Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (2): 283-300.DOI: 10.12211/2096-8280.2022-070
• Invited Review • Previous Articles Next Articles
Merging the frontiers: synthetic biology for advanced bacteriophage design
CHEN Qingli, TONG Yigang
- College of Life Science and Technology,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2022-12-06Revised:2022-12-30Online:2023-04-27Published:2023-04-30 -
Contact:TONG Yigang
工程噬菌体的合成生物学“智造”
陈青黎, 童贻刚
- 北京化工大学生命科学与技术学院 北京 100029
-
通讯作者:童贻刚 -
作者简介:陈青黎 (2000—),女,硕士研究生。研究方向为合成生物学改造工程噬菌体。E-mail:ql.chen@buct.edu.cn童贻刚 (1966—),男,研究员,博士生导师。研究方向为噬菌体学、微生物学、高通量测序、生物信息学研究、合成生物学等。E-mail:tongyigang@mail.buct.edu.cn -
基金资助:国家自然科学基金(32001834);国家重点研发计划(2018Y FA 0903000);军事生物安全研究计划(20SWAQX27)
CLC Number:
Cite this article
CHEN Qingli, TONG Yigang. Merging the frontiers: synthetic biology for advanced bacteriophage design[J]. Synthetic Biology Journal, 2023, 4(2): 283-300.
陈青黎, 童贻刚. 工程噬菌体的合成生物学“智造”[J]. 合成生物学, 2023, 4(2): 283-300.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-070
| 策略 | 菌株 | 具体方法 | 年份 | 出处 |
|---|---|---|---|---|
| 随机诱变 | T4噬菌体 | 紫外线诱变获取突变体 | 1966 | [ |
| T3、T7、肠道沙门氏菌噬菌体 | 改善噬菌体的热稳定性 | 2020 | [ | |
| 经典同源重组 | T2噬菌体 | 改造尾丝蛋白结构 | 2009 | [ |
| T4噬菌体 | 改变或扩大宿主T4样噬菌体宿主范围 | 2017 | [ | |
| T7噬菌体 | T7噬菌体基因组的单碱基替换和全基因替换 | 2020 | [ | |
| T7噬菌体 | 获得新的RBP结构 | 2021 | [ | |
| BRED | 分歧杆菌噬菌体Giles | 非必需基因缺失、读码框内缺失、点突变、无义突变、外源基因精确插入 | 2008 | [ |
| 肠杆菌科噬菌体P1 | 去除IS元件 | 2012 | [ | |
| 沙门氏菌噬菌体 | 实现溶原和裂解性质转换 | 2014 | [ | |
| 克雷伯氏菌噬菌体 | 建立了克雷伯氏菌噬菌体基因组的重组系统 | 2017 | [ | |
| 分枝杆菌噬菌体BPs、ZoeJ | 将治疗性温和噬菌体转化为烈性噬菌体 | 2019 | [ | |
| CRISPR-Cas | 链球菌噬菌体2972 | Ⅱ-A型CRISPR-Cas实现特定点突变和大片段删除 | 2014 | [ |
| T7噬菌体 | Ⅰ-E型CRISPR-Cas系统编辑T7基因组 | 2014 | [ | |
| 乳酸乳球菌烈性噬菌体P2 | 点突变,基因缺失和替换 | 2017 | [ | |
| 克雷伯氏菌噬菌体 | 点突变,基因缺失和替换 | 2018 | [ | |
| 金黄色葡萄球菌噬菌体 | Ⅲ-A型CRISPR-Cas系统CRISPR-Cas10 | 2019 | [ | |
| T4噬菌体 | Ⅴ型CRISPR-Cas12a系统构建包含缺失和插入的重组T4 | 2021 | [ | |
| T5噬菌体 | Ⅱ-A型CRISPR-Cas9系统效果不稳定,提出基于Retron的重组方案 | 2021 | [ | |
| 需钠弧菌噬菌体TT4 | 使用CRISPR-Cas9基因的缺失和替换 | 2022 | [ | |
| 大肠杆菌噬菌体 | CRISPR-Cas13a | 2022 | [ | |
| ФKZ、OMKO1和PaMx41噬菌体 | CRISPR-Cas13a+正向选择基因acrVIA1 | 2022 | [ | |
| Genome Reboot | 大肠杆菌、克雷伯氏菌噬菌体 | YAC | 2015 | [ |
| PICIs | YAC | 2020 | [ | |
| 铜绿假单胞菌噬菌体 | YAC | 2021 | [ | |
| 大肠杆菌噬菌体 | 电转重启 | 2019 | [ | |
| 沙门氏菌噬菌体 | 电转重启 | 2022 | [ | |
| MS2噬菌体 | 无细胞体系 | 1996 | [ | |
| T7噬菌体 | 无细胞体系 | 2012 | [ | |
| T4噬菌体 | 无细胞体系 | 2018 | [ | |
| 耶尔森氏菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 克雷伯氏菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 抗酸性分枝杆菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 李斯特氏菌、芽孢杆菌、 葡萄球菌噬菌体 | L-forms | 2018 | [ |
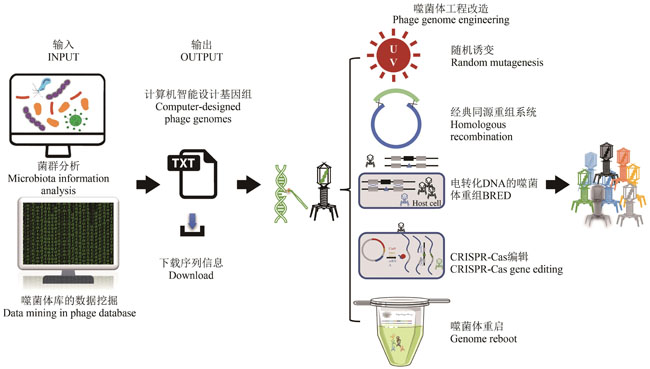
Table 1 Applications of engineered phages through genetic modifications
| 策略 | 菌株 | 具体方法 | 年份 | 出处 |
|---|---|---|---|---|
| 随机诱变 | T4噬菌体 | 紫外线诱变获取突变体 | 1966 | [ |
| T3、T7、肠道沙门氏菌噬菌体 | 改善噬菌体的热稳定性 | 2020 | [ | |
| 经典同源重组 | T2噬菌体 | 改造尾丝蛋白结构 | 2009 | [ |
| T4噬菌体 | 改变或扩大宿主T4样噬菌体宿主范围 | 2017 | [ | |
| T7噬菌体 | T7噬菌体基因组的单碱基替换和全基因替换 | 2020 | [ | |
| T7噬菌体 | 获得新的RBP结构 | 2021 | [ | |
| BRED | 分歧杆菌噬菌体Giles | 非必需基因缺失、读码框内缺失、点突变、无义突变、外源基因精确插入 | 2008 | [ |
| 肠杆菌科噬菌体P1 | 去除IS元件 | 2012 | [ | |
| 沙门氏菌噬菌体 | 实现溶原和裂解性质转换 | 2014 | [ | |
| 克雷伯氏菌噬菌体 | 建立了克雷伯氏菌噬菌体基因组的重组系统 | 2017 | [ | |
| 分枝杆菌噬菌体BPs、ZoeJ | 将治疗性温和噬菌体转化为烈性噬菌体 | 2019 | [ | |
| CRISPR-Cas | 链球菌噬菌体2972 | Ⅱ-A型CRISPR-Cas实现特定点突变和大片段删除 | 2014 | [ |
| T7噬菌体 | Ⅰ-E型CRISPR-Cas系统编辑T7基因组 | 2014 | [ | |
| 乳酸乳球菌烈性噬菌体P2 | 点突变,基因缺失和替换 | 2017 | [ | |
| 克雷伯氏菌噬菌体 | 点突变,基因缺失和替换 | 2018 | [ | |
| 金黄色葡萄球菌噬菌体 | Ⅲ-A型CRISPR-Cas系统CRISPR-Cas10 | 2019 | [ | |
| T4噬菌体 | Ⅴ型CRISPR-Cas12a系统构建包含缺失和插入的重组T4 | 2021 | [ | |
| T5噬菌体 | Ⅱ-A型CRISPR-Cas9系统效果不稳定,提出基于Retron的重组方案 | 2021 | [ | |
| 需钠弧菌噬菌体TT4 | 使用CRISPR-Cas9基因的缺失和替换 | 2022 | [ | |
| 大肠杆菌噬菌体 | CRISPR-Cas13a | 2022 | [ | |
| ФKZ、OMKO1和PaMx41噬菌体 | CRISPR-Cas13a+正向选择基因acrVIA1 | 2022 | [ | |
| Genome Reboot | 大肠杆菌、克雷伯氏菌噬菌体 | YAC | 2015 | [ |
| PICIs | YAC | 2020 | [ | |
| 铜绿假单胞菌噬菌体 | YAC | 2021 | [ | |
| 大肠杆菌噬菌体 | 电转重启 | 2019 | [ | |
| 沙门氏菌噬菌体 | 电转重启 | 2022 | [ | |
| MS2噬菌体 | 无细胞体系 | 1996 | [ | |
| T7噬菌体 | 无细胞体系 | 2012 | [ | |
| T4噬菌体 | 无细胞体系 | 2018 | [ | |
| 耶尔森氏菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 克雷伯氏菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 抗酸性分枝杆菌噬菌体 | 无细胞体系 | 2022 | [ | |
| 李斯特氏菌、芽孢杆菌、 葡萄球菌噬菌体 | L-forms | 2018 | [ |
| 1 | TWORT F W. An investigation on the nature of ultra-microscopic viruses[J]. The Lancet, 1915, 186(4814): 1241-1243. |
| 2 | D'HERELLE F. On an invisible microbe antagonistic toward dysenteric bacilli: brief note by Mr. F. D'Herelle, presented by Mr. Roux. 1917[J]. Research in Microbiology, 2007, 158(7): 553-554. |
| 3 | OLSZAK T, LATKA A, ROSZNIOWSKI B, et al. Phage life cycles behind bacterial biodiversity[J]. Current Medicinal Chemistry, 2017, 24(36): 3987-4001. |
| 4 | JACOB F, MONOD J. Genetic regulatory mechanisms in the synthesis of proteins[J]. Journal of Molecular Biology, 1961, 3(3): 318-356. |
| 5 | CRICK F H C, BARNETT L, BRENNER S, et al. General nature of the genetic code for proteins[J]. Nature, 1961,192(4809):1227-1232. |
| 6 | ANDERSON J E. Restriction endonucleases and modification methylases[J]. Current Opinion in Structural Biology, 1993, 3(1): 24-30. |
| 7 | PINGOUD A, WILSON G G, WENDE W. TypeⅡrestriction endonucleases—a historical perspective and more[J]. Nucleic Acids Research, 2014, 42(12): 7489-7527. |
| 8 | BORKOTOKY S, MURALI A. The highly efficient T7 RNA polymerase: a wonder macromolecule in biological realm[J]. International Journal of Biological Macromolecules, 2018, 118(Pt A): 49-56. |
| 9 | KORLACH J, BJORNSON K P, CHAUDHURI B P, et al. Real-time DNA sequencing from single polymerase molecules[J]. Methods in Enzymology, 2010, 472: 431-455. |
| 10 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. |
| 11 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 12 | KHAN A, OSTAKU J, ARAS E, et al. Combating infectious diseases with synthetic biology[J]. ACS Synthetic Biology, 2022, 11(2): 528-537. |
| 13 | KORTRIGHT K E, CHAN B K, KOFF J L, et al. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria[J]. Cell Host & Microbe, 2019, 25(2): 219-232. |
| 14 | UYTTEBROEK S, CHEN B X, ONSEA J, et al. Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review[J]. The Lancet Infectious Diseases, 2022, 22(8): e208-e220. |
| 15 | WEI J W, PENG N, LIANG Y X, et al. Phage Therapy: consider the Past, Embrace the Future[J]. 2020, 10(21), 7654. |
| 16 | LUONG T, SALABARRIA A C, EDWARDS R A, et al. Standardized bacteriophage purification for personalized phage therapy[J]. Nature Protocols, 2020, 15(9): 2867-2890. |
| 17 | LIU D, VAN BELLEGHEM J D, DE VRIES C R, et al. The safety and toxicity of phage therapy: a review of animal and clinical studies[J]. Viruses, 2021, 13(7): 1268. |
| 18 | TAN Y, SHEN J T, SI T, et al. Engineered live biotherapeutics: progress and challenges[J]. Biotechnology Journal, 2020, 15(10): e2000155. |
| 19 | SUN X Y, ILCA S L, HUISKONEN J T, et al. Dual role of a viral polymerase in viral genome replication and particle self-assembly[J]. mBio, 2018, 9(5): e01242-e01218. |
| 20 | VENTER J C, GLASS J I, HUTCHISON C A III, et al. Synthetic chromosomes, genomes, viruses, and cells[J]. Cell, 2022, 185(15): 2708-2724. |
| 21 | R A Ⅲ WHITE. The future of virology is synthetic[J]. mSystems, 2021, 6(4): e0077021. |
| 22 | WEYNBERG K D, JASCHKE P R. Building better bacteriophage with biofoundries to combat antibiotic-resistant bacteria[J]. PHAGE, 2020, 1(1): 23-26. |
| 23 | KILCHER S, LOESSNER M J. Engineering bacteriophages as versatile biologics[J]. Trends in Microbiology, 2019, 27(4): 355-367. |
| 24 | DEDRICK R M, GUERRERO-BUSTAMANTE C A, GARLENA R A, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus [J]. Nature Medicine, 2019, 25(5): 730-733. |
| 25 | CHEN Y B, BATRA H, DONG J H, et al. Genetic engineering of bacteriophages against infectious diseases[J]. Frontiers in Microbiology, 2019, 10: 954. |
| 26 | VIERTEL T M, RITTER K, HORZ H P. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens[J]. Journal of Antimicrobial Chemotherapy, 2014, 69(9): 2326-2336. |
| 27 | MEILE S, DU J M, DUNNE M, et al. Engineering therapeutic phages for enhanced antibacterial efficacy[J]. Current Opinion in Virology, 2022, 52: 182-191. |
| 28 | MCEWEN S A, COLLIGNON P J. Antimicrobial resistance: a one health perspective[J]. Microbiology Spectrum, 2018, 6(2): 6.2.10. |
| 29 | ASLAM B, KHURSHID M, ARSHAD M I, et al. Antibiotic resistance: one health one world outlook[J]. Frontiers in Cellular and Infection Microbiology, 2021, 11: 771510. |
| 30 | HOWARD-VARONA C, HARGREAVES K R, ABEDON S T, et al. Lysogeny in nature: mechanisms, impact and ecology of temperate phages[J]. The ISME Journal, 2017, 11(7): 1511-1520. |
| 31 | HARRISON E, BROCKHURST M A. Ecological and evolutionary benefits of temperate phage: what does or doesn't kill you makes you stronger[J]. BioEssays, 2017, 39(12): 1700112. |
| 32 | CHEN Y B, YANG L, YANG D, et al. Specific integration of temperate phage decreases the pathogenicity of host bacteria[J]. Frontiers in Cellular and Infection Microbiology, 2020, 10: 14. |
| 33 | SOUSA J A M, PFEIFER E, TOUCHON M, et al. Genome diversification via genetic exchanges between temperate and virulent bacteriophages[EB/OL]. bioRxiv, 2020[2020-04]. . |
| 34 | DE SOUSA J A M, PFEIFER E, TOUCHON M, et al. Causes and consequences of bacteriophage diversification via genetic exchanges across lifestyles and bacterial taxa[J]. Molecular Biology and Evolution, 2021, 38(6): 2497-2512. |
| 35 | AL-ANANY A M, FATIMA R, HYNES A P. Temperate phage-antibiotic synergy eradicates bacteria through depletion of lysogens[J]. Cell Reports, 2021, 35(8): 109172. |
| 36 | SCHROVEN K, AERTSEN A, LAVIGNE R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation[J]. FEMS Microbiology Reviews, 2021, 45(1): fuaa041. |
| 37 | LABRIE S J, MOINEAU S. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages[J].Journal of Bacteriology, 2007,189(4):1482-1487. |
| 38 | AMARILLAS L, RUBÍ-RANGEL L, CHAIDEZ C, et al. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli [J]. Frontiers in Microbiology, 2017, 8: 1355. |
| 39 | CHAUDHARY N, MOHAN B, MAVUDURU R S, et al. Characterization, genome analysis and in vitro activity of a novel phage vB_EcoA_RDN8.1 active against multi-drug resistant and extensively drug-resistant biofilm-forming uropathogenic Escherichia coli isolates, India[J]. Journal of Applied Microbiology, 2022, 132(4): 3387-3404. |
| 40 | BAO H D, SHAHIN K, ZHANG Q Y, et al. Morphologic and genomic characterization of a broad host range Salmonella enterica serovar Pullorum lytic phage vB_SPuM_SP116[J]. Microbial Pathogenesis, 2019, 136: 103659. |
| 41 | MASUDA Y, KAWABATA S, UEDOI T, et al. Construction of leaderless-bacteriocin-producing bacteriophage targeting E. coli and neighboring gram-positive pathogens[J]. Microbiology Spectrum, 2021, 9(1): e0014121. |
| 42 | NICK J A, DEDRICK R M, GRAY A L, et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection[J]. Cell, 2022, 185(11): 1860-1874.e12. |
| 43 | JIN M L, CHEN J C, ZHAO X Y, et al. An engineered λ phage enables enhanced and strain-specific killing of enterohemorrhagic Escherichia coli [J]. Microbiology Spectrum, 2022, 10(4): e01271-e01222. |
| 44 | LIU G W, LIN Q P, JIN S, et al. The CRISPR-Cas toolbox and gene editing technologies[J]. Molecular Cell, 2022, 82(2): 333-347. |
| 45 | YADAV R, KUMAR V, BAWEJA M, et al. Gene editing and genetic engineering approaches for advanced probiotics: a review[J]. Critical Reviews in Food Science and Nutrition, 2018, 58(10): 1735-1746. |
| 46 | ARROYO-OLARTE R D, BRAVO RODRÍGUEZ R, MORALES-RÍOS E. Genome editing in bacteria: CRISPR-cas and beyond[J]. Microorganisms, 2021, 9(4): 844. |
| 47 | ABABI M, TRIDGETT M, OSGERBY A, et al. Scarless recombineering of phage in lysogenic state[J]. Methods in Molecular Biology, 2022, 2479: 1-9. |
| 48 | TRIDGETT M, ABABI M, JARAMILLO A. Lambda red recombineering of bacteriophage in the lysogenic state[J]. Methods in Molecular Biology, 2022, 2479: 11-19. |
| 49 | DRAKE J W. Ultraviolet mutagenesis in bacteriophage T-4. I. Irradiation of extracellular phage particles[J]. Journal of Bacteriology, 1966, 91(5): 1775-1780. |
| 50 | MEISTRICH M L, SHULMAN R G. Mutagenic effect of sensitized irradiation of bacteriophage T4[J]. Journal of Molecular Biology, 1969, 46(1): 157-167. |
| 51 | FAVOR A H, LLANOS C D, YOUNGBLUT M D, et al. Optimizing bacteriophage engineering through an accelerated evolution platform[J]. Scientific Reports, 2020, 10(1): 13981. |
| 52 | LANIGAN T M, KOPERA H C, SAUNDERS T L. Principles of genetic engineering[J]. Genes, 2020, 11(3): 291. |
| 53 | MAHLER M, COSTA A R, VAN BELJOUW S P B, et al. Approaches for bacteriophage genome engineering[J/OL]. Trends in Biotechnology, 2022[2022-12-01].. |
| 54 | POUILLOT F, BLOIS H, IRIS F. Genetically engineered virulent phage banks in the detection and control of emergent pathogenic bacteria[J]. Biosecurity and Bioterrorism: biodefense Strategy, Practice, and Science, 2010, 8(2): 155-169. |
| 55 | JENSEN J D, PARKS A R, ADHYA S, et al. λ Recombineering used to engineer the genome of phage T7[J]. Antibiotics, 2020, 9(11): 805. |
| 56 | MØLLER-OLSEN C, STANLEY HO S F, DEV SHUKLA R, et al. Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells[J]. Scientific Reports, 2018, 8: 17559. |
| 57 | SHITRIT D, HACKL T, LAURENCEAU R, et al. Genetic engineering of marine cyanophages reveals integration but not lysogeny in T7-like cyanophages[J]. The ISME Journal, 2022, 16(2): 488-499. |
| 58 | MARINELLI L J, PIURI M, SWIGONOVÁ Z, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes[J]. PLoS One, 2008, 3(12): e3957. |
| 59 | MARINELLI L J, PIURI M, HATFULL G F. Genetic manipulation of lytic bacteriophages with BRED: bacteriophage recombineering of electroporated DNA[J]. Methods in Molecular Biology, 2019, 1898: 69-80. |
| 60 | FEHÉR T, KARCAGI I, BLATTNER F R, et al. Bacteriophage recombineering in the lytic state using the lambda red recombinases[J]. Microbial Biotechnology, 2012, 5(4): 466-476. |
| 61 | SHIN H, LEE J H, YOON H, et al. Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC[J]. Applied and Environmental Microbiology, 2014, 80(1): 374-384. |
| 62 | PAN Y J, LIN T L, CHEN C C, et al. Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types[J]. Journal of Virology, 2017, 91(6): e02457-e02416. |
| 63 | WETZEL K S, GUERRERO-BUSTAMANTE C A, DEDRICK R M, et al. CRISPY-BRED and CRISPY-BRIP: efficient bacteriophage engineering[J]. Scientific Reports, 2021, 11: 6796. |
| 64 | MARTEL B, MOINEAU S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages[J]. Nucleic Acids Research, 2014, 42(14): 9504-9513. |
| 65 | KIRO R, SHITRIT D, QIMRON U. Efficient engineering of a bacteriophage genome using the type Ⅰ-E CRISPR-Cas system[J]. RNA Biology, 2014, 11(1): 42-44. |
| 66 | LEMAY M L, TREMBLAY D M, MOINEAU S. Genome engineering of virulent lactococcal phages using CRISPR-Cas9[J]. ACS Synthetic Biology, 2017, 6(7): 1351-1358. |
| 67 | LEMAY M L, RENAUD A C, ROUSSEAU G M, et al. Targeted genome editing of virulent phages using CRISPR-Cas9[J]. Bio-protocol, 2018, 8(1): e2674. |
| 68 | SHEN J T, ZHOU J J, CHEN G Q, et al. Efficient genome engineering of a virulent klebsiella bacteriophage using CRISPR-Cas9[J]. Journal of Virology, 2018, 92(17): e00534-e00518. |
| 69 | NAYEEMUL BARI S M, HATOUM-ASLAN A. CRISPR-Cas10 assisted editing of virulent staphylococcal phages[J]. Methods in Enzymology, 2019, 616: 385-409. |
| 70 | DONG J H, CHEN C, LIU Y P, et al. Engineering T4 bacteriophage for in vivo display by typeⅤCRISPR-Cas genome editing[J]. ACS Synthetic Biology, 2021, 10(10): 2639-2648. |
| 71 | ZHANG X L, ZHANG C H, LIANG C J, et al. CRISPR-Cas9 based bacteriophage genome editing[J]. Microbiology Spectrum, 2022, 10(4): e0082022. |
| 72 | GUAN J W, OROMÍ-BOSCH A, MENDOZA S D, et al. Bacteriophage genome engineering with CRISPR-Cas13a[J]. Nature Microbiology, 2022, 7(12): 1956-1966. |
| 73 | ADLER B A, HESSLER T, CRESS B F, et al. Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing[J]. Nature Microbiology, 2022, 7(12): 1967-1979. |
| 74 | GRIGONYTE A M, HARRISON C, MACDONALD P R, et al. Comparison of CRISPR and marker-based methods for the engineering of phage T7[J]. Viruses, 2020, 12(2): 193. |
| 75 | RAMIREZ-CHAMORRO L, BOULANGER P, ROSSIER O. Strategies for bacteriophage T5 mutagenesis: expanding the toolbox for phage genome engineering[J]. Frontiers in Microbiology, 2021, 12: 667332. |
| 76 | MERRYMAN C, GIBSON D G. Methods and applications for assembling large DNA constructs[J]. Metabolic Engineering, 2012, 14(3): 196-204. |
| 77 | RAMSAY M. Yeast artificial chromosome cloning[J]. Molecular Biotechnology, 1994, 1(2): 181-201. |
| 78 | ANDO H, LEMIRE S, PIRES D P, et al. Engineering modular viral scaffolds for targeted bacterial population editing[J]. Cell Systems, 2015, 1(3): 187-196. |
| 79 | PIRES D P, MONTEIRO R, MIL-HOMENS D, et al. Designing P. aeruginosa synthetic phages with reduced genomes[J]. Scientific Reports, 2021, 11: 2164. |
| 80 | RODRIGO I C, HAAG ANDREAS F, PEDRO D M, et al. Rebooting synthetic phage-inducible chromosomal islands: one method to forge them all[J]. BioDesign Research, 2020, 2020: 5783064. |
| 81 | PRYOR J M, POTAPOV V, BILOTTI K, et al. Rapid 40 kb genome construction from 52 parts through data-optimized assembly design[J]. ACS Synthetic Biology, 2022, 11(6): 2036-2042. |
| 82 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 83 | PULKKINEN E M, HINKLEY T C, NUGEN S R. Utilizing in vitro DNA assembly to engineer a synthetic T7 Nanoluc reporter phage for Escherichia coli detection[J]. Integrative Biology, 2019, 11(3): 63-68. |
| 84 | JANEŽ N, MEGLIČ S H, FLISAR K, et al. Introduction of phage genome into Escherichia coli by electroporation[J]. Methods in Molecular Biology, 2019, 1898: 51-56. |
| 85 | MILHO C, SILLANKORVA S. Implication of a gene deletion on a Salmonella Enteritidis phage growth parameters[J]. Virus Research, 2022, 308: 198654. |
| 86 | RUSTAD M, EASTLUND A, JARDINE P, et al. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction[J]. Synthetic Biology, 2018, 3(1): ysy002. |
| 87 | GARENNE D, THOMPSON S, BRISSON A, et al. The all-E. coliTXTL toolbox 3.0: new capabilities of a cell-free synthetic biology platform[J]. Synthetic Biology, 2021, 6(1): ysab017. |
| 88 | EMSLANDER Q, VOGELE K, BRAUN P, et al. Cell-free production of personalized therapeutic phagestargeting multidrug-resistant bacteria[J]. Cell Chemical Biology, 2022, 29(9): 1434-1445.e7. |
| 89 | KATANAEV V L, SPIRIN A S, REUSS M, et al. Formation of bacteriophage MS2 infectious units in a cell-free translation system[J]. FEBS Letters, 1996, 397(2/3): 143-148. |
| 90 | SMITH H O, HUTCHISON C A III, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: ΦX174 bacteriophage from synthetic oligonucleotides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| 91 | SHIN J, JARDINE P, NOIREAUX V. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction[J]. ACS Synthetic Biology, 2012, 1(9): 408-413. |
| 92 | MITSUNAKA S, YAMAZAKI K, PRAMONO A K, et al. Synthetic engineering and biological containment of bacteriophages[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(48): e2206739119. |
| 93 | SPENCER A C, TORRE P, MANSY S S. The encapsulation of cell-free transcription and translation machinery in vesicles for the construction of cellular mimics[J]. Journal of Visualized Experiments: JoVE, 2013(80): e51304. |
| 94 | ALLAN E J, HOISCHEN C, GUMPERT J. Bacterial L-forms[J]. Advances in Applied Microbiology, 2009, 68: 1-39. |
| 95 | ERRINGTON J. Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective[J]. Biochemical Society Transactions, 2017, 45(2): 287-295. |
| 96 | KILCHER S, STUDER P, MUESSNER C, et al. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(3): 567-572. |
| 97 | CHENG L, DENG Z Q, TAO H R, et al. Harnessing stepping-stone hosts to engineer, select, and reboot synthetic bacteriophages in one pot[J]. Cell Reports Methods, 2022, 2(5): 100217. |
| 98 | MAHICHI F, SYNNOTT A J, YAMAMICHI K, et al. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity[J]. FEMS Microbiology Letters, 2009, 295(2): 211-217. |
| 99 | CHEN M M, ZHANG L, ABDELGADER S A, et al. Alterations in gp37 expand the host range of a T4-like phage[J]. Applied and Environmental Microbiology, 2017, 83(23): e01576-e01517. |
| 100 | AVRAMUCZ Á, MØLLER-OLSEN C, GRIGONYTE A M, et al. Analysing parallel strategies to alter the host specificity of bacteriophage T7[J]. Biology, 2021, 10(6): 556. |
| 101 | HYMAN P, ABEDON S T. Bacteriophage host range and bacterial resistance[J]. Advances in Applied Microbiology, 2010, 70: 217-248. |
| 102 | GORDILLO ALTAMIRANO F L, BARR J J. Unlocking the next generation of phage therapy: the key is in the receptors[J]. Current Opinion in Biotechnology, 2021, 68: 115-123. |
| 103 | YOICHI M, ABE M, MIYANAGA K, et al. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157: H7[J]. Journal of Biotechnology, 2005, 115(1): 101-107. |
| 104 | YOSEF I, GOREN M G, GLOBUS R, et al. Extending the host range of bacteriophage particles for DNA transduction[J]. Molecular Cell, 2017, 66(5): 721-728.e3. |
| 105 | YEHL K, LEMIRE S, YANG A C, et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis[J]. Cell, 2019, 179(2): 459-469.e9. |
| 106 | HOLTZMAN T, GLOBUS R, MOLSHANSKI-MOR S, et al. A continuous evolution system for contracting the host range of bacteriophage T7[J]. Scientific Reports, 2020, 10: 307. |
| 107 | DION M B, OECHSLIN F, MOINEAU S. Phage diversity, genomics and phylogeny[J]. Nature Reviews Microbiology, 2020, 18(3): 125-138. |
| 108 | CHEVALLEREAU A, PONS B J, VAN HOUTE S, et al. Interactions between bacterial and phage communities in natural environments[J]. Nature Reviews Microbiology, 2022, 20(1): 49-62. |
| 109 | KAVAGUTTI V S, ANDREI A Ş, MEHRSHAD M, et al. Phage-centric ecological interactions in aquatic ecosystems revealed through ultra-deep metagenomics[J]. Microbiome, 2019, 7(1): 135. |
| 110 | AL-SHAYEB B, SACHDEVA R, CHEN L X, et al. Clades of huge phages from across Earth's ecosystems[J]. Nature, 2020, 578(7795): 425-431. |
| 111 | NERI U, WOLF Y I, ROUX S, et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages[J]. Cell, 2022, 185(21): 4023-4037.e18. |
| 112 | KAUFFMAN K M, CHANG W K, BROWN J M, et al. Resolving the structure of phage-bacteria interactions in the context of natural diversity[J]. Nature Communications, 2022, 13(1): 372. |
| 113 | YOU L L, SHI J, SHEN L Q, et al. Structural basis for transcription antitermination at bacterial intrinsic terminator[J]. Nature Communications, 2019, 10: 3048. |
| 114 | SHI J, WEN A J, ZHAO M X, et al. Structural basis of σ appropriation[J]. Nucleic Acids Research, 2019, 47(17): 9423-9432. |
| 115 | SHI J, WEN A J, JIN S, et al. Transcription activation by a sliding clamp[J]. Nature Communications, 2021, 12: 1131. |
| 116 | LEAVITT A, YIRMIYA E, AMITAI G, et al. Viruses inhibit TIR gcADPR signalling to overcome bacterial defence[J]. Nature, 2022, 611(7935): 326-331. |
| 117 | MIRZAEI M K, MAURICE C F. Ménage à trois in the human gut: interactions between host, bacteria and phages[J]. Nature Reviews Microbiology, 2017, 15(7): 397-408. |
| 118 | CAMARILLO-GUERRERO L F, ALMEIDA A, RANGEL-PINEROS G, et al. Massive expansion of human gut bacteriophage diversity[J]. Cell, 2021, 184(4): 1098-1109.e9. |
| 119 | BENLER S, YUTIN N, ANTIPOV D, et al. Thousands of previously unknown phages discovered in whole-community human gut metagenomes[J]. Microbiome, 2021, 9(1): 78. |
| 120 | HSU B B, GIBSON T E, YELISEYEV V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model[J]. Cell Host & Microbe, 2019, 25(6): 803-814.e5. |
| 121 | BASSO-BLANDIN A, DELAPLACE F. Towards a behavioral-matching based compilation of synthetic biology functions[J]. Acta Biotheoretica, 2015, 63(3): 325-339. |
| 122 | MENOR-FLORES M, VEGA-RODRÍGUEZ M A, MOLINA F. Computational design of phage cocktails based on phage-bacteria infection networks[J]. Computers in Biology and Medicine, 2022, 142: 105186. |
| 123 | MOLINA F, SIMANCAS A, RAMÍREZ M, et al. A new pipeline for designing phage cocktails based on phage-bacteria infection networks[J]. Frontiers in Microbiology, 2021, 12: 564532. |
| 124 | DÍAZ-GALIÁN M V, VEGA-RODRÍGUEZ M A, MOLINA F. PhageCocktail: An R package to design phage cocktails from experimental phage-bacteria infection networks[J]. Computer Methods and Programs in Biomedicine, 2022, 221: 106865. |
| 125 | MADSEN C, MCLAUGHLIN J A, MıSıRLı G, et al. The SBOL stack: a platform for storing, publishing, and sharing synthetic biology designs[J]. ACS Synthetic Biology, 2016, 5(6): 487-497. |
| 126 | NIELSEN A A K, DER B S, SHIN J, et al. Genetic circuit design automation[J]. Science, 2016, 352(6281): aac7341. |
| 127 | JONES T S, OLIVEIRA S M D, MYERS C J, et al. Genetic circuit design automation with Cello 2.0[J]. Nature Protocols, 2022, 17(4): 1097-1113. |
| 128 | 袁盛建, 马迎飞. 噬菌体合成生物学研究进展和应用[J]. 合成生物学, 2020, 1(6): 635-655. |
| YUAN S J, MA Y F. Advances and applications of phage synthetic biology[J]. Synthetic Biology Journal, 2020, 1(6): 635-655. | |
| 129 | Global burden of bacterial antimicrobial resistance in 2019 : a systematic analysis[J]. The Lancet, 2022, 399(10325): 629-655. |
| 130 | CREDENCE R C O. Bacteriosphage Market By Product (Phage Probiotics, Phage Therapeutics) By Route of Administration (Oral, Topical, Others) By Application (Gastroenterology, Respiratory Infection Treatment, Skin Infection Treatment, Wound Prophylaxis, Urogenital Infection Treatment, Others) By Distribution Channel (Retail Pharmacies, Hospital Pharmacies, Online Pharmacies)-Growth, Future Prospects & Competitive Analysis, 2016-2028[R]. India: 2022. |
| 131 | PIRNAY J P. Phage therapy in the year 2035[J]. Frontiers in Microbiology, 2020, 11: 1171. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||