Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (2): 274-286.DOI: 10.12211/2096-8280.2020-078
• Invited Review • Previous Articles Next Articles
Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi
XIAO Han, LIU Yixin
- Joint International Research Laboratory of Metabolic and Developmental Sciences,State Key Laboratory of Microbial Metabolism,School of Life Science & Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2020-10-06Revised:2020-12-22Online:2021-04-30Published:2021-04-30 -
Contact:XIAO Han
CRISPR-Cas系统编辑丝状真菌的进展与挑战
肖晗, 刘宜欣
- 上海交通大学生命科学技术学院,微生物代谢国家重点实验室,教育部代谢与发育科学国际合作联合实验室,上海 200240
-
通讯作者:肖晗 -
作者简介:肖晗(1985—),女,博士,副研究员,主要从事合成生物学、基因编辑和代谢工程研究。E-mail:smallhan@sjtu.edu.cn -
基金资助:国家重点研发计划(2018YFA0900600);国家自然科学基金(319713144);上海市自然科学基金(17ZR1448900)
CLC Number:
Cite this article
XIAO Han, LIU Yixin. Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi[J]. Synthetic Biology Journal, 2021, 2(2): 274-286.
肖晗, 刘宜欣. CRISPR-Cas系统编辑丝状真菌的进展与挑战[J]. 合成生物学, 2021, 2(2): 274-286.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-078

Fig. 1 CRISPR-Cas system assisted gene editing for filamentous fungi(RNP—ribonucleoprotein; BM-RNP—biomimetic mineralized RNP; PEG—polyethylene glycerol; AMT—agrobacterium-mediated transformation; DSB—double stranded break; NHEJ—non-homologous end joining; HR—homologous recombination; APOBEC1—apolipoprotein B mRNA editing enzyme; VPR—VP64-p65-Rta, a tripartite transcriptional activator domain; CRISPRa—CRISPR activation)

Fig. 2 Strategies for in vivo expression of the CRISPR-Cas system in filamentous fungi(For in vivo expression of Cas protein, strategies include codon-optimization, adopting constitutive promoter for driving the expression of Cas protein, and incorporating intron sequence at the 5' end of cas gene. For in vivo expression of gRNA, strategies include adopting polymerase II promoter together with the self-cleaving ribozymes for gRNA expression, adopting polymerase III promoter for gRNA expression, and adopting polymerase III promoter together with the self-cleaving ribozyme for gRNA expression)
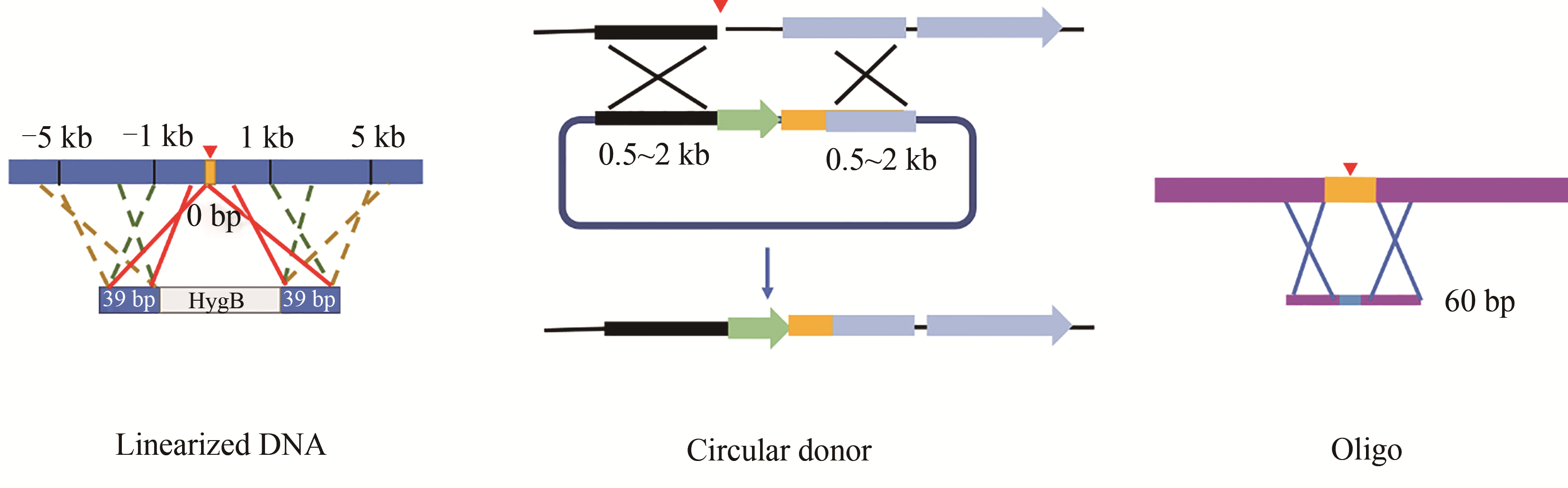
Fig. 3 Design of homologous recombination donor(For linearized DNA, it is flanked by 39 bp HR arms, which are within 5 kb from DSB as generated by CRIPSR-Cas. For circular DNA, it is flanked by HR arms with the size from 0.5 to 2 kb, and a selective marker is usually contained in the middle. For oligo, the intended mutation and PAM sequence are interspaced by no more than 7 bp)
| 菌株 | CRISPR-Cas存在形式 | 递送方式 | 表达策略 | 同源重组供体 | 宿主改造 | 编辑方式和效率 |
|---|---|---|---|---|---|---|
| 稻瘟病菌(Magnaporthe oryzae) | RNP | 利用磷酸钙矿化的纳米颗粒承载RNP与原生质体共培养 | 体外表达并组装成有功能的RNP | — | — | NHEJ,中断编码Sytalone dehydratase的sdh基因的效率为20%[ |
| 水稻恶苗病菌(Fusarium fujikuroi) | 质粒 | PEG介导的原生质体转化 | 内源表达水稻恶苗病菌密码子优化的Cas9,并用RNA聚合酶II型启动子加上自我剪切核酶的方式、RNA聚合酶III型U6或5S rRNA启动子分别表达gRNA | — | — | NHEJ, RNA聚合酶II型启动子加上自我剪切核酶的方式表达gRNA时,不能获得Fusarium cyclin C1编码基因fcc1的基因编辑突变体;用RNA聚合酶III型U6和5S rRNA启动子表达gRNA时, fcc1中断效率分别是37.5%和79.2%[ |
| 嗜热毁丝霉(Myceliophthora thermophila) | PCR产物 | PEG介导的原生质体转化 | 内源表达密码子优化的Cas12a并用U6启动子驱动gRNA的表达 | PCR产物;两端靠近CRISPR-Cas切口处分别有500~600 bp的同源臂,中间包含抗性筛选标记基因表达盒 | — | HDR,编辑单基因的效率为90%;编辑多个基因时,单基因发生编辑的效率为13%~41%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用可自主复制并丢失的质粒装载CRISPR-Cas系统,RNA聚合酶II型启动子加核酶的形式表达gRNA | 质粒;插入片段两侧有CRISPR-Cas在基因组上产生切口附近的同源臂,同源臂两侧有和CRIPSR-Cas在基因组上识别区域一致的spacer | pyrG基因中断菌株 | 每转1 μg DNA获得2个有正确整合形式的转化子[ |
| 黑曲霉(Aspergillus niger) | 质粒 | — | 融合表达大鼠的胞嘧啶脱氨酶rAPOBEC1、黑曲霉密码子优化的nCas9和尿嘧啶糖基化抑制酶,用米曲霉U6启动子驱动gRNA的表达 | — | — | 将靶基因中特定位置的胞嘧啶转变为胸腺嘧啶,效率47.36%~100%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用黑曲霉密码子优化的Cas9和内源5S rRNA基因启动子驱动gRNA的表达 | PCR产物;两端和CRISPR-Cas切口处分别有40 bp的同源臂 | kusA缺陷型菌株 | HDR,效率为33.3%~100%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用可自主复制的质粒装载CRISPR-Cas系统,内源表达Cas9,并用tRNA基因启动子驱动gRNA的表达 | 60 bp的寡核苷酸链,突变位点和PAM的距离不超过15 bp | kusA缺陷型菌株 | HDR,效率为80%[ |
| 烟曲霉(Aspergillus fumigatus) | 质粒和RNA | PEG介导的原生质体转化 | 内源表达人密码子优化的Cas9和体外转录gRNA | PCR产物;两端靠近CRISPR-Cas切口处分别有35 bp的同源臂 | — | HDR,效率为95%~100%[ |
| 里氏木霉(Trichoderma reesei) | 质粒和RNA | PEG介导的原生质体转化 | 内源表达里氏木霉优化的Cas9和体外转录成熟的gRNA | 质粒;两端和CRISPR-Cas切口处分别有200 bp的同源臂 | 以表达Cas9的菌株为出发菌株 | HDR,编辑内源一个假定的甲基转移酶基因lae1的效率为93%[ |
| 灵芝 (Ganoderma lucidum) | 质粒 | PEG介导的原生质体转化 | 利用灵芝密码子优化的Cas9和内源U6启动子驱动gRNA的表达,在gRNA 3′端加入HDV | PCR产物;两端和CRISPR-Cas切口处分别有1 kb的同源臂,中间缺失包括PAM在内的8 bp并引入终止密码子TGA | — | HDR,编辑细胞色素P450编码基因cyp5150l8的效率为每转化3×107个原生质体获得2个基因编辑菌株[ |
Tab. 1 Examples of CRISPR-Cas system assisted gene editing in filamentous fungi.
| 菌株 | CRISPR-Cas存在形式 | 递送方式 | 表达策略 | 同源重组供体 | 宿主改造 | 编辑方式和效率 |
|---|---|---|---|---|---|---|
| 稻瘟病菌(Magnaporthe oryzae) | RNP | 利用磷酸钙矿化的纳米颗粒承载RNP与原生质体共培养 | 体外表达并组装成有功能的RNP | — | — | NHEJ,中断编码Sytalone dehydratase的sdh基因的效率为20%[ |
| 水稻恶苗病菌(Fusarium fujikuroi) | 质粒 | PEG介导的原生质体转化 | 内源表达水稻恶苗病菌密码子优化的Cas9,并用RNA聚合酶II型启动子加上自我剪切核酶的方式、RNA聚合酶III型U6或5S rRNA启动子分别表达gRNA | — | — | NHEJ, RNA聚合酶II型启动子加上自我剪切核酶的方式表达gRNA时,不能获得Fusarium cyclin C1编码基因fcc1的基因编辑突变体;用RNA聚合酶III型U6和5S rRNA启动子表达gRNA时, fcc1中断效率分别是37.5%和79.2%[ |
| 嗜热毁丝霉(Myceliophthora thermophila) | PCR产物 | PEG介导的原生质体转化 | 内源表达密码子优化的Cas12a并用U6启动子驱动gRNA的表达 | PCR产物;两端靠近CRISPR-Cas切口处分别有500~600 bp的同源臂,中间包含抗性筛选标记基因表达盒 | — | HDR,编辑单基因的效率为90%;编辑多个基因时,单基因发生编辑的效率为13%~41%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用可自主复制并丢失的质粒装载CRISPR-Cas系统,RNA聚合酶II型启动子加核酶的形式表达gRNA | 质粒;插入片段两侧有CRISPR-Cas在基因组上产生切口附近的同源臂,同源臂两侧有和CRIPSR-Cas在基因组上识别区域一致的spacer | pyrG基因中断菌株 | 每转1 μg DNA获得2个有正确整合形式的转化子[ |
| 黑曲霉(Aspergillus niger) | 质粒 | — | 融合表达大鼠的胞嘧啶脱氨酶rAPOBEC1、黑曲霉密码子优化的nCas9和尿嘧啶糖基化抑制酶,用米曲霉U6启动子驱动gRNA的表达 | — | — | 将靶基因中特定位置的胞嘧啶转变为胸腺嘧啶,效率47.36%~100%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用黑曲霉密码子优化的Cas9和内源5S rRNA基因启动子驱动gRNA的表达 | PCR产物;两端和CRISPR-Cas切口处分别有40 bp的同源臂 | kusA缺陷型菌株 | HDR,效率为33.3%~100%[ |
| 黑曲霉(Aspergillus niger) | 质粒 | PEG介导的原生质体转化 | 利用可自主复制的质粒装载CRISPR-Cas系统,内源表达Cas9,并用tRNA基因启动子驱动gRNA的表达 | 60 bp的寡核苷酸链,突变位点和PAM的距离不超过15 bp | kusA缺陷型菌株 | HDR,效率为80%[ |
| 烟曲霉(Aspergillus fumigatus) | 质粒和RNA | PEG介导的原生质体转化 | 内源表达人密码子优化的Cas9和体外转录gRNA | PCR产物;两端靠近CRISPR-Cas切口处分别有35 bp的同源臂 | — | HDR,效率为95%~100%[ |
| 里氏木霉(Trichoderma reesei) | 质粒和RNA | PEG介导的原生质体转化 | 内源表达里氏木霉优化的Cas9和体外转录成熟的gRNA | 质粒;两端和CRISPR-Cas切口处分别有200 bp的同源臂 | 以表达Cas9的菌株为出发菌株 | HDR,编辑内源一个假定的甲基转移酶基因lae1的效率为93%[ |
| 灵芝 (Ganoderma lucidum) | 质粒 | PEG介导的原生质体转化 | 利用灵芝密码子优化的Cas9和内源U6启动子驱动gRNA的表达,在gRNA 3′端加入HDV | PCR产物;两端和CRISPR-Cas切口处分别有1 kb的同源臂,中间缺失包括PAM在内的8 bp并引入终止密码子TGA | — | HDR,编辑细胞色素P450编码基因cyp5150l8的效率为每转化3×107个原生质体获得2个基因编辑菌株[ |
| 1 | WARD O P. Production of recombinant proteins by filamentous fungi[J]. Biotechnology Advances, 2012, 30(5): 1119-1139. |
| 2 | 刘剑飞, 胡留杰, 廖敦秀, 等. 食用菌生物修复重金属污染研究进展[J]. 应用生态学报,2011,22 (2): 543-548. |
| LIU J F, HU L J, LIAO D X, et al. Bioremediation of heavy metal pollution by edible fungi: a review[J]. Chinese Journal of Applied Ecology, 2011, 22(2): 543-548. | |
| 3 | LAICH F, FIERRO F, MARTIN J F. Production of penicillin by fungi growing on food products: identification of a complete penicillin gene cluster in Penicillium griseofulvum and a truncated cluster in Penicillium verrucosum [J]. Applied and Environmental Microbiology, 2002, 68(3): 1211-1219. |
| 4 | BORUTA T, BIZUKOJC M. Production of lovastatin and itaconic acid by Aspergillus terreus: a comparative perspective[J]. World Journal of Microbiology and Biotechnology, 2017, 33(2): 34. |
| 5 | XU Y N, ZHONG J J. Impacts of calcium signal transduction on the fermentation production of antitumor ganoderic acids by medicinal mushroom Ganoderma lucidum [J]. Biotechnology Advances, 2012, 30(6): 1301-1308. |
| 6 | WANG Q, ZHONG C, XIAO H. Genetic engineering of filamentous fungi for efficient protein expression and secretion[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 293. |
| 7 | BRAKHAGE A A, SCHROECKH V. Fungal secondary metabolites-strategies to activate silent gene clusters[J]. Fungal Genetics and Biology, 2011, 48(1): 15-22. |
| 8 | KIM S, HA B S, RO H S. Current technologies and related issues for mushroom transformation[J]. Mycobiology, 2015, 43(1): 1-8. |
| 9 | QIN H, XIAO H, ZOU G, et al. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species[J]. Process Biochemistry, 2017, 56: 57-61. |
| 10 | XU J W, XU N, ZHONG J J. Enhancement of ganoderic acid accumulation by overexpression of an N-Terminally truncated 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in the basidiomycete Ganoderma lucidum [J]. Applied and Environmental Microbiology, 2012, 78(22): 7968-7976. |
| 11 | BAO Z H, XIAO H, LANG J, et al. Homology-Integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(5): 585-594. |
| 12 | JIANG Y, CHEN B, DUAN C L, et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 System[J]. Applied and Environmental Microbiology, 2015, 81(7): 2506-2514. |
| 13 | NISSIM L, PERLI S D, FRIDKIN A, et al. Multiplexed and programmable regulation of gene networks with an Integrated RNA and CRISPR/Cas toolkit in human cells[J]. Molecular Cell, 2014, 54(4): 698-710. |
| 14 | LIU R, CHEN L, JIANG Y, et al. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system[J]. Cell Discovery, 2015, 1: 15007. |
| 15 | WANG P A, XIAO H, ZHONG J J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum [J]. Applied and Environmental Microbiology, 2020, 104(4): 1661-1671. |
| 16 | SONG R, ZHAI Q, SUN L, et al. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective[J]. Appled Microbiology and Biotechnology, 2019, 103(17): 6919-6932. |
| 17 | YU X, JI S L, HE Y L, et al. Development of an expression plasmid and its use in genetic manipulation of Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (Higher Basidiomycetes)[J]. International Journal of Medicinal Mushrooms, 2014, 16(2): 161-168. |
| 18 | SAYARI M, Nest M A VAN DER, STEENKAMP E T, et al. Agrobacterium-mediated transformation of Ceratocystis albifundus [J]. Microbiological Research, 2019, 226: 55-64. |
| 19 | KWON M J, SCHÜTZE T, SPOHNER S, et al. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi[J]. Fungal Biology and Biotechnology, 2019, 6(1):15. |
| 20 | POHL C, KIEL J A K W, DRIESSEN A J M, et al. CRISPR/Cas9 based genome editing of Penicillium chrysogenum [J]. ACS Synthetic Biology, 2016, 5(7): 754-764. |
| 21 | HAO Z, SU X. Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex[J]. BMC Biotechnol, 2019, 19(1): 2. |
| 22 | WANG Q, COBINE P A, COLEMAN J J. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes[J]. Fungal Genetics and Biology, 2018, 117: 21-29. |
| 23 | LI S, SONG Z, LIU C, et al. Biomimetic mineralization-based CRISPR/Cas9 ribonucleoprotein nanoparticles for gene editing[J]. ACS Applied Materials and Interfaces, 2019, 11(51): 47762-47770. |
| 24 | LU S, SHEN X, CHEN B. Development of an efficient vector system for gene knock-out and near in-cis gene complementation in the Sugarcane smut fungus[J]. Scientific Reports, 2017, 7(1): 3113. |
| 25 | SARKARI P, MARX H, BLUMHOFF M L, et al. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger [J]. Bioresource Technology, 2017, 245: 1327-1333. |
| 26 | ARAZOE T, MIYOSHI K, YAMATO T, et al. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus[J]. Biotechnology and Bioengineering, 2015, 112(12): 2543-2549. |
| 27 | MATSU-URA T, BAEK M, KWON J, et al. Efficient gene editing in Neurospora crassa with CRISPR technology[J]. Fungal Biology and Biotechnology, 2015, 2(1): 4. |
| 28 | LIU K, SUN B, YOU H, et al. Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system[J]. Microbial Biotechnology, 2020, 13(2): 386-396. |
| 29 | KATAYAMA T, NAKAMURA H, ZHANG Y, et al. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae [J]. Applied and Environmental Microbiology, 2019, 85(3): |
| 30 | WEBER J, VALIANTE V, NØDVIG C S, et al. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing[J]. ACS Synthetic Biology, 2017, 6(1): 62-68. |
| 31 | HUANG L, DONG H, ZHENG J, et al. Highly efficient single base editing in Aspergillus niger with CRISPR/Cas9 cytidine deaminase fusion[J]. Microbiological Research, 2019, 223/224/225: 44-50. |
| 32 | ROUX I, WOODCRAFT C, HU J, et al. CRISPR-mediated activation of biosynthetic gene clusters for bioactive molecule discovery in filamentous fungi[J]. ACS Synthetic Biology, 2020, 9(7): 1843-1854. |
| 33 | JIMÉNEZ A, HOFF B, REVUELTA J L. Multiplex genome editing in Ashbya gossypii using CRISPR-Cpf1[J]. New Biotechnology, 2020, 57: 29-33. |
| 34 | VANEGAS K G, JARCZYNSKA Z D, STRUCKO T, et al. Cpf1 enables fast and efficient genome editing in Aspergilli [J]. Fungal Biology and Biotechnology, 2019, 6(1): 6. |
| 35 | LIU Q, ZHANG Y, LI F, et al. Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila [J]. Biotechnology for Biofuels, 2019, 12: 293. |
| 36 | KUIVANEN J, WANG Y M J, RICHARD P. Engineering Aspergillus niger for galactaric acid production: Elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9[J]. Microbial Cell Factories, 2016, 15(1): 210. |
| 37 | ZHENG Y M, LIN F L, GAO H, et al. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology[J]. Scientific Reports, 2017, 7(1):9250. |
| 38 | SHI T Q, GAO J, WANG W J, et al. CRISPR/Cas9-Based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production[J]. ACS Synthetic Biology, 2019, 8(2): 445-454. |
| 39 | NøDVIG C S, NIELSEN J B, KOGLE M E, et al. A CRISPR-Cas9 system for genetic engineering of filamentous fungi[J]. PLoS One, 2015, 10(7): e0133085. |
| 40 | WENDEROTH M, PINECKER C, VOß B, et al. Establishment of CRISPR/Cas9 in Alternaria alternata [J]. Fungal Genetics and Biology, 2017, 101: 55-60. |
| 41 | KUJOTH G C, SULLIVAN T D, MERKHOFER R, et al. CRISPR/Cas9-mediated gene disruption reveals the importance of zinc metabolism for fitness of the dimorphic fungal pathogen Blastomyces dermatitidis [J]. mBio, 2018, 9(2): e00412-18. |
| 42 | ZHANG Y, OUYANG L, NAN Y, et al. Efficient gene deletion and replacement in Aspergillus niger by modified in vivo CRISPR/Cas9 systems[J]. Bioresources and Bioprocessing, 2019, 6(1): 4. |
| 43 | WANG Y, WEi D, ZHU X, et al. A 'suicide' CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans [J]. Scientific Reports, 2016, 6: 31145. |
| 44 | SCHUSTER M, SCHWEIZER G, KAHMANN R. Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes[J]. Fungal Genetics and Biology, 2018, 112: 21-30. |
| 45 | CHUTRAKUL C, PANCHANAWAPORN S, JEENNOR S, et al. Functional characterization of novel U6 RNA polymerase III promoters: Their implication for CRISPR-Cas9-mediated gene editing in Aspergillus oryzae [J]. Current Microbiology, 2019, 76(12): 1443-1451. |
| 46 | WU C, CHEN Y, QIU Y, et al. A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription[J]. Biotechnology Letters, 2020, 42(7): 1203-1210. |
| 47 | ZHENG X, ZHENG P, ZHANG K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger [J]. ACS Synthetic Biology, 2018, 8(7): 1568-1574. |
| 48 | SONG L, OUEDRAOGO J P, KOLBUSZ M, et al. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger [J]. PLoS One, 2018, 13(8): e0202868. |
| 49 | LEYNAUD-KIEFFER L M C, CURRAN S C, KIM I, et al. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable[J]. PLoS One, 2019, 14(1): e0210243. |
| 50 | DONG L, YU D, LIN X, et al. Improving expression of thermostable trehalase from Myceliophthora sepedonium in Aspergillus niger mediated by the CRISPR/Cas9 tool and its purification, characterization[J]. Protein Expression and Purification, 2020, 165: 105482. |
| 51 | XIE S, WANG Y, WEI W, et al. The Bax inhibitor UvBI-1, a negative regulator of mycelial growth and conidiation, mediates stress response and is critical for pathogenicity of the rice false smut fungus Ustilaginoidea virens [J]. Current Genetics, 2019, 65(5): 1185-1197. |
| 52 | YAMATO T, HANDA A, ARAZOE T, et al. Single crossover-mediated targeted nucleotide substitution and knock-in strategies with CRISPR/Cas9 system in the rice blast fungus[J]. Scientific Reports, 2019, 9(1): 7427. |
| 53 | ZHANG C, MENG X, WEI X, et al. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus [J]. Fungal Genetics and Biology, 2016, 86: 47-57. |
| 54 | DONG H, ZHENG J, YU D, et al. Efficient genome editing in Aspergillus niger with an improved recyclable CRISPR-HDR toolbox and its application in introducing multiple copies of heterologous genes[J]. Journal of Microbiological Methods, 2019, 163: 105655. |
| 55 | NøDVIG C S, HOOF J B, KOGLE M E, et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli [J]. Fungal Genetics and Biology, 2018, 115: 78-89. |
| 56 | KUN R S, MENG J, SALAZAR-CEREZO S, et al. CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass[J]. Enzyme and Microbial Technology, 2020, 136: 109508. |
| 57 | ZHANG L, ZHENG X, CAIRNS T C, et al. Disruption or reduced expression of the orotidine-5'-decarboxylase gene pyrG increases citric acid production: A new discovery during recyclable genome editing in Aspergillus niger [J]. Microbial Cell Factories, 2020, 19(1): 76. |
| 58 | JIANG D W, ZHU W, WANG Y C, et al. Molecular tools for functional genomics in filamentous fungi: Recent advances and new strategies[J]. Biotechnology Advances, 2013, 31(8): 1562-1574. |
| 59 | HAN X, LIU Z B, JO M C, et al. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation[J]. Science Advances, 2015, 1(7): e1500454. |
| 60 | EASTWOOD D, GREEN J, GROGAN H, et al. Viral agents causing brown cap mushroom disease of Agaricus bisporus [J]. Applied and Environmental Microbiology, 2015, 81(20): 7125-7134. |
| 61 | VERDOES J C, PUNT P J, BERG P VAN DER, et al. Characterization of an efficient gene cloning strategy for Aspergillus niger based on an autonomously replicating plasmid: cloning of the nicB gene of A. niger[J]. Gene, 1994, 146(2): 159-165. |
| 62 | SU X, SCHMITZ G, ZHANG M, et al. Heterologous gene expression in filamentous fungi[J]. Advances in Applied Microbiology, 2012, 81: 1-61. |
| 63 | CAIRNS T C, FEURSTEIN C, ZHENG X, et al. A quantitative image analysis pipeline for the characterization of filamentous fungal morphologies as a tool to uncover targets for morphology engineering: A case study using aplD in Aspergillus niger [J]. Biotechnology for Biofuels, 2019, 12(1): 149. |
| 64 | WEYDA I, YANG L, VANG J, et al. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius [J]. Journal of Microbiological Methods, 2017, 135: 26-34. |
| 65 | KUIVANEN J, KORJA V, HOLMSTRÖM S, et al. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes[J]. Fungal Biology and Biotechnology, 2019, 6(1): 3. |
| 66 | COLOT H V, PARK G, TURNER G E, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103 (27): 10352-10357. |
| [1] | DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems [J]. Synthetic Biology Journal, 2025, 6(1): 105-117. |
| [2] | REN Jiawei, ZHANG Jinpeng, XU Guoqiang, ZHANG Xiaomei, XU Zhenghong, ZHANG Xiaojuan. Effect of terminators on the downstream transcript unit with gene expression in Escherichiacoli [J]. Synthetic Biology Journal, 2025, 6(1): 213-227. |
| [3] | LI Geng, SHEN Xiaolin, SUN Xinxiao, WANG Jia, YUAN Qipeng. Research progress in recombinant expression and application of peroxidases [J]. Synthetic Biology Journal, 2024, 5(6): 1498-1517. |
| [4] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| [5] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| [6] | DU Yao, GAO Hongdan, LIU Jiakun, LIU Xiaorong, XING Zhihao, ZHANG Tao, MA Dongli. Research progress of the CRISPR-Cas system in the detecting pathogen nucleic acids [J]. Synthetic Biology Journal, 2024, 5(1): 202-216. |
| [7] | XU Zhimeng, XIE Zhen. Research progress and biotechnological applications of the prime editing [J]. Synthetic Biology Journal, 2024, 5(1): 1-15. |
| [8] | CHEN Yaru, CAO Yingxiu, SONG Hao. Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms [J]. Synthetic Biology Journal, 2023, 4(6): 1281-1299. |
| [9] | MA Mengdan, SHANG Mengyu, LIU Yuchen. Application and prospect of CRISPR-Cas9 system in tumor biology [J]. Synthetic Biology Journal, 2023, 4(4): 703-719. |
| [10] | LIN Jicong, ZOU Gen, LIU Hongmin, WEI Yongjun. Application of CRISPR/Cas genome editing technology in the synthesis of secondary metabolites of filamentous fungi [J]. Synthetic Biology Journal, 2023, 4(4): 738-755. |
| [11] | CHEN Jiawen, HUANG Jiandong, SUN Haitao. Current developments in the use of engineered bacteria for cancer therapy [J]. Synthetic Biology Journal, 2023, 4(4): 690-702. |
| [12] | PAN Jiahao, PAN Weisong, QIU Jian, XIE Donling, ZOU Qi, WU Chuan. Research progress on recombinant collagen expression system [J]. Synthetic Biology Journal, 2023, 4(4): 808-823. |
| [13] | MA Mengdan, LIU Yuchen. Potential application of synthetic biology in disease information recording and real-time monitoring [J]. Synthetic Biology Journal, 2023, 4(2): 301-317. |
| [14] | LIU Ke, LIN Guihong, LIU Kun, ZHOU Wei, WANG Fengqing, WEI Dongzhi. Mining, engineering and functional expansion of CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 47-66. |
| [15] | LIANG Liya, LIU Rongming. Protein engineering of DNA targeting type Ⅱ CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 86-101. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||