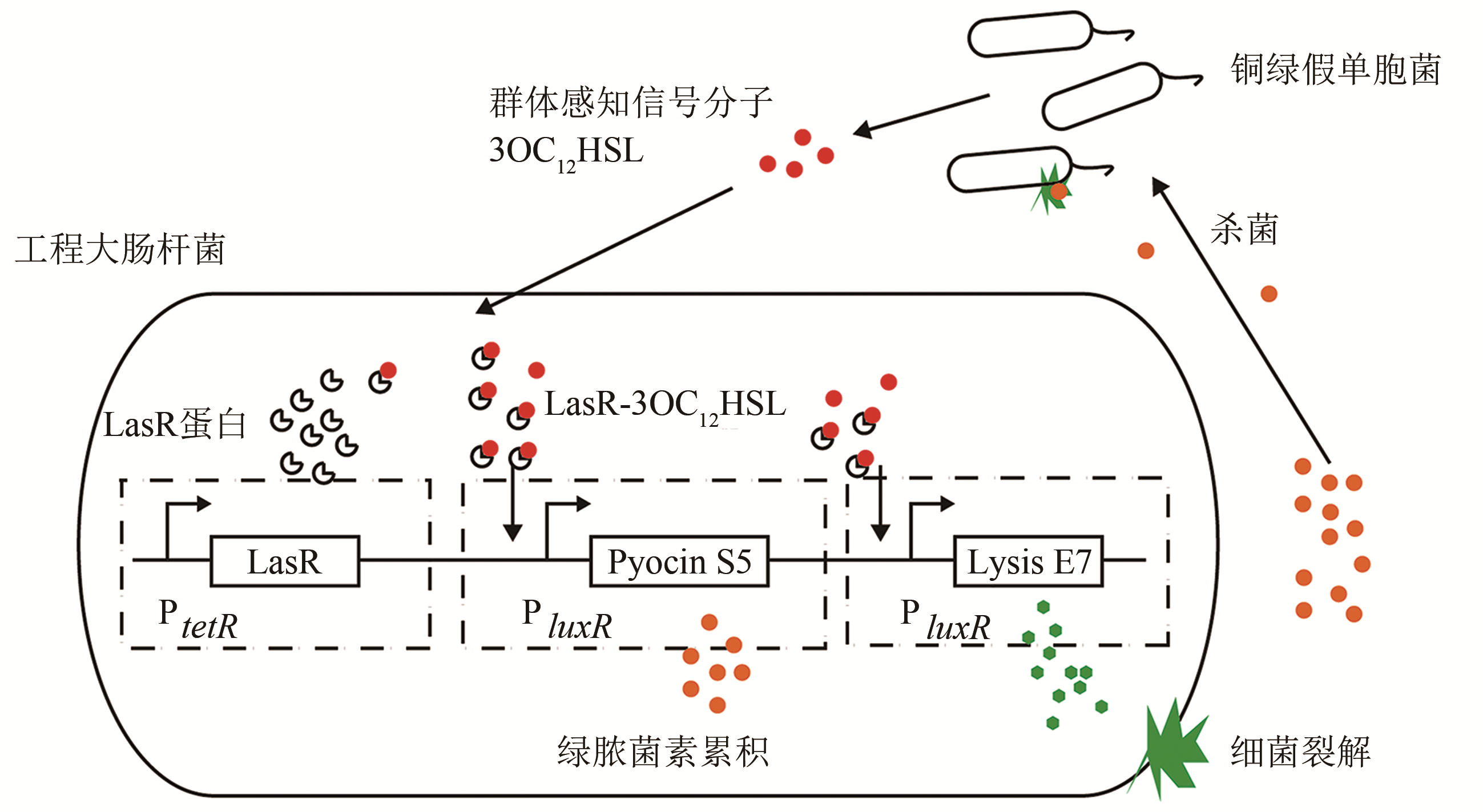
| 50 | SAEIDI N, WONG C K, LO T M, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen [J]. Molecular Systems Biology, 2011, 7(1): 521-531. |
| 51 | GRAY K M, PASSADOR L, IGLEWSKI B H, et al. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa [J]. Journal of Bacteriology, 1994, 176(10): 3076-3080. |
| 52 | PU L, YANG S, XIA A, et al. Optogenetics manipulation enables prevention of biofilm formation of engineered Pseudomonas aeruginosa on surfaces [J]. ACS Synthetic Biology, 2018, 7(1): 200-208. |
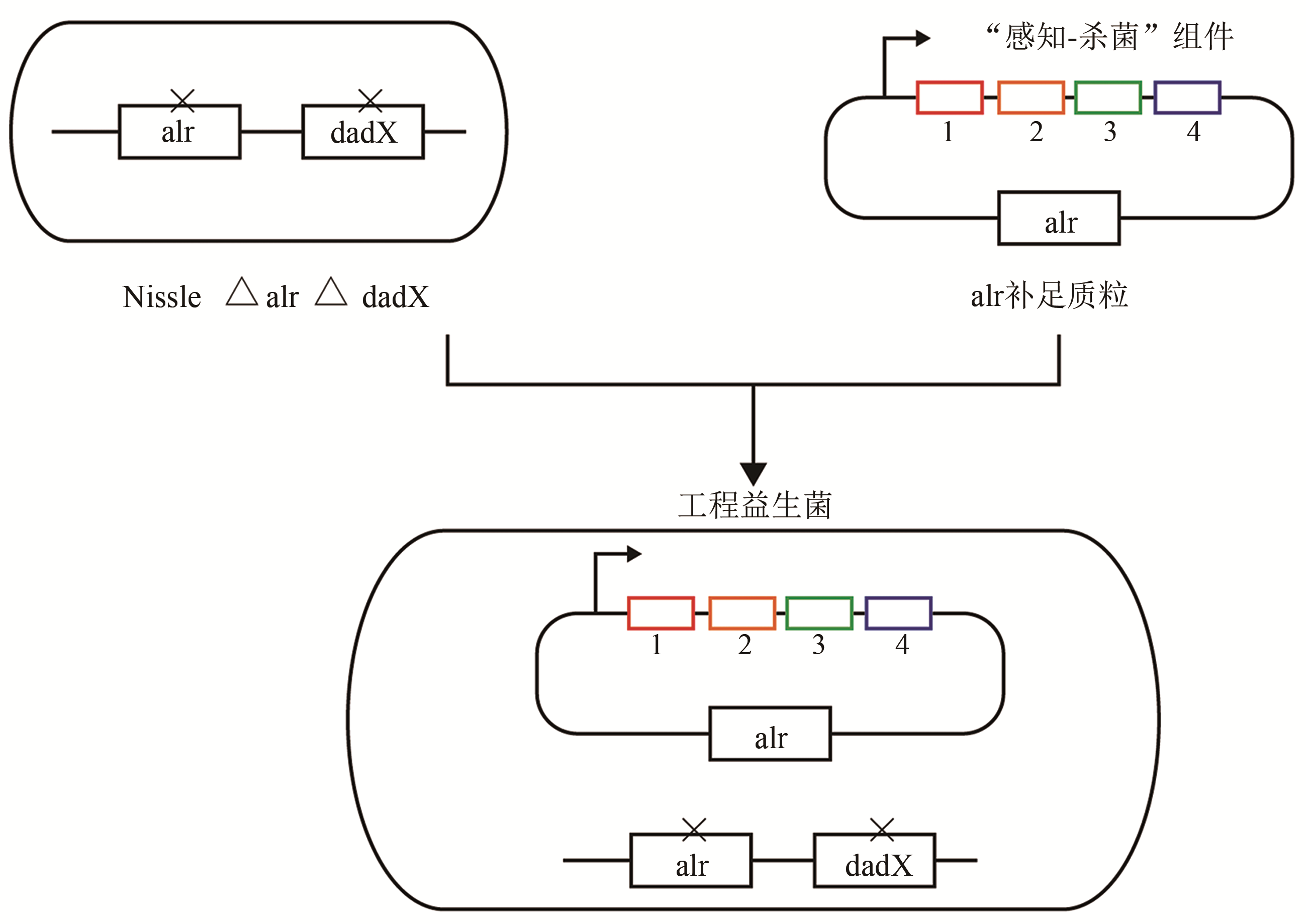
| 53 | OHLENDORF R, VIDAVSKI R R, ELDAR A, et al. From dusk till dawn: one-plasmid systems for light-regulated gene expression [J]. Journal of Molecular Biology, 2012, 416(4): 534-542. |
| 54 | COTTER P A, STIBITZ S. c-di-GMP-mediated regulation of virulence and biofilm formation [J]. Current Opinion in Microbiology, 2007, 10(1): 17-23. |
| 55 | CHUA K J, KWOK W C, AGGARWAL N, et al. Designer probiotics for the prevention and treatment of human diseases [J]. Current Opinion in Chemical Biology, 2017, 40:8-16. |
| 56 | HWANG I Y, KOH E, WONG A, et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models [J]. Nature Communications, 2017, 8(1): 15028. |
| 57 | CHOUDHARY E, THAKUR P, PAREEK M, et al. Gene silencing by CRISPR interference in mycobacteria [J]. Nature Communications, 2015, 6(1): 6267. |
| 58 | SINGH A K, CARETTE X, POTLURI L P, et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system [J]. Nucleic Acids Research, 2016, 44(18): E143. |
| 59 | YU W Y, WANG Y X, MEI J Z, et al. Overview of tuberculosis [M]. Berlin: Springer, 2020: 1-10. |
| 60 | TUMPEY T M, BASLER C F, AGUILAR P V, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus [J]. Science, 2005, 310(5745): 77-80. |
| 61 | WEBER W, FUSSENEGGER M. Emerging biomedical applications of synthetic biology [J]. Nature Reviews Genetics, 2012, 13(1): 21-35. |
| 62 | TUMPEY T M, MAINES T R, HOEVEN N VAN, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission [J]. Science, 2007, 315(5812): 655-659. |
| 63 | SMITH G J, VIJAYKRISHNA D, BAHL J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic [J]. Nature, 2009, 459(7250): 1122-1125. |
| 64 | WIMMER E, MUELLER S, TUMPEY T M, et al. Synthetic viruses: a new opportunity to understand and prevent viral disease [J]. Nature Biotechnology, 2009, 27(12): 1163. |
| 65 | PERLMAN S, NETLAND J. Coronaviruses post-SARS: update on replication and pathogenesis [J]. Nature Reviews Microbiology, 2009, 7(6): 439-450. |
| 66 | BECKER M M, GRAHAM R L, DONALDSON E F, et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice [J]. PNAS, 2008, 105(50): 19944-19999. |
| 67 | BURBELO P D, CHING K H, BUSH E R, et al. Antibody-profiling technologies for studying humoral responses to infectious agents [J]. Expert Review of Vaccines, 2010, 9(6): 567-578. |
| 68 | CHANDRA A, WORMSER G P, MARQUES A R, et al. Anti-Borrelia burgdorferi antibody profile in post-Lyme disease syndrome [J]. Clinical and Vaccine Immunology, 2011, 18(5): 767-771. |
| 69 | BURBELO P D, BREN K E, CHING K H, et al. LIPS arrays for simultaneous detection of antibodies against partial and whole proteomes of HCV, HIV and EBV [J]. Molecular BioSystems, 2011, 7(5): 1453-1462. |
| 70 | DASH P K, KAMINSKI R, BELLA R, et al. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice [J]. Nature Communications, 2019, 10(1): 2753. |
| 71 | XU L, WANG J, LIU Y, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia [J]. New England Journal of Medicine, 2019, 381(13): 1240-1247. |
| 72 | LIN S R, YANG H C, KUO Y T, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo [J]. Molecular Therapy - Nucleic Acids, 2014, 3:E186. |
| 73 | PRICE A A, SAMPSON T R, RATNER H K, et al. Cas9-mediated targeting of viral RNA in eukaryotic cells [J]. PNAS, 2015, 112(19): 6164-6169. |
| 74 | KINDSMÜLLER K, WAGNER R. Synthetic biology: impact on the design of innovative vaccines [J]. Human Vaccines, 2011, 7(6): 658-662. |
| 75 | SUBBARAMAN N. Science snipes at Oxitec transgenic-mosquito trial [M]. London: Nature Publishing Group, 2011. |
| 76 | PEDROLLI D B, RIBEIRO N V, SQUIZATO P N, et al. Engineering microbial living therapeutics: the synthetic biology toolbox [J]. Trends in Biotechnology, 2019, 37(1): 100-115. |
| 77 | MILLER M B, BASSLER B L. Quorum sensing in bacteria [J]. Annual Reviews in Microbiology, 2001, 55(1): 165-199. |
| 78 | DUAN F, MARCH J C. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model [J]. PNAS, 2010, 107(25): 11260-11264. |
| 79 | AKAHATA W, YANG Z-Y, ANDERSEN H, et al. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection [J]. Nature Medicine, 2010, 16(3): 334. |
| 80 | COLEMAN J R, PAPAMICHAIL D, SKIENA S, et al. Virus attenuation by genome-scale changes in codon pair bias [J]. Science, 2008, 320(5884): 1784-1787. |
| 81 | AMIDI M, DE RAAD M, DE GRAAUW H, et al. Optimization and quantification of protein synthesis inside liposomes [J]. Journal of Liposome Research, 2010, 20(1): 73-83. |
| 82 | DREESEN I A, CHARPIN-EL HAMRI G, FUSSENEGGER M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection [J]. Journal of Biotechnology, 2010, 145(3): 273-280. |
| 83 | SI L, XU H, ZHOU X, et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines [J]. Science, 2016, 354(6316): 1170-1173. |
| 84 | LI P, KE X, WANG T, et al. Zika Virus attenuation by codon pair deoptimization induces sterilizing immunity in mouse models [J]. Journal of Virology, 2018, 92(17): E00701- E00718. |
| 85 | WANG L, WANG X, BI K, et al. Oral vaccination with attenuated salmonella typhimurium-delivered TsPmy DNA vaccine elicits protective immunity against trichinella spiralis in BALB/c Mice [J]. PLoS Negl. Trop. Dis., 2016, 10(9): E0004952. |
| 86 | STARKS H, BRUHN K W, SHEN H, et al. Listeria monocytogenes as a vaccine vector: virulence attenuation or existing antivector immunity does not diminish therapeutic efficacy [J]. The Journal of Immunology, 2004, 173(1): 420-427. |
| 87 | HEGAZY W A H, XU X, METELITSA L, et al. Evaluation of Salmonella enterica Type III secretion system effector proteins as carriers for heterologous vaccine antigens [J]. Infection and Immunity, 2012, 80(3): 1193-1202. |
| 88 | VALENZUELA P, MEDINA A, RUTTER W J, et al. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast [J]. Nature, 1982, 298(5872): 347-350. |
| 89 | BLANQUET S, ANTONELLI R, LAFORET L, et al. Living recombinant Saccharomyces cerevisiae secreting proteins or peptides as a new drug delivery system in the gut [J]. Journal of Biotechnology, 2004, 110(1): 37-49. |
| 90 | H-L KWAG, KIM H J, CHANG D Y, et al. The production and immunogenicity of human papillomavirus type 58 virus-like particles produced in Saccharomyces cerevisiae [J]. Journal of Microbiology, 2012, 50(5): 813-820. |
| 91 | HALLER A A, LAUER G M, KING T H, et al. Whole recombinant yeast-based immunotherapy induces potent T cell responses targeting HCV NS3 and Core proteins [J]. Vaccine, 2007, 25(8): 1452-1463. |
| 92 | WANG X, XIAO X, ZHAO M, et al. EV71 virus-like particles produced by co-expression of capsid proteins in yeast cells elicit humoral protective response against EV71 lethal challenge [J]. BMC Research Notes, 2016, 9(1): 42. |
| 93 | RAMASAMY V, ARORA U, SHUKLA R, et al. A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice [J]. PLoS Negl. Trop. Dis., 2018, 12(1): E0006191. |
| 94 | ATHMARAM T N, SARASWAT S, SANTHOSH S R, et al. Yeast expressed recombinant Hemagglutinin protein of novel H1N1 elicits neutralising antibodies in rabbits and mice [J]. Virology Journal, 2011, 8(1): 524. |
| 95 | HEPLER R W, KELLY R, MCNEELY T B, et al. A recombinant 63-kDa form of Bacillus anthracis protective antigen produced in the yeast Saccharomyces cerevisiae provides protection in rabbit and primate inhalational challenge models of anthrax infection [J]. Vaccine, 2006, 24(10): 1501-1514. |
| 96 | KING T H, SHANLEY C A, GUO Z, et al. GI-19007, a novel Saccharomyces cerevisiae-based therapeutic vaccine against tuberculosis [J]. Clinical and Vaccine Immunology, 2017, 24(12): E00245. |
| 97 | WALCH B, BREINIG T, GEGINAT G, et al. Yeast-based protein delivery to mammalian phagocytic cells is increased by coexpression of bacterial listeriolysin [J]. Microbes and Infection, 2011, 13(11): 908-913. |
| 98 | FU G, CONDON K C, EPTON M J, et al. Female-specific insect lethality engineered using alternative splicing [J]. Nature Biotechnology, 2007, 25(3): 353-357. |
| 99 | FU G, LEES R S, NIMMO D, et al. Female-specific flightless phenotype for mosquito control [J]. PNAS, 2010, 107(10): 4550-4554. |
| 100 | DE VALDEZ M R W, NIMMO D, BETZ J, et al. Genetic elimination of dengue vector mosquitoes [J]. PNAS, 2011, 108(12): 4772-4775. |
| 101 | RITCHIE S A, STAUNTON K M. Reflections from an old queenslander: can rear and release strategies be the next great era of vector control? [J]. Proceedings of the Royal Society B, 2019, 286(1905): 20190973. |
| 102 | WINDBICHLER N, MENICHELLI M, PAPATHANOS P A, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito [J]. Nature, 2011, 473(7346): 212-215. |
| 103 | WANG S, DOS-SANTOS A L A, HUANG W, et al. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria [J]. Science, 2017, 357(6358): 1399-1402. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems [J]. Synthetic Biology Journal, 2025, 6(1): 105-117. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||