Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (3): 464-487.DOI: 10.12211/2096-8280.2023-008
• Invited Review • Previous Articles Next Articles
Research progress of artificial intelligence in desiging protein structures
CHEN Zhihang, JI Menglin, QI Yifei
- School of Pharmacy,Fudan University,Shanghai 201203,China
-
Received:2023-01-13Revised:2023-03-15Online:2023-07-05Published:2023-06-30 -
Contact:QI Yifei
人工智能蛋白质结构设计算法研究进展
陈志航, 季梦麟, 戚逸飞
- 复旦大学药学院,上海 201203
-
通讯作者:戚逸飞 -
作者简介:陈志航 (1998—),男,硕士研究生。研究方向为人工智能蛋白质设计。 E-mail:zhihangchen21@m.fudan.edu.cn季梦麟 (2000—),男,硕士研究生。研究方向为人工智能蛋白质设计。 E-mail:22211030067@m.fudan.edu.cn戚逸飞 (1983—),男,副研究员,硕士生导师。研究方向为生物大分子结构和功能模拟以及人工智能药物设计。 E-mail:yfqi@fudan.edu.cn -
基金资助:国家自然科学基金(22033001)
CLC Number:
Cite this article
CHEN Zhihang, JI Menglin, QI Yifei. Research progress of artificial intelligence in desiging protein structures[J]. Synthetic Biology Journal, 2023, 4(3): 464-487.
陈志航, 季梦麟, 戚逸飞. 人工智能蛋白质结构设计算法研究进展[J]. 合成生物学, 2023, 4(3): 464-487.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-008
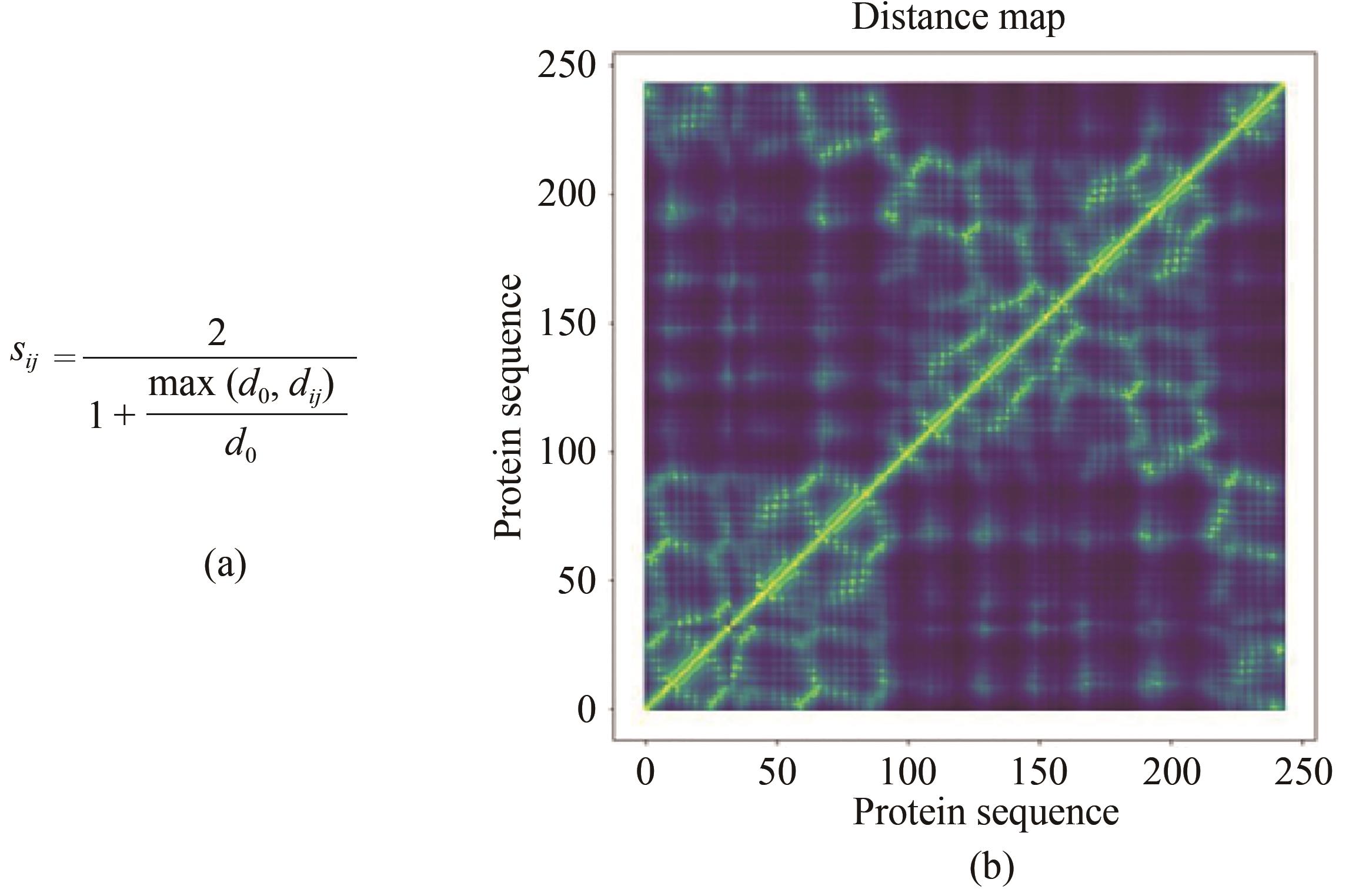
Fig. 1 Calculating the distance of residues in SPROF(a) dij is the distance between the Cα atoms of residues i and j, d0=0.4 nm, and (b) matrix for residue-residue distance of a protein structure.
模型 Models | 恢复率/%(↑) Recovery/% (↑) | 困惑度(↓) Perplexity (↓) |
|---|---|---|
| GraphTrans | 35.82 | 6.63 |
| StructGNN[ | 37.1 | 6.49 |
| GVP-GNN-large | 39.20 | 6.17 |
| GVP-GNN-Transformer | 38.30 | 6.44 |
| GVP-GNN-Transformer+AF2 | 51.60 | 4.01 |
| ProteinMPNN | 45.96 | 4.61 |
| ProDesign | 50.22 | 4.69 |
| PiFold[ | 50.22 | 4.62 |
| LM-DESIGN[ | 55.65 | 4.52 |
Table 1 Sequence recovery rate and perplexity of the fixed-backbone sequence design model on CATH 4.2 test set
模型 Models | 恢复率/%(↑) Recovery/% (↑) | 困惑度(↓) Perplexity (↓) |
|---|---|---|
| GraphTrans | 35.82 | 6.63 |
| StructGNN[ | 37.1 | 6.49 |
| GVP-GNN-large | 39.20 | 6.17 |
| GVP-GNN-Transformer | 38.30 | 6.44 |
| GVP-GNN-Transformer+AF2 | 51.60 | 4.01 |
| ProteinMPNN | 45.96 | 4.61 |
| ProDesign | 50.22 | 4.69 |
| PiFold[ | 50.22 | 4.62 |
| LM-DESIGN[ | 55.65 | 4.52 |
模型类别 Group | 模型 Models | TS50 | TS500 | ||
|---|---|---|---|---|---|
恢复率/%(↑) Recovery/%(↑) | 困惑度(↓) Perplexity(↓) | 恢复率/%(↑) Recovery/%(↑) | 困惑度(↓) Perplexity (↓) | ||
| MLP | SPIN | 30.00 | — | — | — |
| SPIN2 | 34.00 | — | — | — | |
| Wang’s model | 33.00 | — | — | — | |
| CNN | SPROF | 39.80 | — | — | — |
| ProDCoNN | 46.50 | — | — | — | |
| DenseCPD | 50.71 | — | 55.53 | — | |
| GNN | StructGNN | 43.89 | 5.40 | 45.69 | 4.98 |
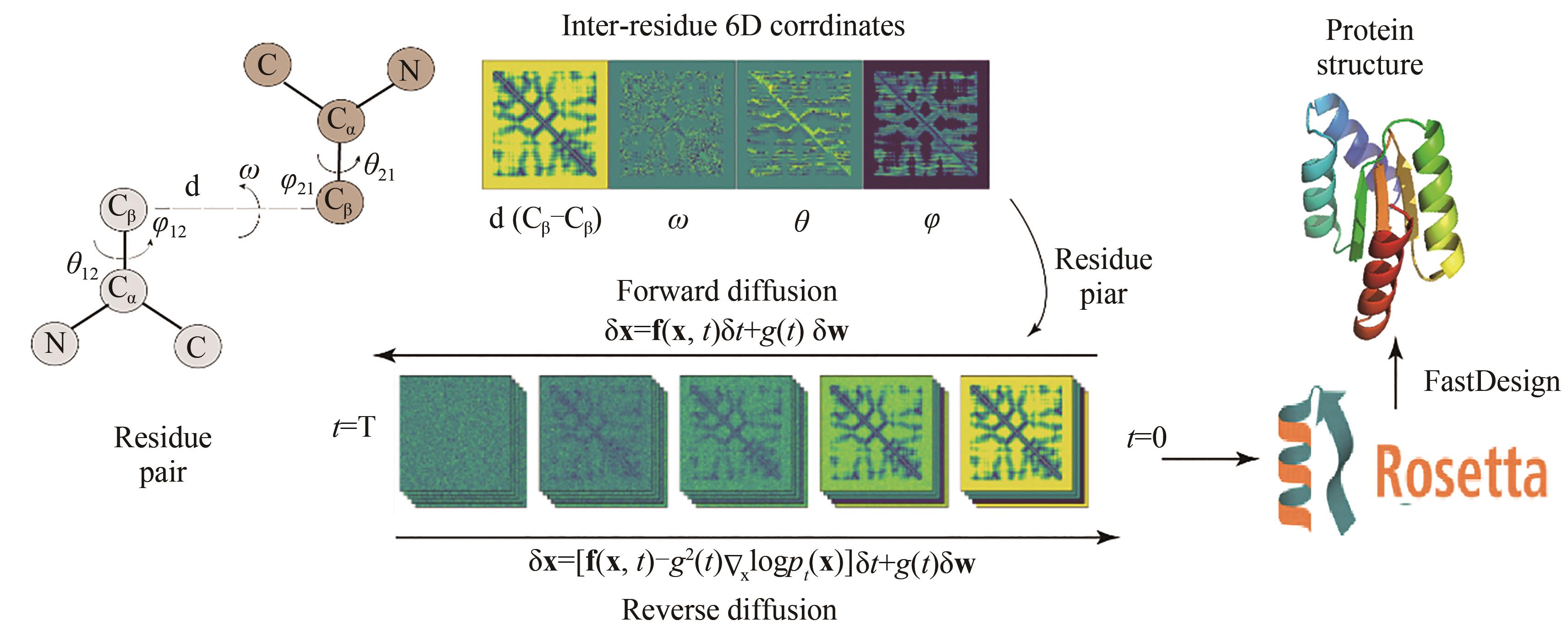
| GraphTrans | 42.20 | 5.60 | 44.66 | 5.16 | |
| GVP-GNN | 44.14 | 4.71 | 49.14 | 4.20 | |
| GCA[ | 47.02 | 5.09 | 47.74 | 4.72 | |
| ADesign[ | 48.36 | 5.25 | 49.23 | 4.93 | |
| ProteinMPNN | 54.43 | 3.93 | 58.08 | 3.53 | |
| PiFold | 58.72 | 3.86 | 60.42 | 3.44 | |
| LM-DESIGN(PiFold) | 57.89 | 3.50 | 67.78 | 3.19 | |
Table 2 Sequence recovery rate and perplexity of the fixed-backbone sequence design model on TS50 &TS500 test sets
模型类别 Group | 模型 Models | TS50 | TS500 | ||
|---|---|---|---|---|---|
恢复率/%(↑) Recovery/%(↑) | 困惑度(↓) Perplexity(↓) | 恢复率/%(↑) Recovery/%(↑) | 困惑度(↓) Perplexity (↓) | ||
| MLP | SPIN | 30.00 | — | — | — |
| SPIN2 | 34.00 | — | — | — | |
| Wang’s model | 33.00 | — | — | — | |
| CNN | SPROF | 39.80 | — | — | — |
| ProDCoNN | 46.50 | — | — | — | |
| DenseCPD | 50.71 | — | 55.53 | — | |
| GNN | StructGNN | 43.89 | 5.40 | 45.69 | 4.98 |
| GraphTrans | 42.20 | 5.60 | 44.66 | 5.16 | |
| GVP-GNN | 44.14 | 4.71 | 49.14 | 4.20 | |
| GCA[ | 47.02 | 5.09 | 47.74 | 4.72 | |
| ADesign[ | 48.36 | 5.25 | 49.23 | 4.93 | |
| ProteinMPNN | 54.43 | 3.93 | 58.08 | 3.53 | |
| PiFold | 58.72 | 3.86 | 60.42 | 3.44 | |
| LM-DESIGN(PiFold) | 57.89 | 3.50 | 67.78 | 3.19 | |
| 1 | HUANG P S, BOYKEN S E, BAKER D. The coming of age of de novo protein design[J]. Nature, 2016, 537(7620): 320-327. |
| 2 | KHERSONSKY O, LIPSH R, AVIZEMER Z, et al. Automated design of efficient and functionally diverse enzyme repertoires[J]. Molecular Cell, 2018, 72(1): 178-186.e5. |
| 3 | GLASGOW A A, HUANG Y M, MANDELL D J, et al. Computational design of a modular protein sense-response system[J]. Science, 2019, 366(6468): 1024-1028. |
| 4 | ANFINSEN C B. Principles that govern the folding of protein chains[J]. Science, 1973, 181(4096): 223-230. |
| 5 | LEAVER-FAY A, O'MEARA M J, TYKA M, et al. Scientific benchmarks for guiding macromolecular energy function improvement[J]. Methods in Enzymology, 2013, 523: 109-143. |
| 6 | LEMAN J K, WEITZNER B D, LEWIS S M, et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks[J]. Nature Methods, 2020, 17(7): 665-680. |
| 7 | NADRA A D, SERRANO L, ALIBÉS A. Chapter one-DNA-binding specificity prediction with FoldX[M]//Methods in enzymology. New York: Academic Press. 2011, 498: 3-18. |
| 8 | HUANG X Q, PEARCE R, ZHANG Y. EvoEF2: accurate and fast energy function for computational protein design[J]. Bioinformatics, 2020, 36(4): 1135-1142. |
| 9 | ALFORD R F, LEAVER-FAY A, JELIAZKOV J R, et al. The Rosetta all-atom energy function for macromolecular modeling and design[J]. Journal of Chemical Theory and Computation, 2017, 13(6): 3031-3048. |
| 10 | KUHLMAN B, DANTAS G, IRETON G C, et al. Design of a novel globular protein fold with atomic-level accuracy[J]. Science, 2003, 302(5649): 1364-1368. |
| 11 | SIEGEL J B, ZANGHELLINI A, LOVICK H M, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction[J]. Science, 2010, 329(5989): 309-313. |
| 12 | SILVA D A, YU S, ULGE U Y, et al. De novo design of potent and selective mimics of IL-2 and IL-15[J]. Nature, 2019, 565(7738): 186-191. |
| 13 | MOHAN K, UEDA G, KIM A R, et al. Topological control of cytokine receptor signaling induces differential effects in hematopoiesis[J]. Science, 2019, 364(6442): eaav7532. |
| 14 | CHEVALIER A, SILVA D A, ROCKLIN G J, et al. Massively parallel de novo protein design for targeted therapeutics[J]. Nature, 2017, 550(7674): 74-79. |
| 15 | CAO L X, GORESHNIK I, COVENTRY B, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors[J]. Science, 2020, 370(6515): 426-431. |
| 16 | LANGAN R A, BOYKEN S E, NG A H, et al. De novo design of bioactive protein switches[J]. Nature, 2019, 572(7768): 205-210. |
| 17 | DAWSON W M, LANG E J M, RHYS G G, et al. Structural resolution of switchable states of a de novo peptide assembly[J]. Nature Communications, 2021, 12: 1530. |
| 18 | SHEN H, FALLAS J A, LYNCH E, et al. De novo design of self-assembling helical protein filaments[J]. Science, 2018, 362(6415): 705-709. |
| 19 | HSIA Y, BALE J B, GONEN S, et al. Design of a hyperstable 60-subunit protein icosahedron[J]. Nature, 2016, 535(7610): 136-139. |
| 20 | ROCKLIN G J, CHIDYAUSIKU T M, GORESHNIK I, et al. Global analysis of protein folding using massively parallel design, synthesis, and testing[J]. Science, 2017, 357(6347): 168-175. |
| 21 | BERMAN H M, WESTBROOK J, FENG Z K, et al. The protein data bank[J]. Nucleic Acids Research, 2000, 28(1): 235-242. |
| 22 | FOX N K, BRENNER S E, CHANDONIA J M. SCOPe: structural classification of proteins—extended, integrating SCOP and ASTRAL data and classification of new structures[J]. Nucleic Acids Research, 2014, 42(D1): D304-D309. |
| 23 | CONSORTIUM T U, BATEMAN A, MARTIN M J, et al. UniProt: the universal protein knowledgebase[J]. Nucleic Acids Research, 2017, 45(D1): D158-D169. |
| 24 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D412-D419. |
| 25 | FRAPPIER V, KEATING A E. Data-driven computational protein design[J]. Current Opinion in Structural Biology, 2021, 69: 63-69. |
| 26 | KWON Y, SHIN W H, KO J, et al. AK-score: accurate protein-ligand binding affinity prediction using an ensemble of 3D-convolutional neural networks[J]. International Journal of Molecular Sciences, 2020, 21(22): 8424. |
| 27 | JIANG D J, HSIEH C Y, WU Z X, et al. InteractionGraphNet: a novel and efficient deep graph representation learning framework for accurate protein-ligand interaction predictions[J]. Journal of Medicinal Chemistry, 2021, 64(24): 18209-18232. |
| 28 | JONES D, KIM H, ZHANG X H, et al. Improved protein-ligand binding affinity prediction with structure-based deep fusion inference[J]. Journal of Chemical Information and Modeling, 2021, 61(4): 1583-1592. |
| 29 | JIMÉNEZ J, ŠKALIČ M, MARTÍNEZ-ROSELL G, et al. KDEEP: protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks[J]. Journal of Chemical Information and Modeling, 2018, 58(2): 287-296. |
| 30 | SLEDZIESKI S, SINGH R, COWEN L, et al. D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions[J]. Cell Systems, 2021, 12(10): 969-982.e6. |
| 31 | BARANWAL M, MAGNER A, SALDINGER J, et al. Struct2Graph: a graph attention network for structure based predictions of protein-protein interactions[J]. BMC Bioinformatics, 2022, 23(1): 370. |
| 32 | WANG S, CHEN W Q, HAN P F, et al. RGN: residue-based graph attention and convolutional network for protein-protein interaction site prediction[J]. Journal of Chemical Information and Modeling, 2022, 62(23): 5961-5974. |
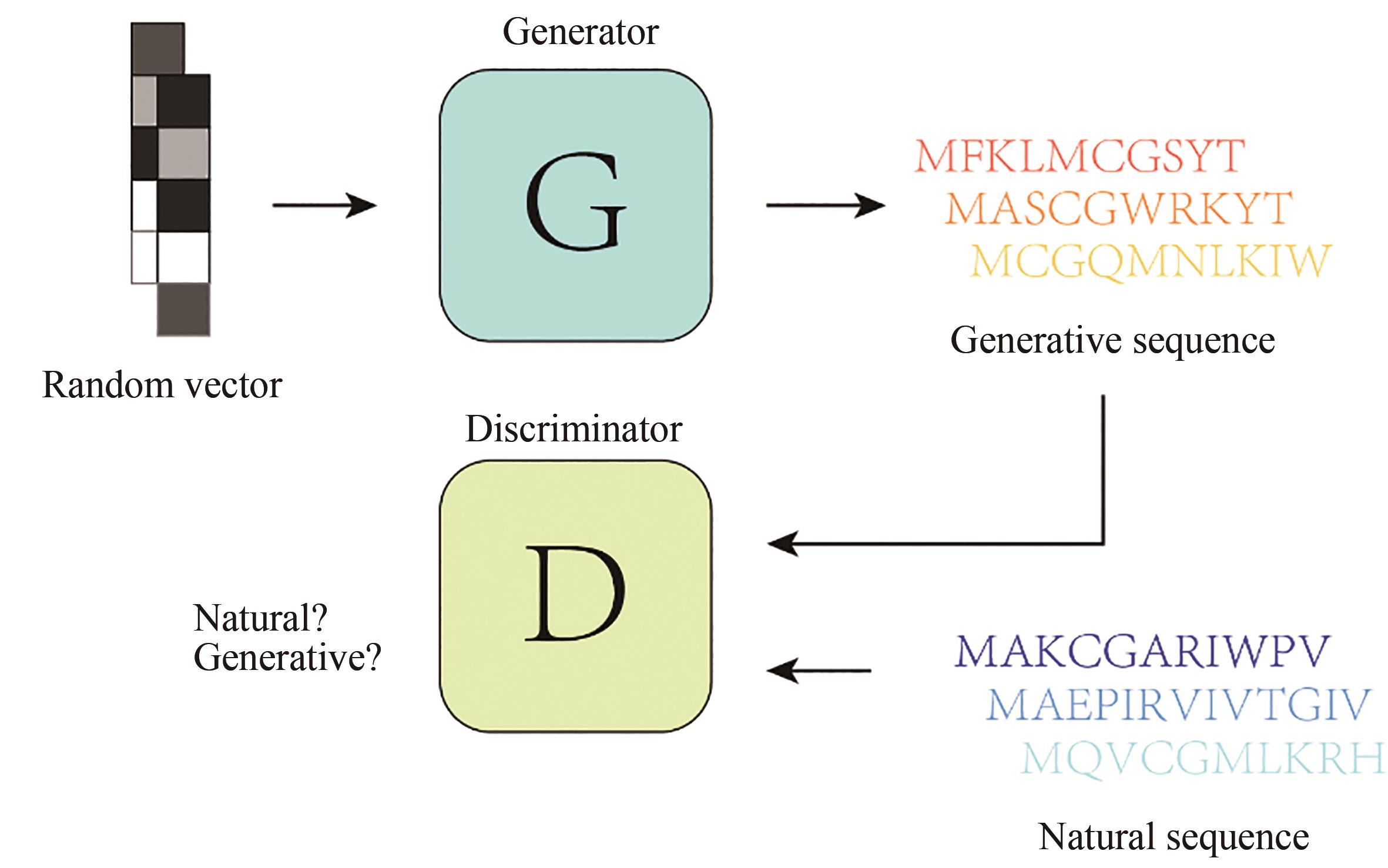
| 33 | SHEN W X, ZENG X, ZHU F, et al. Out-of-the-box deep learning prediction of pharmaceutical properties by broadly learned knowledge-based molecular representations[J]. Nature Machine Intelligence, 2021, 3(4): 334-343. |
| 34 | BUTTON A, MERK D, HISS J A, et al. Automated de novo molecular design by hybrid machine intelligence and rule-driven chemical synthesis[J]. Nature Machine Intelligence, 2019, 1(7): 307-315. |
| 35 | DE CAO N, KIPF T. MolGAN: an implicit generative model for small molecular graphs[EB/OL]. arXiv, 2018: 1805.11973[2023-10-01]. |
| 36 | WINTER R, MONTANARI F, STEFFEN A, et al. Efficient multi-objective molecular optimization in a continuous latent space[J]. Chemical Science, 2019, 10(34): 8016-8024. |
| 37 | DING W Z, NAKAI K T, GONG H P. Protein design via deep learning[J]. Briefings in Bioinformatics, 2022, 23(3): bbac102. |
| 38 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 39 | BAEK M, DIMAIO F, ANISHCHENKO I, et al. Accurate prediction of protein structures and interactions using a three-track neural network[J]. Science, 2021, 373(6557): 871-876. |
| 40 | DAHIYAT B I, MAYO S L. Protein design automation[J]. Protein Science, 1996, 5(5): 895-903. |
| 41 | DAHIYAT B I, MAYO S L. De novo protein design: fully automated sequence selection[J]. Science, 1997, 278(5335): 82-87. |
| 42 | LI Z X, YANG Y D, FARAGGI E, et al. Direct prediction of profiles of sequences compatible with a protein structure by neural networks with fragment-based local and energy-based nonlocal profiles[J]. Proteins: Structure, Function, and Bioinformatics, 2014, 82(10): 2565-2573. |
| 43 | DAI L, YANG Y D, KIM H R, et al. Improving computational protein design by using structure-derived sequence profile[J]. Proteins: Structure, Function, and Bioinformatics, 2010, 78(10): 2338-2348. |
| 44 | YANG Y D, ZHOU Y Q. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions[J]. Protein Science, 2008, 17(7): 1212-1219. |
| 45 | WANG J X, CAO H L, ZHANG J Z H, et al. Computational protein design with deep learning neural networks[J]. Scientific Reports, 2018, 8: 6349. |
| 46 | O'CONNELL J, LI Z X, HANSON J, et al. SPIN2: predicting sequence profiles from protein structures using deep neural networks[J]. Proteins: Structure, Function, and Bioinformatics, 2018, 86(6): 629-633. |
| 47 | CHEN S, SUN Z, LIN L H, et al. To improve protein sequence profile prediction through image captioning on pairwise residue distance map[J]. Journal of Chemical Information and Modeling, 2020, 60(1): 391-399. |
| 48 | KRIZHEVSKY A, SUTSKEVER I, HINTON G E. ImageNet classification with deep convolutional neural networks[J]. Communications of the ACM, 2017, 60(6): 84-90. |
| 49 | ZHANG Y, CHEN Y, WANG C R, et al. ProDCoNN: protein design using a convolutional neural network[J]. Proteins: Structure, Function, and Bioinformatics, 2020, 88(7): 819-829. |
| 50 | ANAND N, EGUCHI R, MATHEWS I I, et al. Protein sequence design with a learned potential[J]. Nature Communications, 2022, 13: 746. |
| 51 | QI Y F, ZHANG J Z H. DenseCPD: improving the accuracy of neural-network-based computational protein sequence design with DenseNet[J]. Journal of Chemical Information and Modeling, 2020, 60(3): 1245-1252. |
| 52 | HUANG G, LIU Z, VAN DER MAATEN L, et al. Densely connected convolutional networks[C]//2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). July 21-26, 2017, Honolulu, HI, USA. IEEE, 2017: 2261-2269. |
| 53 | SHROFF R, COLE A W, DIAZ D J, et al. Discovery of novel gain-of-function mutations guided by structure-based deep learning[J]. ACS Synthetic Biology, 2020, 9(11): 2927-2935. |
| 54 | LU H Y, DIAZ D J, CZARNECKI N J, et al. Machine learning-aided engineering of hydrolases for PET depolymerization[J]. Nature, 2022, 604(7907): 662-667. |
| 55 | NORN C, WICKY B I M, JUERGENS D, et al. Protein sequence design by conformational landscape optimization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(11): e2017228118. |
| 56 | YANG J Y, ANISHCHENKO I, PARK H, et al. Improved protein structure prediction using predicted interresidue orientations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(3): 1496-1503. |
| 57 | WANG X, FLANNERY S T, KIHARA D. Protein docking model evaluation by graph neural networks[J]. Frontiers in Molecular Biosciences, 2021, 8: 647915. |
| 58 | KIPF T N, WELLING M. Semi-supervised classification with graph convolutional networks[EB/OL]. arXiv, 2016: 1609.02907[2023-01-10]. |
| 59 | INGRAHAM J, GARG V K, BARZILAY R, et al. Generative models for graph-based protein design[C/OL]// Advances in Neural Information Processing Systems 32 (NeurIPS 2019), December 2019, Vancouver, Canada, Neural Information Processing Systems Foundation, 2019[2023-01-10]. . |
| 60 | VASWANI A, SHAZEER N, PARMAR N, et al. Attention is all you need[C]//Proceedings of the 31st International Conference on Neural Information Processing Systems. December 4-9, 2017, Long Beach, California, USA. New York: ACM, 2017: 6000-6010. |
| 61 | STROKACH A, BECERRA D, CORBI-VERGE C, et al. Fast and flexible protein design using deep graph neural networks[J]. Cell Systems, 2020, 11(4): 402-411.e4. |
| 62 | JING B, EISMANN S, SURIANA P, et al. Learning from protein structure with geometric vector perceptrons[EB/OL]. arXiv, 2020: 2009.01411[2023-01-10]. . |
| 63 | ORELLANA G A, CACERES-DELPIANO J, IBAÑEZ R, et al. Protein sequence sampling and prediction from structural data[EB/OL]. bioRxiv, 2021[2023-01-10] . |
| 64 | LI A J, LU M, DESTA I, et al. Neural network-derived Potts models for structure-based protein design using backbone atomic coordinates and tertiary motifs[J]. Protein Science, 2023, 32(2): e4554. |
| 65 | ZHENG F, ZHANG J, GRIGORYAN G. Tertiary structural propensities reveal fundamental sequence/structure relationships[J]. Structure, 2015, 23(5): 961-971. |
| 66 | HSU C, VERKUIL R, LIU J, et al. Learning inverse folding from millions of predicted structures[C/OL]//Proceedings of the 39th International Conference on Machine Learning. Proceedings of Machine Learning Research, PMLR. 2022: 8946-8970 [2023-01-10]. . |
| 67 | MCPARTLON M, LAI B, XU J B. A deep SE(3)-equivariant model for learning inverse protein folding[EB/OL]. bioXiv, 202[2023-01-10]. . |
| 68 | ZADEH A, CHEN M, PORIA S, et al. Tensor fusion network for multimodal sentiment analysis[EB/OL]. arXiv, 2017: 1707.07250[2023-01-10]. . |
| 69 | XIONG P, HU X H, HUANG B, et al. Increasing the efficiency and accuracy of the ABACUS protein sequence design method[J]. Bioinformatics, 2020, 36(1): 136-144. |
| 70 | LIU Y F, ZHANG L, WANG W L, et al. Rotamer-free protein sequence design based on deep learning and self-consistency[J]. Nature Computational Science, 2022, 2(7): 451-462. |
| 71 | XIONG P, WANG M, ZHOU X Q, et al. Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability[J]. Nature Communications, 2014, 5: 5330. |
| 72 | RONEY J P, OVCHINNIKOV S. State-of-the-art estimation of protein model accuracy using AlphaFold[J]. Physical Review Letters, 2022, 129(23): 238101. |
| 73 | DAUPARAS J, ANISHCHENKO I, BENNETT N, et al. Robust deep learning-based protein sequence design using ProteinMPNN[J]. Science, 2022, 378(6615): 49-56. |
| 74 | HUANG B, FAN T W, WANG K Y, et al. Accurate and efficient protein sequence design through learning concise local environment of residues[J]. Bioinformatics, 2023: btad122. |
| 75 | ZHENG Z, DENG Y, XUE D, et al. Structure-informed language models are protein designers[EB/OL]. arXiv, 2023: 2302.01649[2023-02-10]. . |
| 76 | INGRAHAM J, GARG V K, BARZILAY R, et al. Generative models for graph-based protein design[C]// Proceedings of the 33rd International Conference on Neural Information Processing Systems, 8-14 December 2019, Vancouver, Canada, Curran Associates Inc, 2019:1417[2023-01-10]. . |
| 77 | GAO Z Y, TAN C, LI S Z. ProDesign: toward effective and efficient protein design[EB/OL]. arXiv, 2022[2023-01-10]. . |
| 78 | TAN C, GAO Z Y, XIA J, et al. Generative de novo protein design with global context[EB/OL]. arXiv, 2022[2023-01-10]. . |
| 79 | GAO Z Y, TAN C, LI S Z. AlphaDesign: a graph protein design method and benchmark on AlphaFoldDB[EB/OL]. arXiv, 2022[2023-01-10]. . |
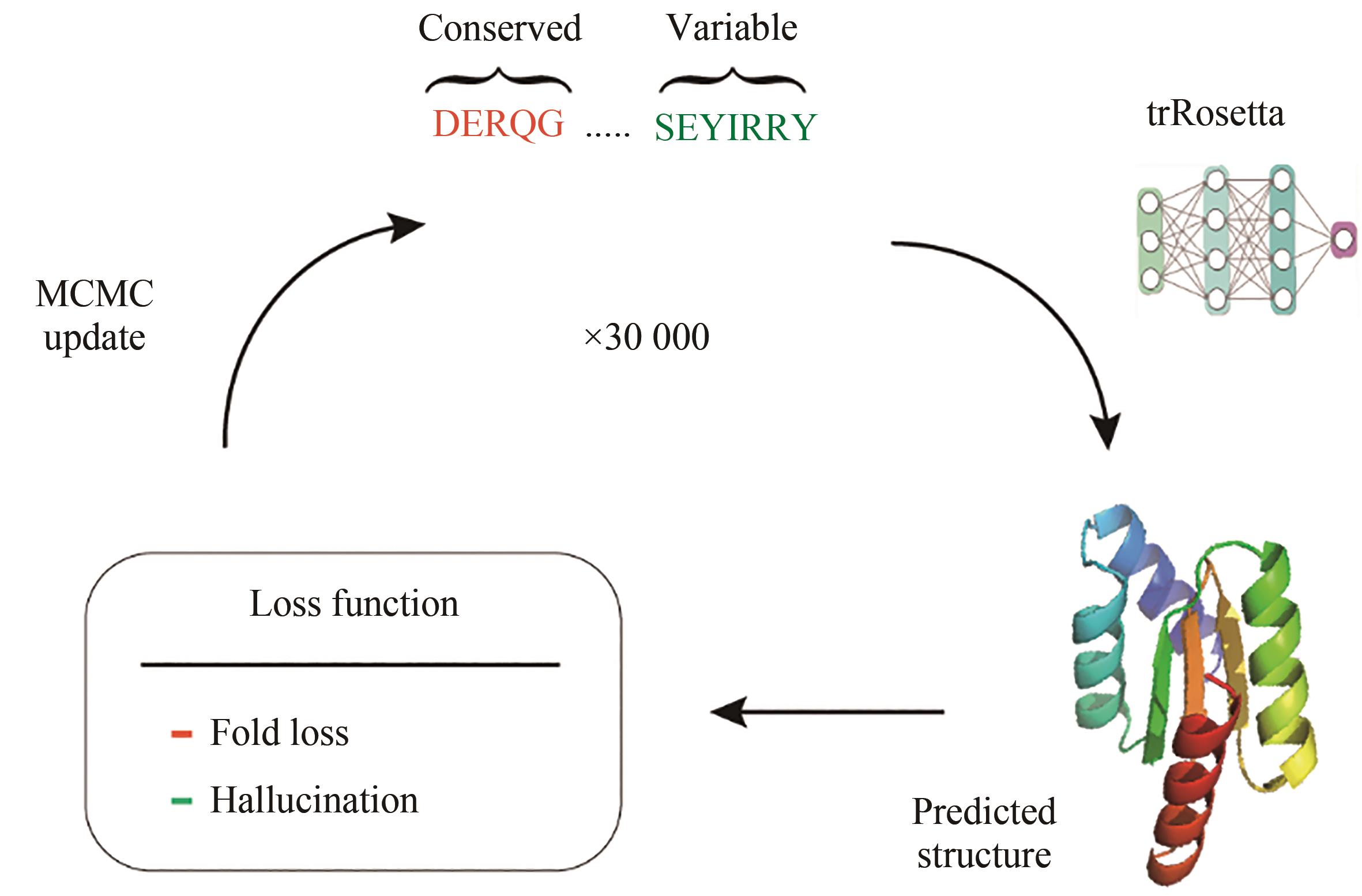
| 80 | ANISHCHENKO I, PELLOCK S J, CHIDYAUSIKU T M, et al. De novo protein design by deep network hallucination[J]. Nature, 2021, 600(7889): 547-552. |
| 81 | TISCHER D, LISANZA S, WANG J, et al. Design of proteins presenting discontinuous functional sites using deep learning[EB/OL]. bioXiv, 2020[2023-01-10]. . |
| 82 | WANG J, LISANZA S, JUERGENS D, et al. Scaffolding protein functional sites using deep learning[J]. Science, 2022, 377(6604): 387-394. |
| 83 | ZHANG S H, XU Y J, PEI J F, et al. AutoFoldFinder: an automated adaptive optimization toolkit for de novo protein fold design[EB/OL]. 2021[2023-01-10]. . |
| 84 | YEH A H W, NORN C, KIPNIS Y, et al. De novo design of luciferases using deep learning[J]. Nature, 2023, 614(7949): 774-780. |
| 85 | DOU J Y, VOROBIEVA A A, SHEFFLER W, et al. De novo design of a fluorescence-activating β-barrel[J]. Nature, 2018, 561(7724): 485-491. |
| 86 | CAO L X, COVENTRY B, GORESHNIK I, et al. Design of protein-binding proteins from the target structure alone[J]. Nature, 2022, 605(7910): 551-560. |
| 87 | HUANG B, XU Y, HU X H, et al. A backbone-centred energy function of neural networks for protein design[J]. Nature, 2022, 602(7897): 523-528. |
| 88 | LIANG S D, LI Z X, ZHAN J, et al. De novo protein design by an energy function based on series expansion in distance and orientation dependence[J]. Bioinformatics, 2021, 38(1): 86-93. |
| 89 | LIANG S D, ZHENG D D, ZHANG C, et al. Fast and accurate prediction of protein side-chain conformations[J]. Bioinformatics, 2011, 27(20): 2913-2914. |
| 90 | LIANG S D, ZHOU Y Q, GRISHIN N, et al. Protein side chain modeling with orientation-dependent atomic force fields derived by series expansions[J]. Journal of Computational Chemistry, 2011, 32(8): 1680-1686. |
| 91 | LIANG S D, ZHANG C, ZHOU Y Q. LEAP: highly accurate prediction of protein loop conformations by integrating coarse-grained sampling and optimized energy scores with all-atom refinement of backbone and side chains[J]. Journal of Computational Chemistry, 2014, 35(4): 335-341. |
| 92 | ANAND N, HUANG P S. Generative modeling for protein structures[C/OL]// 6th International Conference on Learning Representations, Vancouver, BC, Canada, April 30-May 3, 2018[2023-01-10]. . |
| 93 | ANAND N, EGUCHI R, HUANG P S. Fully differentiable full-atom protein backbone generation[EB/OL]. ICLR 2019 Workshop on Deep Generative Models for Highly Structured Data, 2019[2023-01-10]. . |
| 94 | EGUCHI R R, CHOE C A, HUANG P S. Ig-VAE: generative modeling of protein structure by direct 3D coordinate generation[J]. PLoS Computational Biology, 2022, 18(6): e1010271. |
| 95 | LAI B Q, MCPARTLON M, XU J B. End-to-End deep structure generative model for protein design[EB/OL]. bioRxiv, 2022[2023-01-10]. . |
| 96 | GUO X J, DU Y Q, TADEPALLI S, et al. Generating tertiary protein structures via interpretable graph variational autoencoders[J]. Bioinformatics Advances, 2021, 1(1): vbab036. |
| 97 | HARTEVELD Z, SOUTHERN J, LOUKAS A, et al. Deep sharpening of topological features for de novo protein design[EB/OL]. ICLR 2022 Machine Learning for Drug Discovery, 2022 [2023-01-10]. . |
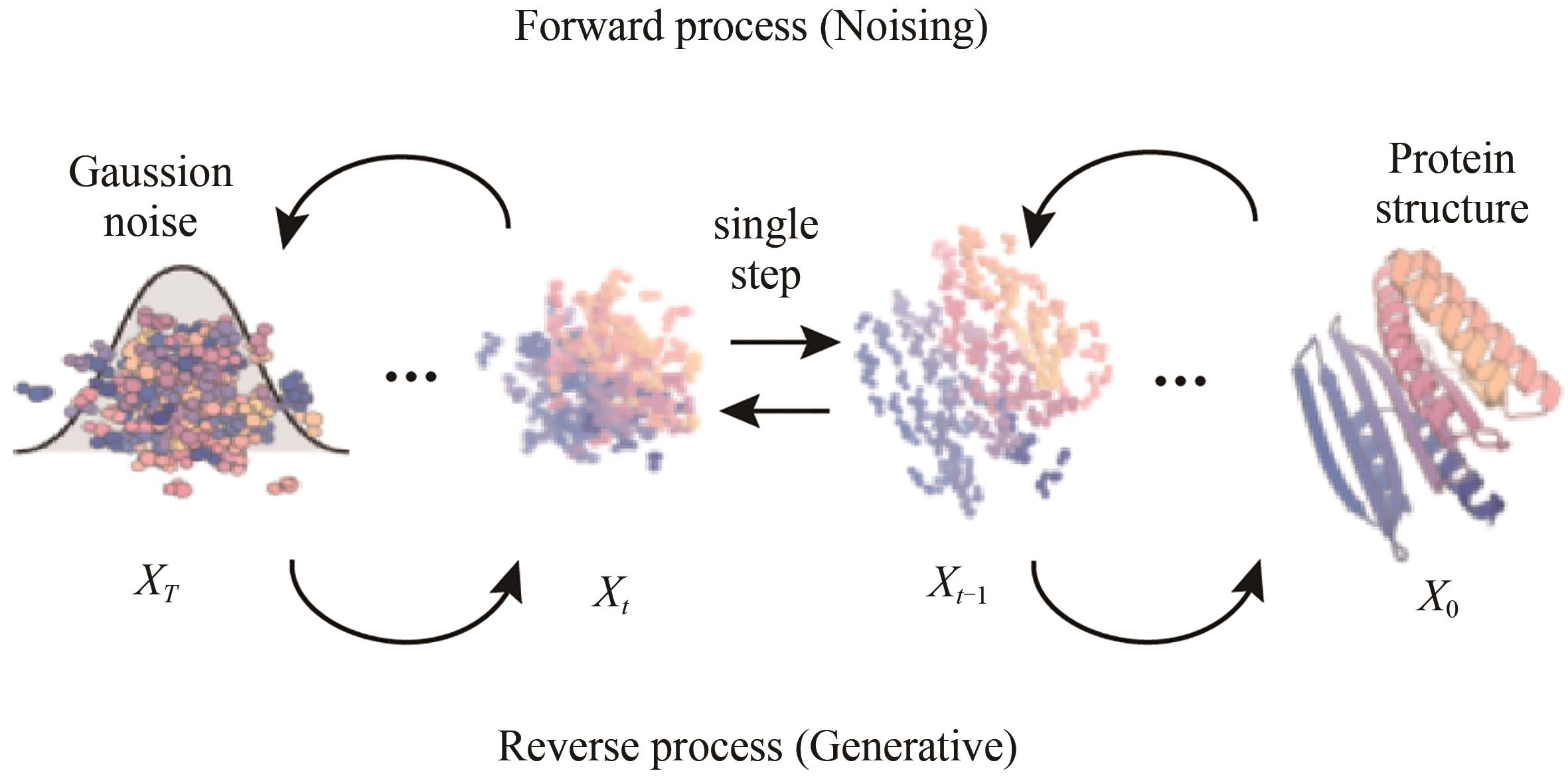
| 98 | HO J, JAIN A, ABBEEL P. Denoising diffusion probabilistic models[EB/OL]. arXiv, 2020: 2006.11239. . |
| 99 | SOHL-DICKSTEIN J, WEISS E A, MAHESWARANATHAN N, et al. Deep unsupervised learning using nonequilibrium thermodynamics[C]//Proceedings of the 32nd International Conference on International Conference on Machine Learning-Volume 37. July 6-11, 2015, Lille, France. New York: ACM, 2015: 2256-2265. |
| 100 | WATSON J L, JUERGENS D, BENNETT N R, et al. Broadly applicable and accurate protein design by integrating structure prediction networks and diffusion generative models[EB/OL]. bioXiv, 2022[2023-01-10]. . |
| 101 | TRIPPE B L, YIM J, TISCHER D, et al. Diffusion probabilistic modeling of protein backbones in 3D for the motif-scaffolding problem[EB/OL]. arXiv, 2022: 2206.04119[2023-01-10]. . |
| 102 | WU K E, YANG K K, BERG R V D, et al. Protein structure generation via folding diffusion[EB/OL]. arXiv, 2022: 2209.15611[2023-01-10]. . |
| 103 | LEE J S, KIM P. ProteinSGM: score-based generative modeling for de novo protein design[EB/OL]. 2022[2023-01-10]. . |
| 104 | LEAVER-FAY A, TYKA M, LEWIS S M, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules[J]. Methods in Enzymology, 2011, 487: 545-574. |
| 105 | INGRAHAM J, BARANOV M, COSTELLO Z, et al. Illuminating protein space with a programmable generative model[EB/OL]. bioXiv, 2022[2023-01-10]. . |
| 106 | ANAND N, ACHIM T. Protein structure and sequence generation with equivariant denoising diffusion probabilistic models[EB/OL]. arXiv, 2022: 2205.15019[2023-01-10]. . |
| 107 | DEVLIN J, CHANG M, LEE K, et al. BERT: Pre-training of deep bidirectional transformers for language understanding[EB/OL]. arXiv, 2018: 1810.04805[2023-01-10]. . |
| 108 | DE BORTOLI V, MATHIEU E, HUTCHINSON M, et al. Riemannian score-based generative modelling[EB/OL]. arXiv, 2022: 2202.02763[2023-01-10]. . |
| 109 | LEACH A, SCHMON S M, DEGIACOMI M T, et al. Denoising diffusion probabilistic models on SO(3) for rotational alignment[EB/OL]. ICLR 2022 Workshop on Geometrical and Topological Representation Learning, 2022[2023-01-10]. . |
| 110 | LIU Y F, CHEN L H, LIU H Y. De novo protein backbone generation based on diffusion with structured priors and adversarial training[EB/OL]. bioRxiv, 2022[2023-01-10]. . |
| 111 | RIVES A, MEIER J, SERCU T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(15): e2016239118. |
| 112 | LUO S T, SU Y F, PENG X G, et al. Antigen-specific antibody design and optimization with diffusion-based generative models for protein structures[EB/OL]. bioXiv, 2022[2023-01-10]. . |
| 113 | REPECKA D, JAUNISKIS V, KARPUS L, et al. Expanding functional protein sequence spaces using generative adversarial networks[J]. Nature Machine Intelligence, 2021, 3(4): 324-333. |
| 114 | MADANI A, MCCANN B, NAIK N, et al. ProGen: Language modeling for protein generation[EB/OL]. arXiv, 2020: 2004.03497[2023-01-10]. . |
| 115 | ELNAGGAR A, HEINZINGER M, DALLAGO C, et al. ProtTrans: towards cracking the language of lifes code through self-supervised deep learning and high performance computing[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2021, 44(10), 7112-7127. |
| 116 | GLIGORIJEVIĆ V, BERENBERG D, RA S, et al. Function-guided protein design by deep manifold sampling[EB/OL]. bioRxiv, 2021[2023-01-10]. . |
| 117 | MOFFAT L, KANDATHIL S M, JONES D T. Design in the DARK: learning deep generative models for de novo protein design[EB/OL]. bioRxiv, 2022[2023-01-10]. . |
| 118 | FERRUZ N, SCHMIDT S, HÖCKER B. ProtGPT2 is a deep unsurprised language model for protein design[J]. Nature Communications, 2022,13(1): 4348. |
| 119 | HESSLOW D, ZANICHELLI N, NOTIN P, et al. RITA: a study on scaling up generative protein sequence models[EB/OL]. arXiv, 2022: 2205.05789[2023-01-10]. . |
| 120 | NIJKAMP E, RUFFOLO J, WEINSTEIN E N, et al. ProGen2: exploring the boundaries of protein language models[EB/OL]. arXiv, 2022[2023-01-10]. . |
| 121 | LI Z X, YANG Y D, ZHAN J, et al. Energy functions in de novo protein design: current challenges and future prospects[J]. Annual Review of Biophysics, 2013, 42: 315-335. |
| [1] | DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems [J]. Synthetic Biology Journal, 2025, 6(1): 105-117. |
| [2] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [3] | WANG Ziyuan, YANG Lirong, WU Jianping, ZHENG Wenlong. A review on enzyme-catalyzed synthesis of chiral amino acids [J]. Synthetic Biology Journal, 2024, 5(6): 1319-1349. |
| [4] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [5] | LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids [J]. Synthetic Biology Journal, 2024, 5(3): 507-526. |
| [6] | ZHU Jingyong, LI Junxiang, LI Xuhui, ZHANG Jin, WU Wenjing. Advances in applications of deep learning for predicting sequence-based protein interactions [J]. Synthetic Biology Journal, 2024, 5(1): 88-106. |
| [7] | Liqi KANG, Pan TAN, Liang HONG. Enzyme engineering in the age of artificial intelligence [J]. Synthetic Biology Journal, 2023, 4(3): 524-534. |
| [8] | Qiaozhen MENG, Fei GUO. Applications of foldability in intelligent enzyme engineering and design: take AlphaFold2 for example [J]. Synthetic Biology Journal, 2023, 4(3): 571-589. |
| [9] | Qilong LAI, Shuai YAO, Yuguo ZHA, Hong BAI, Kang NING. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials [J]. Synthetic Biology Journal, 2023, 4(3): 611-627. |
| [10] | Yidong SONG, Qianmu YUAN, Yuedong YANG. Application of deep learning in protein function prediction [J]. Synthetic Biology Journal, 2023, 4(3): 488-506. |
| [11] | WANG Sheng, WANG Zechen, CHEN Weihua, CHEN Ke, PENG Xiangda, OU Fafen, ZHENG Liangzhen, SUN Jinyuan, SHEN Tao, ZHAO Guoping. Design of synthetic biology components based on artificial intelligence and computational biology [J]. Synthetic Biology Journal, 2023, 4(3): 422-443. |
| [12] | Liya LIANG, Rongming LIU. Protein engineering of DNA targeting type Ⅱ CRISPR/Cas systems [J]. Synthetic Biology Journal, 2023, 4(1): 86-101. |
| [13] | Jingwei LYU, Zixin DENG, Qi ZHANG, Wei DING. Identification of RiPPs precursor peptides and cleavage sites based on deep learning [J]. Synthetic Biology Journal, 2022, 3(6): 1262-1276. |
| [14] | Yanping QI, Jin ZHU, Kai ZHANG, Tong LIU, Yajie WANG. Recent development of directed evolution in protein engineering [J]. Synthetic Biology Journal, 2022, 3(6): 1081-1108. |
| [15] | Jingjing LI, Chao MA, Fan WANG, Hongjie ZHANG, Kai LIU. Biosynthesis of high-performance protein materials and their applications [J]. Synthetic Biology Journal, 2022, 3(4): 638-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||