Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (2): 301-317.DOI: 10.12211/2096-8280.2022-058
• Invited Review • Previous Articles Next Articles
Potential application of synthetic biology in disease information recording and real-time monitoring
MA Mengdan1,2,3, LIU Yuchen1,2
- 1.The first Affiliated Hospital of Shenzhen University,Shenzhen Second People’s Hospital,Shenzhen Institute of Translational Medicine,Shenzhen 518035,Guangdong,China
2.Guangdong Provincial Key Laboratory of Systems Biology and Synthetic Biology for Urogenital Tumors,Shenzhen 518035,Guangdong,China
3.Shantou University Medical College,Shantou 515041,Guangdong,China
-
Received:2022-10-21Revised:2022-12-29Online:2023-04-27Published:2023-04-30 -
Contact:LIU Yuchen
合成生物学在疾病信息记录与实时监测中的应用潜力
马孟丹1,2,3, 刘宇辰1,2
- 1.深圳大学第一附属医院,深圳市第二人民医院,深圳转化医学研究院,广东 深圳 518035
2.广东省泌尿生殖肿瘤系统生物学与合成生物学重点实验室,广东 深圳 518035
3.汕头大学医学院,广东 汕头 515041
-
通讯作者:刘宇辰 -
作者简介:马孟丹 (1997—),女,硕士研究生。主要研究方向为医学合成生物学。E-mail:mamengdan7@163.com刘宇辰 (1988—),男,副研究员,研究生导师、博士后合作导师。主要研究方向:(1)肿瘤生物治疗和医学合成生物学;(2)创新肿瘤治疗新方法;(3)构建人工基因线路,肿瘤精准治疗。E-mail:liuyuchenmdcg@163.com -
基金资助:国家重点研发计划(2021YFA0911600)
CLC Number:
Cite this article
MA Mengdan, LIU Yuchen. Potential application of synthetic biology in disease information recording and real-time monitoring[J]. Synthetic Biology Journal, 2023, 4(2): 301-317.
马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-058
| 记录装置系统 | 种群分布vs单细胞记录 | 写入周期 | 记录能力 | 发生顺序 | 持续时间 | 灵敏度 | 保真度 | |
|---|---|---|---|---|---|---|---|---|
| 双稳态开关 | 种群 | 短 | 一般 | 否 | 是 | 低 | 低 | |
| DNA重组酶技术 | 种群 | 长 | 一般 | 否 | 否 | 低 | 低 | |
| ssDNA编辑技术 | HiSCRIBE[ | 种群 | 长 | 强 | 否 | 是 | 一般 | 高 |
| CRISPR系统 | Record-seq[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 |
| CAMERA[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 较高 | |
| DNA typewriter[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 | |
| LINNAEUS[ | 种群 | 短 | 弱 | 否 | 否 | 一般 | 低 | |
| mSCRIBE[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| iTracer[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| DOMINO[ | 种群 | 长 | 强 | 否 | 否 | 一般 | 一般 | |
Table 1 Summary for real-time monitoring and recording systems in cell
| 记录装置系统 | 种群分布vs单细胞记录 | 写入周期 | 记录能力 | 发生顺序 | 持续时间 | 灵敏度 | 保真度 | |
|---|---|---|---|---|---|---|---|---|
| 双稳态开关 | 种群 | 短 | 一般 | 否 | 是 | 低 | 低 | |
| DNA重组酶技术 | 种群 | 长 | 一般 | 否 | 否 | 低 | 低 | |
| ssDNA编辑技术 | HiSCRIBE[ | 种群 | 长 | 强 | 否 | 是 | 一般 | 高 |
| CRISPR系统 | Record-seq[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 |
| CAMERA[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 较高 | |
| DNA typewriter[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 | |
| LINNAEUS[ | 种群 | 短 | 弱 | 否 | 否 | 一般 | 低 | |
| mSCRIBE[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| iTracer[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| DOMINO[ | 种群 | 长 | 强 | 否 | 否 | 一般 | 一般 | |
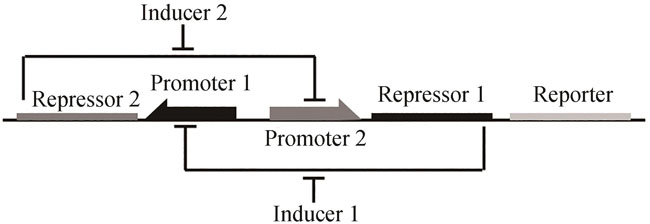
Fig. 1 Design for toggle switch[48]Repressors 1 and 2 inhibit transcription driven by Promoters 1 and 2, respectively, which is induced by Inducers 1 and 2 correspondingly
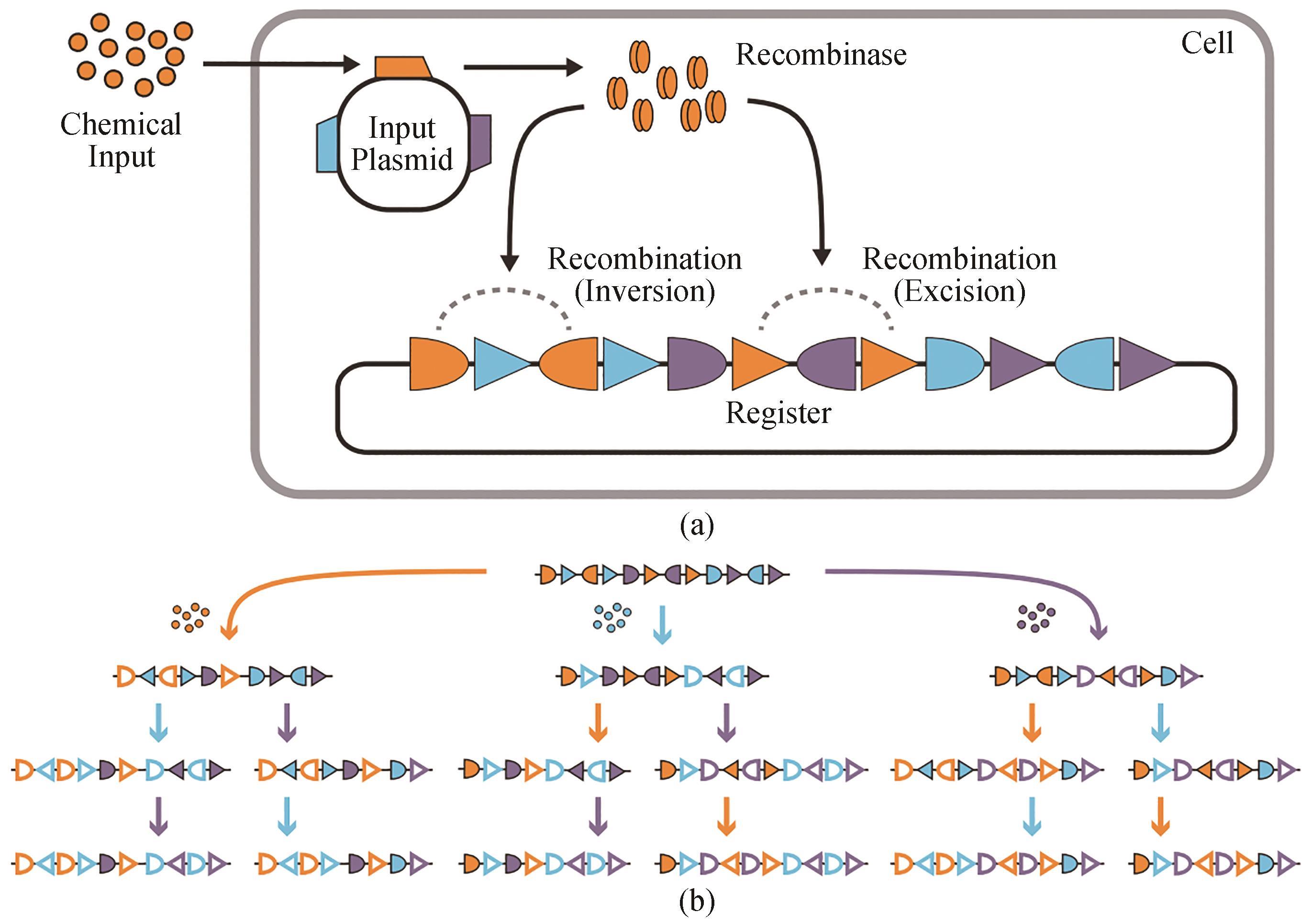
Fig. 2 Summary for three-input and 16-state RSM[55](a) RSM mechanism. A chemical input induces the expression of a recombinase encoded by a gene on the input plasmid, which modifies a DNA register with overlapping and orthogonal recombinase recognition sites. Specific recombinases can be controlled by corresponding inputs. Each of these recombinases can target multiple orthogonal pairs of their cognate recognition sites (shown as triangles and half-ovals) to catalyze inversion (when the sites are anti-aligned) or excision (when the sites are aligned). (b) The register is designed to adopt a specific DNA state for every identity and order of inputs. Three different inputs are represented by colored arrows (orange, blue, and purple), each of which expresses a specific recombinase. Unrecombined recognition sites are shown by solid symbols, and symbols without filling highlight recombined recognition sites
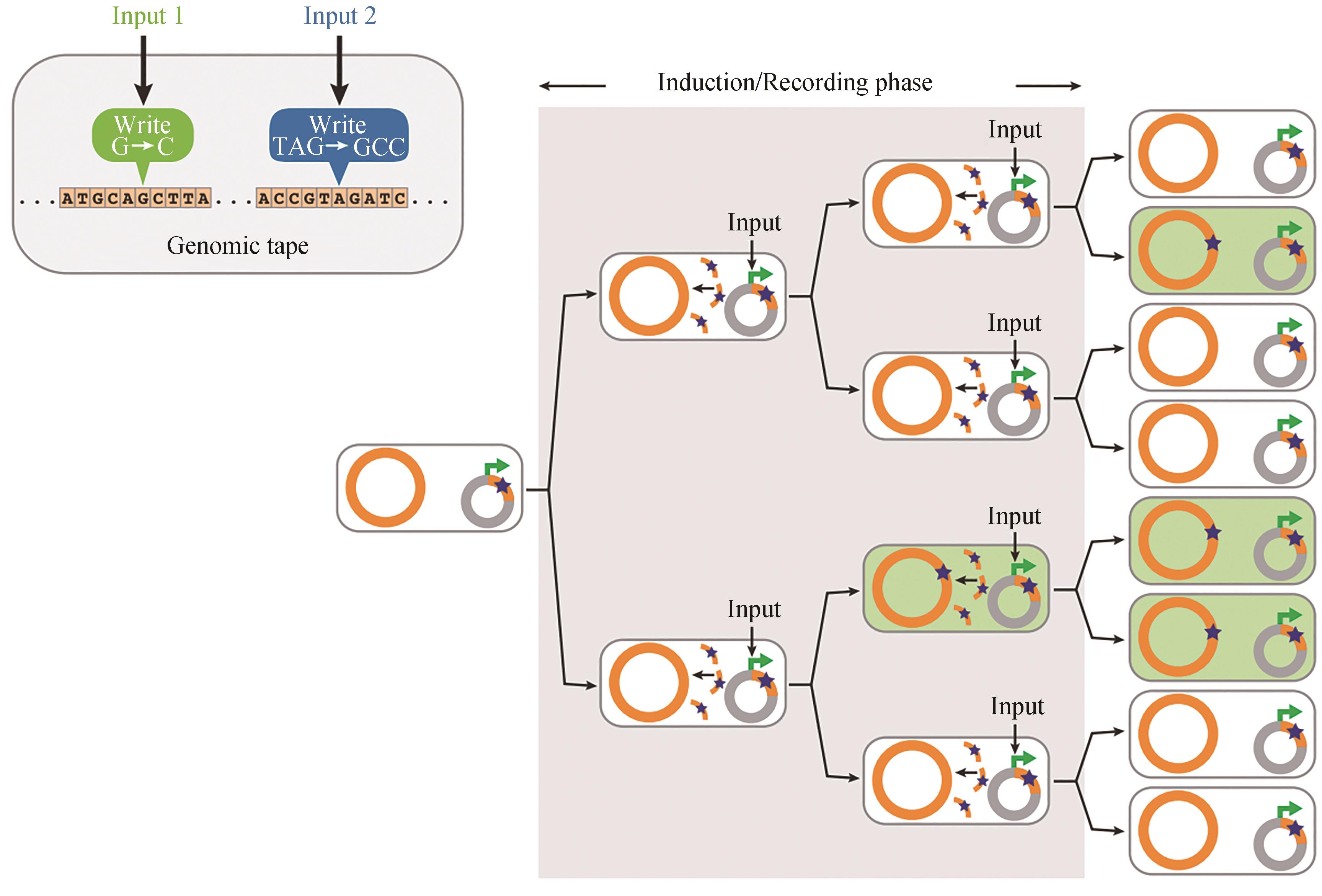
Fig. 3 SCRIBE-based distributed encoded memory at genome levels [25]In the presence of an input, ssDNAs (orange curved lines) are produced from a plasmid-borne cassette (gray circles) and recombined into specific genomic loci (orange circles) that are targeted on the basis of sequence homology. This results in the accumulation of precise mutations (stars in green cells) as a function of the magnitude and duration of exposure to the input
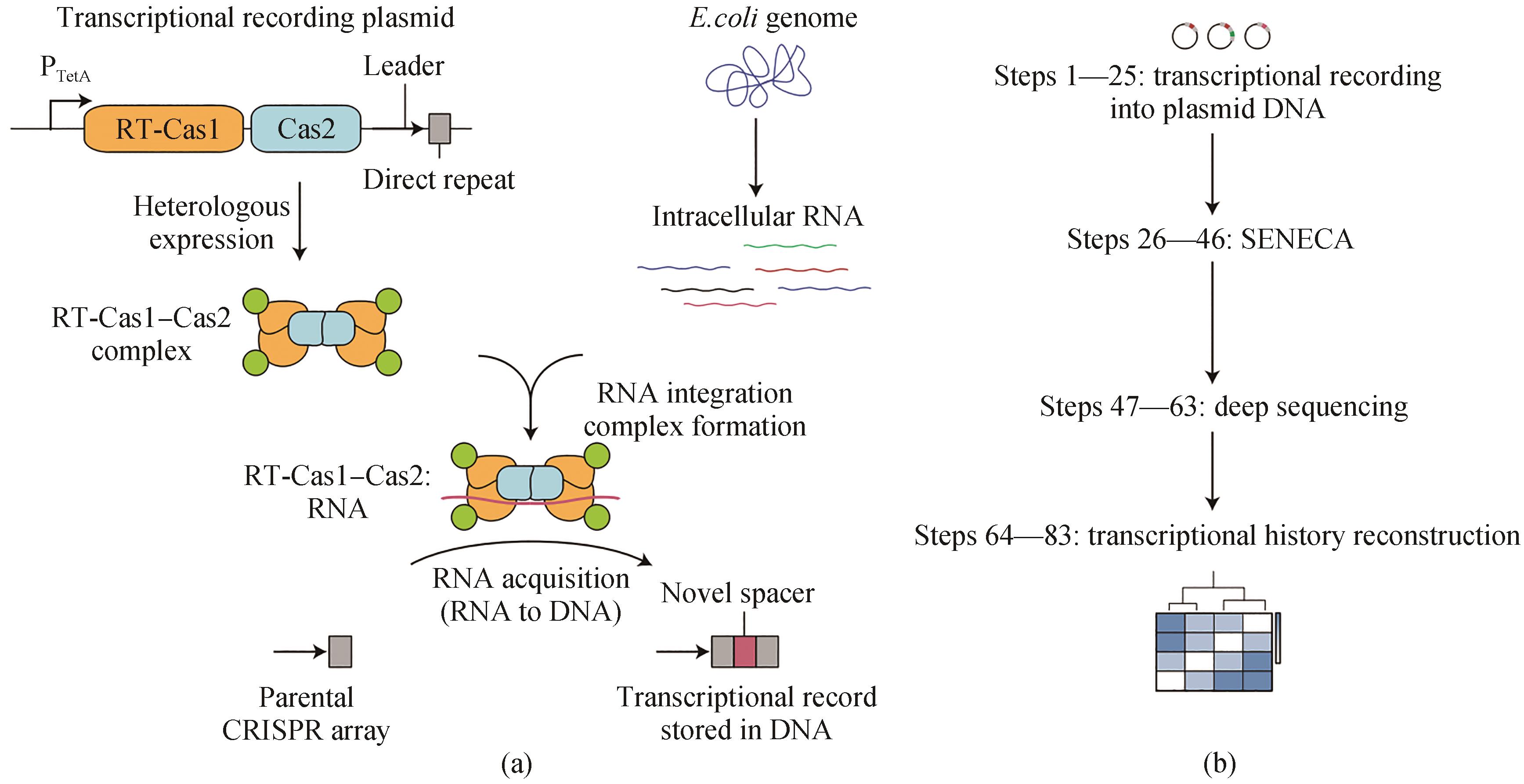
Fig. 4 Transcriptional record of RNA extracted from CRISPR spacer acquisition[59](a) Record-seq uses the RNA-acquiring RT-Cas1-Cas2 complex from Fusicatenibacter saccharivorans to encode transcriptional information into plasmid-borne CRISPR arrays. The transcriptional record is generated by CRISPR spacer acquisition directly from intracellular RNAs followed by reverse transcription of RNA protospacers through the RT domain of FsRT-Cas1-Cas2. (b) Extraction of plasmid DNA followed by the selective amplification of expanded CRISPR arrays (SENECA) and deep sequencing enable the reconstruction of transcriptional histories
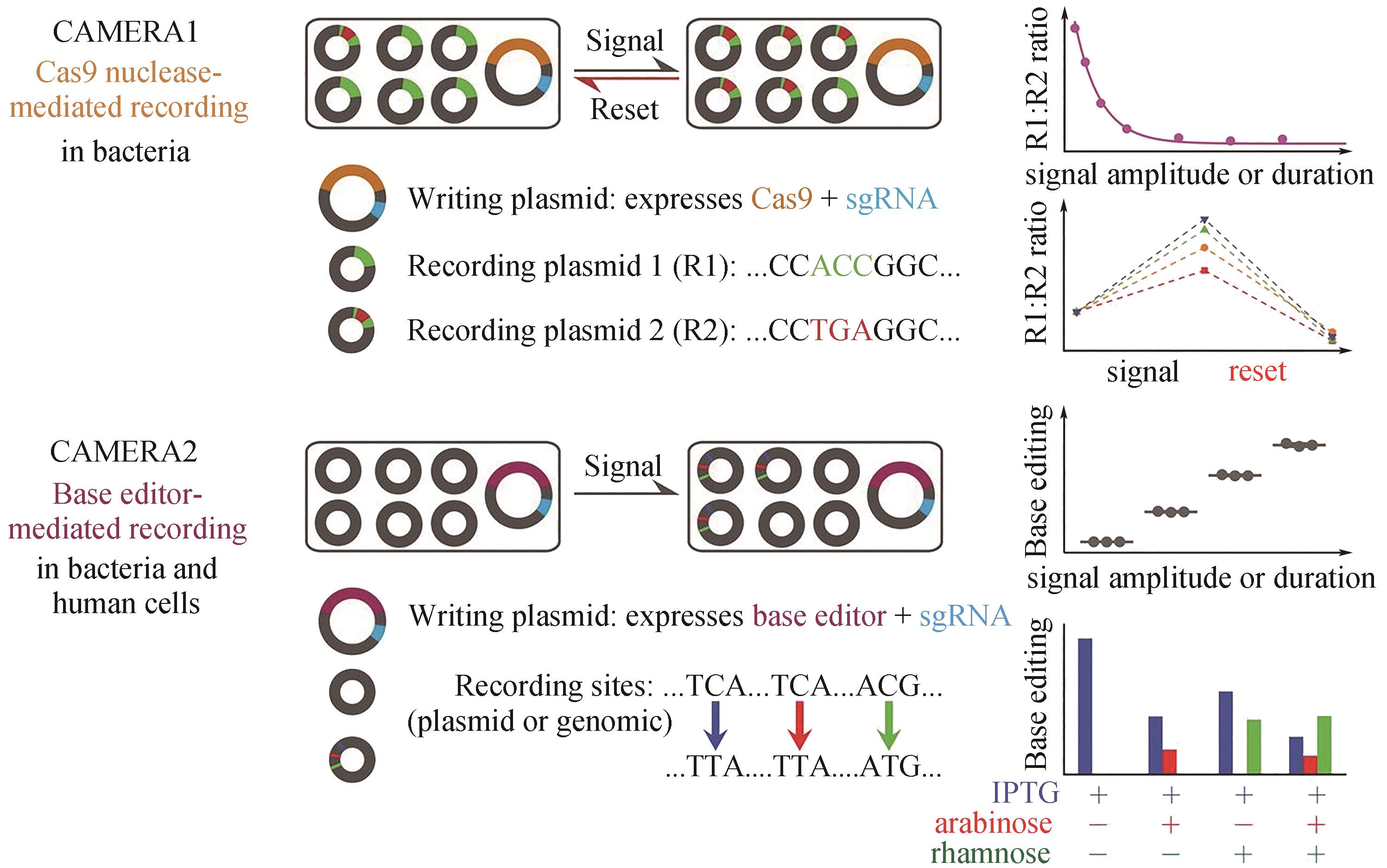
Fig. 5 Multiple analog of cellular recording by CAMERA systems in bacteria and mammalian cells[39]CAMERA 1 records stimuli as changes in the ratio of mutually exclusive DNA sequences. CAMERA 2 uses base editors to record the duration or amplitude of signals as single-nucleotide changes. Both systems can be repeatedly used to independently record multiple events, including exposure to antibiotics, nutrients, viruses, and light, as well as Wnt signaling
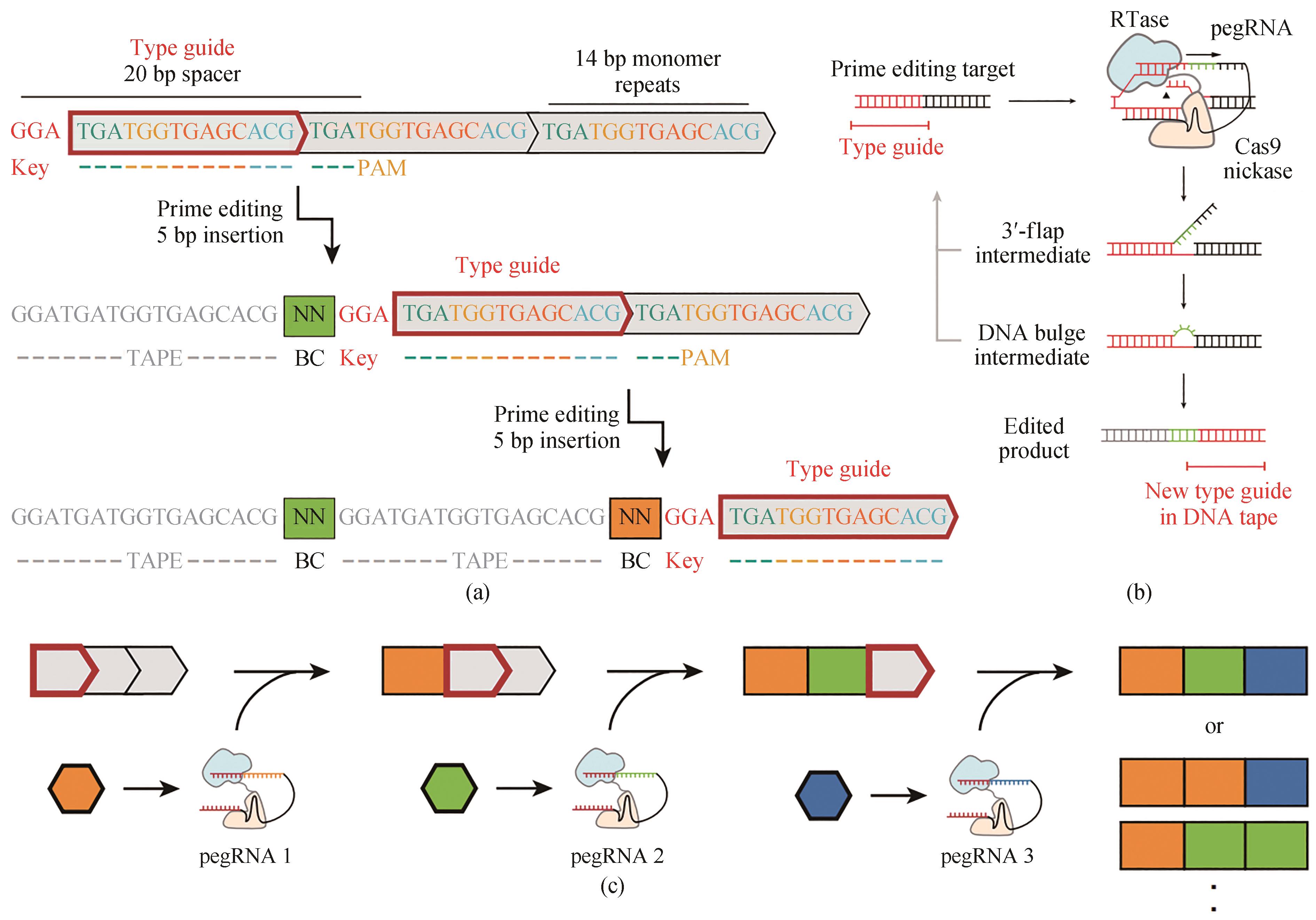
Fig. 6 Sequential genome editing with DNA typewriter[40](a) Schematic of two successive editing events at the type guide, which shifts in position with each editing event. The DNA tape consists of a tandem array of CRISPR–Cas9 target sites (grey boxes), all but the first of which are truncated at their 5′ ends and therefore inactive. The 5-bp insertion includes a 2-bp pegRNA-specific barcode as well as a 3-bp key that activates the next monomer. Because genome editing is sequential in this scheme, the temporal order of recorded events can simply be read out by their physical order along the array. (b) Schematic of prime editing with DNA typewriter. Prime editing recognizes a CRISPR–Cas9 target and modifies it with the edit specified by the pegRNA. With DNA typewriter, an insertional editing event generates a new prime editing target at the subsequent monomer. (c) Schematic of ordered recording via DNA typewriter. Individual pegRNAs are potentially event-driven or constitutively expressed, together with the PE2 enzyme
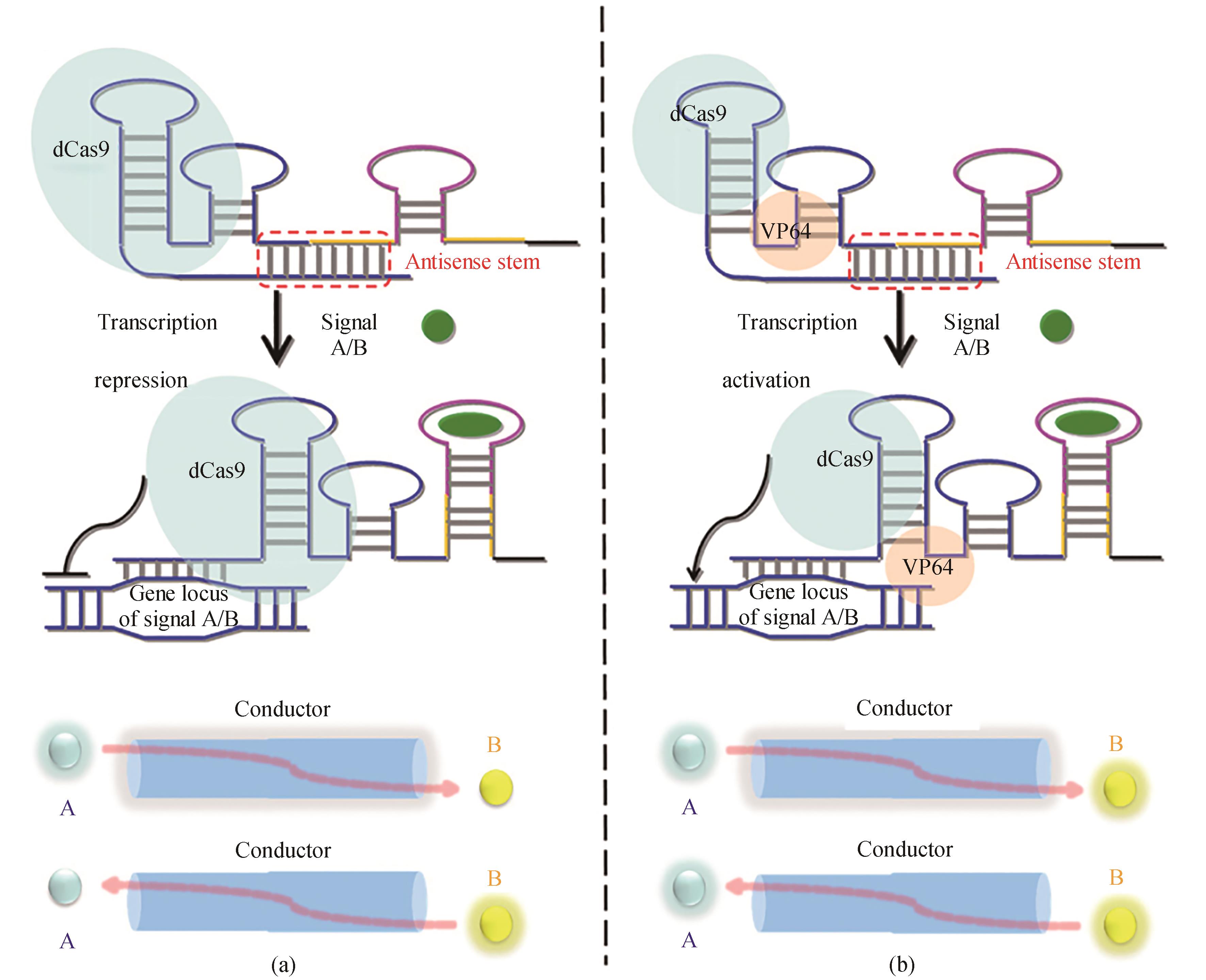
Fig. 7 Design of the CRISPR-Cas9-based signal conductor to link one signal with another[70]General illustration of the strand-displacement mechanism by which the redesigned sgRNA acts to deactivate (a) or activate (b) gene expression. The antisense sequence of the sgRNA is shown in blue, and the aptamer stem is shown in yellow. In the absence of signal A/B, the guide region of sgRNA is paired within the antisense stem and the sgRNA is in the 'off' state. In the presence of signal A/B, the conformation of the redesigned sgRNA is switched to the 'on' state. In this state, the guide region of the sgRNA binds to its target DNA, and thus turns the production of signal B/A on and off through the dCas9-VP64 fusion protein and the dCas9 protein, respectively.
| 70 | LIU Y C, ZHAN Y H, CHEN Z C, et al. Directing cellular information flow via CRISPR signal conductors[J]. Nature Methods, 2016, 13(11): 938-944. |
| 71 | SLOMOVIC S, PARDEE K, COLLINS J J. Synthetic biology devices for in vitro and in vivo diagnostics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(47): 14429-14435. |
| 72 | HAELLMAN V, FUSSENEGGER M. Synthetic biology—toward therapeutic solutions[J]. Journal of Molecular Biology, 2016, 428(5 Pt B): 945-962. |
| 73 | WALTER J G, STAHL F. Aptazymes: expanding the specificity of natural catalytic nucleic acids by application of in vitro selected oligonucleotides[J]. Advances in Biochemical Engineering/Biotechnology, 2020, 170: 107-119. |
| 74 | YOKOBAYASHI Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells[J]. Current Opinion in Chemical Biology, 2019, 52: 72-78. |
| 75 | WANG X Y, DONG K L, KONG D Q, et al. A far-red light-inducible CRISPR-Cas12a platform for remote-controlled genome editing and gene activation[J]. Science Advances, 2021, 7(50): eabh2358. |
| 76 | KAVITA K, BREAKER R R. Discovering riboswitches: the past and the future[J]. Trends in Biochemical Sciences, 2023, 48(2): 119-141. |
| 77 | LIU Y C, HAN J H, CHEN Z C, et al. Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors[J]. Nature Communications, 2017, 8: 2095. |
| 78 | KIM T, LU T K. CRISPR/Cas-based devices for mammalian synthetic biology[J]. Current Opinion in Chemical Biology, 2019, 52: 23-30. |
| 79 | WIELAND M, FUSSENEGGER M. Engineering molecular circuits using synthetic biology in mammalian cells[J]. Annual Review of Chemical and Biomolecular Engineering, 2012, 3: 209-234. |
| 80 | INNISS M C, SILVER P A. Building synthetic memory[J]. Current Biology, 2013, 23(17): R812-R816. |
| 81 | ZHAN H J, ZHOU Q, GAO Q J, et al. Multiplexed promoterless gene expression with CRISPReader[J]. Genome Biology, 2019, 20(1): 113. |
| 1 | CAMERON D E, BASHOR C J, COLLINS J J. A brief history of synthetic biology[J]. Nature Reviews Microbiology, 2014, 12(5): 381-390. |
| 2 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 3 | MOON T S, LOU C B, TAMSIR A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491(7423): 249-253. |
| 4 | STRICKER J, COOKSON S, BENNETT M R, et al. A fast, robust and tunable synthetic gene oscillator[J]. Nature, 2008, 456(7221): 516-519. |
| 5 | FRIEDLAND A E, LU T K, WANG X, et al. Synthetic gene networks that count[J]. Science, 2009, 324(5931): 1199-1202. |
| 6 | WEI P, WONG W W, PARK J S, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells[J]. Nature, 2012, 488(7411): 384-388. |
| 7 | ROYBAL K T, RUPP L J, MORSUT L, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits[J]. Cell, 2016, 164(4): 770-779. |
| 8 | MAYS Z J, NAIR N U. Synthetic biology in probiotic lactic acid bacteria: at the frontier of living therapeutics[J]. Current Opinion in Biotechnology, 2018, 53: 224-231. |
| 9 | SLOMOVIC S, COLLINS J J. DNA sense-and-respond protein modules for mammalian cells[J]. Nature Methods, 2015, 12(11): 1085-1090. |
| 10 | COURBET A, ENDY D, RENARD E, et al. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates[J]. Science Translational Medicine, 2015, 7(289): 289ra83. |
| 11 | FENNO L E, MATTIS J, RAMAKRISHNAN C, et al. Targeting cells with single vectors using multiple-feature Boolean logic[J]. Nature Methods, 2014, 11(7): 763-772. |
| 12 | BOGORAD I W, LIN T S, LIAO J C. Synthetic non-oxidative glycolysis enables complete carbon conservation[J]. Nature, 2013, 502(7473): 693-697. |
| 13 | BERNABÉ-ORTS J M, QUIJANO-RUBIO A, VAZQUEZ-VILAR M, et al. A memory switch for plant synthetic biology based on the phage ϕC31 integration system[J]. Nucleic Acids Research, 2020, 48(6): 3379-3394. |
| 14 | JIN S, BAE J Y, SONG Y, et al. Synthetic biology on acetogenic bacteria for highly efficient conversion of C1 gases to biochemicals[J]. International Journal of Molecular Sciences, 2020, 21(20): 7639. |
| 15 | BIRD L J, KUNDU B B, TSCHIRHART T, et al. Engineering wired life: synthetic biology for electroactive bacteria[J]. ACS Synthetic Biology, 2021, 10(11): 2808-2823. |
| 16 | CUBILLOS-RUIZ A, GUO T X, SOKOLOVSKA A, et al. Engineering living therapeutics with synthetic biology[J]. Nature Reviews Drug Discovery, 2021, 20(12): 941-960. |
| 17 | LIU Z H, WANG J Y, NIELSEN J. Yeast synthetic biology advances biofuel production[J]. Current Opinion in Microbiology, 2022, 65: 33-39. |
| 18 | SHENDURE J, JI H. Next-generation DNA sequencing[J]. Nature Biotechnology, 2008, 26(10): 1135-1145. |
| 19 | HEATHER J M, CHAIN B. The sequence of sequencers: the history of sequencing DNA[J]. Genomics, 2016, 107(1): 1-8. |
| 20 | FARZADFARD F, LU T K. Emerging applications for DNA writers and molecular recorders[J]. Science, 2018, 361(6405): 870-875. |
| 21 | SONG X, REIF J. Nucleic acid databases and molecular-scale computing[J]. ACS Nano, 2019, 13(6): 6256-6268. |
| 22 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456-466. |
| 23 | HAM T S, LEE S K, KEASLING J D, et al. Design and construction of a double inversion recombination switch for heritable sequential genetic memory[J]. PLoS One, 2008, 3(7): e2815. |
| 24 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337(6102): 1628. |
| 82 | ANZALONE A V, KOBLAN L W, LIU D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| 83 | LIENERT F, LOHMUELLER J J, GARG A, et al. Synthetic biology in mammalian cells: next generation research tools and therapeutics[J]. Nature Reviews Molecular Cell Biology, 2014, 15(2): 95-107. |
| 84 | LU T K, COLLINS J J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(12): 4629-4634. |
| 85 | ORTIZ M E, ENDY D. Engineered cell-cell communication via DNA messaging[J]. Journal of Biological Engineering, 2012, 6(1): 16. |
| 86 | ZHANG Y, PTACIN J L, FISCHER E C, et al. A semi-synthetic organism that stores and retrieves increased genetic information[J]. Nature, 2017, 551(7682): 644-647. |
| 87 | HO J M L, BENNETT M R. Improved memory devices for synthetic cells[J]. Science, 2018, 360(6385): 150-151. |
| 25 | FARZADFARD F, LU T K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations[J]. Science, 2014, 346(6211): 1256272. |
| 26 | OISHI K, KLAVINS E. Framework for engineering finite state machines in gene regulatory networks[J]. ACS Synthetic Biology, 2014, 3(9): 652-665. |
| 27 | AALIPOUR A, CHUANG H Y, MURTY S, et al. Engineered immune cells as highly sensitive cancer diagnostics[J]. Nature Biotechnology, 2019, 37(5): 531-539. |
| 28 | JIANG K Y, KOOB J, DAWN CHEN X, et al. Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR[J]. Nature Biotechnology, 2022: 1-10. |
| 29 | 杨璐, 吴楠, 白茸茸, 等. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080. |
| YANG L, WU N, BAI R R, et al. Design, optimization and application of whole-cell microbial biosensors with engineered genetic circuits[J]. Synthetic Biology Journal, 2022, 3(6): 1061-1080. | |
| 30 | KRETZSCHMAR K, WATT F M. Lineage tracing[J]. Cell, 2012, 148(1/2): 33-45. |
| 31 | WAGNER D E, KLEIN A M. Lineage tracing meets single-cell omics: opportunities and challenges[J]. Nature Reviews Genetics, 2020, 21(7): 410-427. |
| 32 | MEACHAM C E, MORRISON S J. Tumour heterogeneity and cancer cell plasticity[J]. Nature, 2013, 501(7467): 328-337. |
| 33 | WEINREB C, RODRIGUEZ-FRATICELLI A, CAMARGO F D, et al. Lineage tracing on transcriptional landscapes links state to fate during differentiation[J]. Science, 2020, 367(6479): eaaw3381. |
| 34 | SNIPPERT H J, VAN DER FLIER L G, SATO T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells[J]. Cell, 2010, 143(1): 134-144. |
| 35 | MCKENNA A, FINDLAY G M, GAGNON J A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing[J]. Science, 2016, 353(6298): aaf7907. |
| 36 | MO C Y, GUO J X, QIN J C, et al. Single-cell transcriptomics of LepR-positive skeletal cells reveals heterogeneous stress-dependent stem and progenitor pools[J]. The EMBO Journal, 2022, 41(4): e108415 |
| 37 | HE L J, LI Y, LI Y, et al. Enhancing the precision of genetic lineage tracing using dual recombinases[J]. Nature Medicine, 2017, 23(12): 1488-1498. |
| 38 | TANNA T, SCHMIDT F, CHEREPKOVA M Y, et al. Recording transcriptional histories using Record-seq[J]. Nature Protocols, 2020, 15(2): 513-539. |
| 39 | TANG W X, LIU D R. Rewritable multi-event analog recording in bacterial and mammalian cells[J]. Science, 2018, 360(6385): eaap8992. |
| 40 | CHOI J, CHEN W, MINKINA A, et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing[J]. Nature, 2022, 608(7921): 98-107. |
| 41 | SPANJAARD B, HU B, MITIC N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars[J]. Nature Biotechnology, 2018, 36(5): 469-473. |
| 42 | PERLI S D, CUI C H, LU T K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells[J]. Science, 2016, 353(6304): aag0511. |
| 43 | HE Z S, MAYNARD A, JAIN A, et al. Lineage recording in human cerebral organoids[J]. Nature Methods, 2022, 19(1): 90-99. |
| 44 | FARZADFARD F, GHARAEI N, HIGASHIKUNI Y, et al. Single-nucleotide-resolution computing and memory in living cells[J]. Molecular Cell, 2019, 75(4): 769-780.e4. |
| 45 | SHETH R U, WANG H H. DNA-based memory devices for recording cellular events[J]. Nature Reviews Genetics, 2018, 19(11): 718-732. |
| 46 | AUSLÄNDER S, FUSSENEGGER M. Synthetic biology. Dynamic genome engineering in living cells[J]. Science, 2014, 346(6211): 813-814. |
| 47 | ATKINSON M R, SAVAGEAU M A, MYERS J T, et al. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli [J]. Cell, 2003, 113(5): 597-607. |
| 48 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 49 | BIANCALANI T, ASSAF M. Genetic toggle switch in the absence of cooperative binding: exact results[J]. Physical Review Letters, 2015, 115(20): 208101. |
| 50 | BHOMKAR P, MATERI W, WISHART D S. The bacterial nanorecorder: engineering E. coli to function as a chemical recording device[J]. PLoS One, 2011, 6(11): e27559. |
| 51 | KOTULA J W, KERNS S J, SHAKET L A, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(13): 4838-4843. |
| 52 | MIMEE M, TUCKER A C, VOIGT CA, et al. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota[J]. Cell Systems, 2015, 1(1): 62-71. |
| 53 | BONNET J, YIN P, ORTIZ M E, et al. Amplifying genetic logic gates[J]. Science, 2013, 340(6132): 599-603. |
| 54 | SIUTI P, YAZBEK J, LU T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature Biotechnology, 2013, 31(5): 448-452. |
| 55 | ROQUET N, SOLEIMANY A P, FERRIS A C, et al. Synthetic recombinase-based state machines in living cells[J]. Science, 2016, 353(6297): aad8559. |
| 56 | LAMPSON B C, INOUYE M, INOUYE S. Retrons, msDNA, and the bacterial genome[J]. Cytogenetic and Genome Research, 2005, 110(1/2/3/4): 491-499. |
| 57 | LIM D B, MAAS W K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B[J]. Cell, 1989, 56(5): 891-904. |
| 58 | SCHMIDT F, CHEREPKOVA M Y, PLATT R J. Transcriptional recording by CRISPR spacer acquisition from RNA[J]. Nature, 2018, 562(7727): 380-385. |
| 59 | SCHMIDT F, ZIMMERMANN J, TANNA T, et al. Noninvasive assessment of gut function using transcriptional recording sentinel cells[J]. Science, 2022, 376(6594): eabm6038. |
| 60 | AMITAI G, SOREK R. CRISPR-Cas adaptation: insights into the mechanism of action[J]. Nature Reviews Microbiology, 2016, 14(2): 67-76. |
| 61 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 62 | DOUDNA J A, CHARPENTIER E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| 63 | MANGHWAR H, LINDSEY K, ZHANG X L, et al. CRISPR/cas system: recent advances and future prospects for genome editing[J]. Trends in Plant Science, 2019, 24(12): 1102-1125. |
| 64 | ZHAN H J, XIAO L L, LI A L, et al. Engineering cellular signal sensors based on CRISPR-sgRNA reconstruction approaches[J]. International Journal of Biological Sciences, 2020, 16(8): 1441-1449. |
| 65 | NISSIM L, PERLI S D, FRIDKIN A, et al. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells[J]. Molecular Cell, 2014, 54(4): 698-710. |
| 66 | ZHAN H J, LI A L, CAI Z M, et al. Improving transgene expression and CRISPR-Cas9 efficiency with molecular engineering-based molecules[J]. Clinical and Translational Medicine, 2020, 10(6): e194. |
| 67 | NIU Q W, LIN S S, REYES J L, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance[J]. Nature Biotechnology, 2006, 24(11): 1420-1428. |
| 68 | HUANG X B, CHEN Z C, LIU Y C. RNAi-mediated control of CRISPR functions[J]. Theranostics, 2020, 10(15): 6661-6673. |
| 69 | LIU Y C, ZENG Y Y, LIU L, et al. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells[J]. Nature Communications, 2014, 5: 5393. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems [J]. Synthetic Biology Journal, 2025, 6(1): 105-117. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||