Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (5): 904-915.DOI: 10.12211/2096-8280.2023-026
• Invited Review • Previous Articles Next Articles
The recent progresses and applications of in-parallel fermentation technology
BAI Zhonghu1, REN He1, NIE Jianqi1, SUN Yang2
- 1.National Engineering Research Center of Cereal Fermentation and Food Biomanufacturing,Jiangnan University,Wuxi 214122,Jiangsu,China
2.School of Life Science,Henan University,Kaifeng 475004,Henan,China
-
Received:2023-03-30Revised:2023-06-25Online:2023-11-15Published:2023-10-31 -
Contact:BAI Zhonghu
高通量平行发酵技术的发展与应用
白仲虎1, 任和1, 聂简琪1, 孙杨2
- 1.江南大学粮食发酵与食品生物制造国家工程研究中心,江苏 无锡 214122
2.河南大学生命科学学院,河南 开封 475004
-
通讯作者:白仲虎 -
作者简介:白仲虎 (1965—),男,教授,博士生导师。研究方向为发酵工程、生物反应器、生物医药过程工程。E-mail:baizhonghu@jiangnan.edu.cn -
基金资助:国家高技术研究发展计划(2015A020802)
CLC Number:
Cite this article
BAI Zhonghu, REN He, NIE Jianqi, SUN Yang. The recent progresses and applications of in-parallel fermentation technology[J]. Synthetic Biology Journal, 2023, 4(5): 904-915.
白仲虎, 任和, 聂简琪, 孙杨. 高通量平行发酵技术的发展与应用[J]. 合成生物学, 2023, 4(5): 904-915.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-026
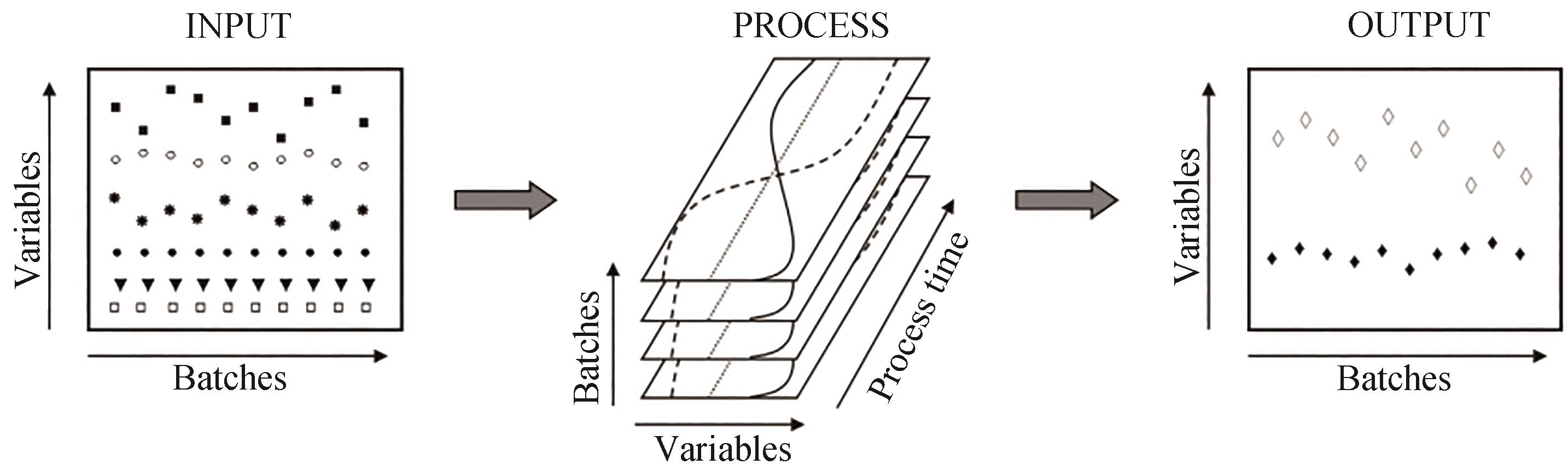
Fig. 3 Schematic diagram of data collection from bioprocesses[37][Inputs to the process parameters are operating within predetermined controlled set points. Changes in these parameters may result in significant changes in the output (yield or quality) of the culture target parameters.]
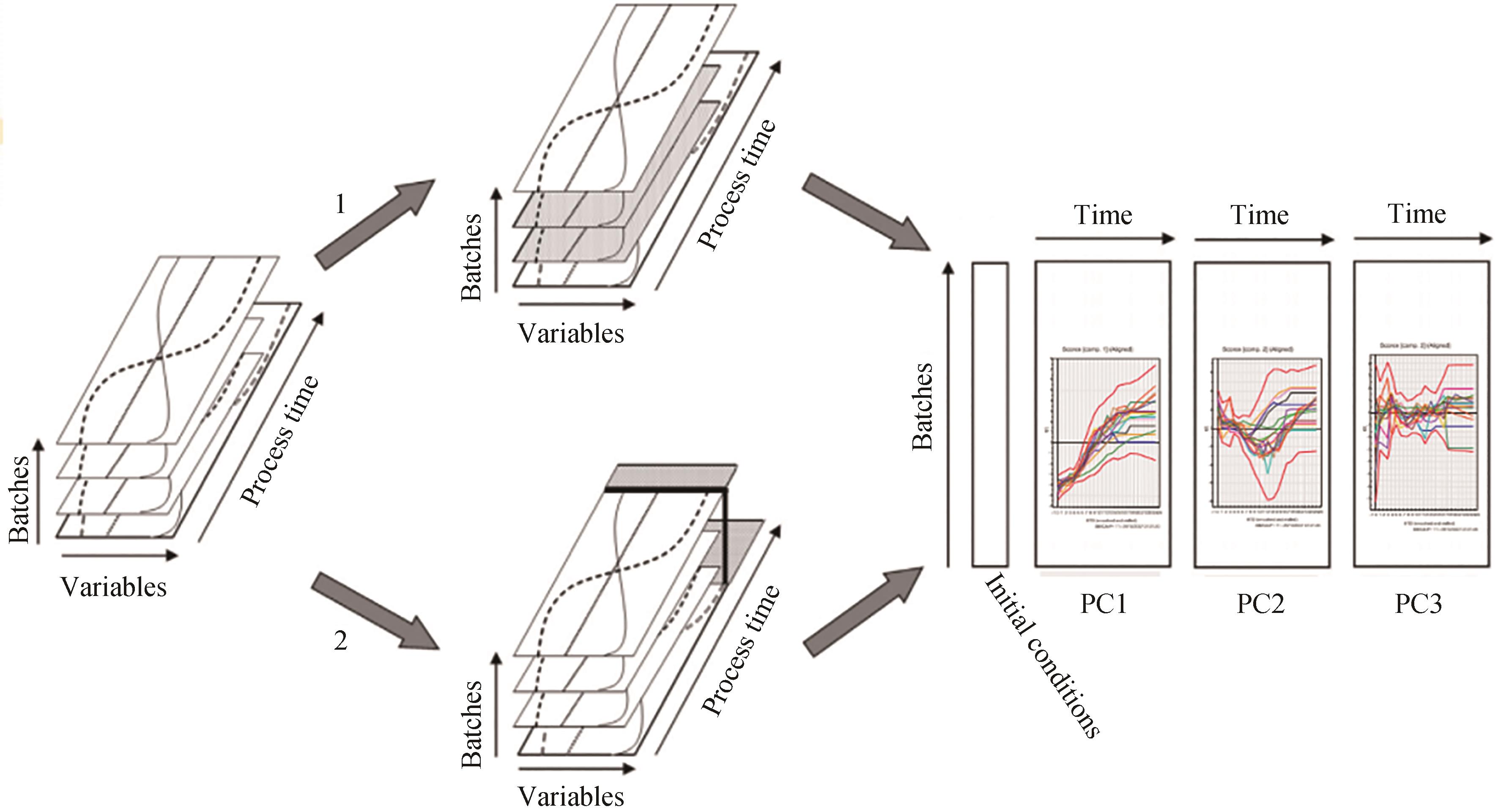
Fig. 4 Schematic diagram of unfolding and comparing the batch models of the processes of "good" and "bad" batches[37](In option 1, only the so-called "good" batches that meet the requirements of the target parameters are retained. In option 2, all batches exhibit the so-called "bad" batches that do not meet the target parameters. The PLS-MVA batch model can be built based on the good batches, and by bringing the process parameters of the bad batches into the batch model, the causes of the problems of the bad batches can be identified.)
| 1 | RIENZO M, JACKSON S J, CHAO L K, et al. High-throughput screening for high-efficiency small-molecule biosynthesis[J]. Metabolic Engineering, 2021, 63: 102-125. |
| 2 | AKKERMANS S, NIMMEGEERS P, VAN IMPE J F. Comparing design of experiments and optimal experimental design techniques for modelling the microbial growth rate under static environmental conditions[J]. Food Microbiology, 2018, 76: 504-512. |
| 3 | WALSH I, MYINT M, NGUYEN-KHUONG T, et al. Harnessing the potential of machine learning for advancing "Quality by Design" in biomanufacturing[J]. mAbs, 2022, 14(1): 2013593. |
| 4 | ICH Q8(R 2). Pharmaceutical Development[EB/OL]. 2009[2023-03-01]. . |
| 5 | LIAO X P, MA H W, TANG Y J. Artificial intelligence: a solution to involution of design-build-test-learn cycle[J]. Current Opinion in Biotechnology, 2022, 75: 102712. |
| 6 | ORSI E, BEEKWILDER J, EGGINK G, et al. The transition of Rhodobacter sphaeroides into a microbial cell factory[J]. Biotechnology and Bioengineering, 2021, 118(2): 531-541. |
| 7 | FLETCHER E, CHEN Y, CASPETA L, et al. Editorial: genomic strategies for efficient microbial cell factories[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 962828. |
| 8 | GURDO N, VOLKE D C, MCCLOSKEY D, et al. Automating the design-build-test-learn cycle towards next-generation bacterial cell factories[J]. New Biotechnology, 2023, 74: 1-15. |
| 9 | NAKAZAWA S, IMAICHI O, KOGURE T, et al. History-driven genetic modification design technique using a domain-specific lexical model for the acceleration of DBTL cycles for microbial cell factories[J]. ACS Synthetic Biology, 2021, 10(9): 2308-2317. |
| 10 | ISETT K, GEORGE H, HERBER W, et al. Twenty-four-well plate miniature bioreactor high-throughput system: assessment for microbial cultivations[J]. Biotechnology and Bioengineering, 2007, 98(5): 1017-1028. |
| 11 | MÜHLMANN M, KUNZE M, RIBEIRO J, et al. Cellulolytic RoboLector-towards an automated high-throughput screening platform for recombinant cellulase expression[J]. Journal of Biological Engineering, 2017, 11(1): 1. |
| 12 | VELEZ-SUBERBIE M L, BETTS J P J, WALKER K L, et al. High throughput automated microbial bioreactor system used for clone selection and rapid scale-down process optimization[J]. Biotechnology Progress, 2018, 34(1): 58-68. |
| 13 | HEMMERICH J, NOACK S, WIECHERT W, et al. Microbioreactor systems for accelerated bioprocess development[J]. Biotechnology Journal, 2018, 13(4): 1700141. |
| 14 | ABATE A R, HUNG T, MARY P, et al. High-throughput injection with microfluidics using picoinjectors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(45): 19163-19166. |
| 15 | BROMIG L, VON DEN EICHEN N, WEUSTER-BOTZ D. Control of parallelized bioreactorsⅠ: dynamic scheduling software for efficient bioprocess management in high-throughput systems[J]. Bioprocess and Biosystems Engineering, 2022, 45(12): 1927-1937. |
| 16 | LONG Q, LIU X X, YANG Y K, et al. The development and application of high throughput cultivation technology in bioprocess development[J]. Journal of Biotechnology, 2014, 192 Pt B: 323-338. |
| 17 | SANDNER V, PYBUS L P, MCCREATH G, et al. Scale-down model development in ambr systems: an industrial perspective[J]. Biotechnology Journal, 2019, 14(4): 1700766. |
| 18 | CRUZ BOURNAZOU M N, BARZ T, NICKEL D B, et al. Online optimal experimental re-design in robotic parallel fed-batch cultivation facilities[J]. Biotechnology and Bioengineering, 2017, 114(3): 610-619. |
| 19 | JANSEN R, TENHAEF N, MOCH M, et al. FeedER: a feedback-regulated enzyme-based slow-release system for fed-batch cultivation in microtiter plates[J].Bioprocess and Biosystems Engineering, 2019, 42(11): 1843-1852. |
| 20 | HEMMERICH J, WIECHERT W, OLDIGES M. Automated growth rate determination in high-throughput microbioreactor systems[J].BMC Research Notes, 2017, 10(1): 617. |
| 21 | MORSCHETT H, JANSEN R, NEUENDORF C, et al. Parallelized microscale fed-batch cultivation in online-monitored microtiter plates: implications of media composition and feed strategies for process design and performance[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(1): 35-47. |
| 22 | HEMMERICH J, TENHAEF N, STEFFENS C, et al. Less sacrifice, more insight: repeated low-volume sampling of microbioreactor cultivations enables accelerated deep phenotyping of microbial strain libraries[J]. Biotechnology Journal, 2019, 14(9): 1800428. |
| 23 | UNTHAN S, RADEK A, WIECHERT W, et al. Bioprocess automation on a Mini Pilot Plant enables fast quantitative microbial phenotyping[J]. Microbial Cell Factories, 2015, 14: 32. |
| 24 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| 25 | ZHU X F, DU C C, MOHSIN A, et al. An efficient high-throughput screening of high gentamicin-producing mutants based on titer determination using an integrated computer-aided vision technology and machine learning[J]. Analytical Chemistry, 2022, 94(33): 11659-11669. |
| 26 | KAMMINGA T, SLAGMAN S J, MARTINS DOS SANTOS V A P, et al. Risk-based bioengineering strategies for reliable bacterial vaccine production[J]. Trends in Biotechnology, 2019, 37(8): 805-816. |
| 27 | LIU Y, NIELSEN J. Recent trends in metabolic engineering of microbial chemical factories[J]. Current Opinion in Biotechnology, 2019, 60: 188-197. |
| 28 | SARNAIK A, LIU A, NIELSEN D, et al. High-throughput screening for efficient microbial biotechnology[J]. Current Opinion in Biotechnology, 2020, 64: 141-150. |
| 29 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 30 | RAJ K, VENAYAK N, DIEP P, et al. Automation assisted anaerobic phenotyping for metabolic engineering[J].Microbial Cell Factories, 2021, 20(1): 184. |
| 31 | RIENZO M, LIN K C, MOBILIA K C, et al. High-throughput optofluidic screening for improved microbial cell factories via real-time micron-scale productivity monitoring[J]. Lab on a Chip, 2021, 21(15): 2901-2912. |
| 32 | POLITIS S N, COLOMBO P, COLOMBO G, et al. Design of experiments (DoE) in pharmaceutical development[J]. Drug Development and Industrial Pharmacy, 2017, 43(6): 889-901. |
| 33 | SWAIN S, PARHI R, JENA B R, et al. Quality by design: concept to applications[J]. Current Drug Discovery Technologies, 2019, 16(3): 240-250. |
| 34 | NOLD V, JUNGHANS L, BISGEN L, et al. Applying intensified design of experiments to mammalian cell culture processes[J]. Engineering in Life Sciences, 2022, 22(12): 784-795. |
| 35 | TAI M, LY A, LEUNG I, et al. Efficient high-throughput biological process characterization: definitive screening design with the ambr250 bioreactor system[J]. Biotechnology Progress, 2015, 31(5): 1388-1395. |
| 36 | PEREIRA R F S, DE CARVALHO C C C R. Optimization of multiparameters for increased yields of cytochrome B5 in bioreactors[J]. Molecules, 2021, 26(14): 4148. |
| 37 | MERCIER S M, DIEPENBROEK B, DALM M C F, et al. Multivariate data analysis as a PAT tool for early bioprocess development data[J]. Journal of Biotechnology, 2013, 167(3): 262-270. |
| 38 | MERCIER S M, DIEPENBROEK B, WIJFFELS R H, et al. Multivariate PAT solutions for biopharmaceutical cultivation: current progress and limitations[J]. Trends in Biotechnology, 2014, 32(6): 329-336. |
| 39 | RATHORE A S, MITTAL S, PATHAK M, et al. Guidance for performing multivariate data analysis of bioprocessing data: pitfalls and recommendations[J]. Biotechnology Progress, 2014, 30(4): 967-973. |
| 40 | XU P, CLARK C, RYDER T, et al. Characterization of TAP Ambr 250 disposable bioreactors, as a reliable scale-down model for biologics process development[J]. Biotechnology Progress, 2017, 33(2): 478-489. |
| 41 | NIKITA S, MISHRA S, GUPTA K, et al. Advances in bioreactor control for production of biotherapeutic products[J]. Biotechnology and Bioengineering, 2023, 120(5): 1189-1214. |
| 42 | SÃO PEDRO M N, SILVA T C, PATIL R, et al. White paper on high-throughput process development for integrated continuous biomanufacturing[J]. Biotechnology and Bioengineering, 2021, 118(9): 3275-3286. |
| 43 | KHANAL O, LENHOFF A M. Developments and opportunities in continuous biopharmaceutical manufacturing[J]. mAbs, 2021, 13(1): 1903664. |
| 44 | ANANE E, GARCÍA Á C, HABY B, et al. A model-based framework for parallel scale-down fed-batch cultivations in mini-bioreactors for accelerated phenotyping[J]. Biotechnology and Bioengineering, 2019, 116(11): 2906-2918. |
| 45 | TAPIA F, VÁZQUEZ-RAMÍREZ D, GENZEL Y, et al. Bioreactors for high cell density and continuous multi-stage cultivations: options for process intensification in cell culture-based viral vaccine production[J]. Applied Microbiology and Biotechnology, 2016, 100(5): 2121-2132. |
| 46 | JANOSCHEK S, SCHULZE M, ZIJLSTRA G, et al. A protocol to transfer a fed-batch platform process into semi-perfusion mode: the benefit of automated small-scale bioreactors compared to shake flasks as scale-down model[J]. Biotechnology Progress, 2019, 35(2): e2757. |
| 47 | LIN H, LEIGHTY R W, GODFREY S, et al. Principles and approach to developing mammalian cell culture media for high cell density perfusion process leveraging established fed-batch media[J]. Biotechnology Progress, 2017, 33(4): 891-901. |
| 48 | XU J L, OU J F, MCHUGH K P, et al. Upstream cell culture process characterization and in-process control strategy development at pandemic speed[J]. mAbs, 2022, 14(1): 2060724. |
| 49 | WOLF M K F, LORENZ V, KARST D J, et al. Development of a shake tube-based scale-down model for perfusion cultures[J]. Biotechnology and Bioengineering, 2018, 115(11): 2703-2713. |
| 50 | MACDONALD M A, NÖBEL M, ROCHE RECINOS D, et al. Perfusion culture of Chinese Hamster Ovary cells for bioprocessing applications[J]. Critical Reviews in Biotechnology, 2022, 42(7): 1099-1115. |
| 51 | KUIPER M, SPENCER C, FÄLDT E, et al. Repurposing fed-batch media and feeds for highly productive CHO perfusion processes[J]. Biotechnology Progress, 2019, 35(4): e2821. |
| 52 | JANAKIRAMAN V, KWIATKOWSKI C, KSHIRSAGAR R, et al. Application of high-throughput mini-bioreactor system for systematic scale-down modeling, process characterization, and control strategy development[J]. Biotechnology Progress, 2015, 31(6): 1623-1632. |
| [1] | LIU Jianming, ZHANG Chijian, ZHANG Bing, ZENG Anping. Clostridium pasteurianum as an industrial chassis for efficient production of 1,3-propanediol: from metabolic engineering to fermentation and product separation [J]. Synthetic Biology Journal, 2024, 5(6): 1386-1403. |
| [2] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [3] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [4] | GUO Xiaojie, JIAN Xingjin, WANG Liyan, ZHANG Chong, XING Xinhui. Progress in bioreactors and instruments for phenotype testing with synthetic biology research [J]. Synthetic Biology Journal, 2024, 5(1): 16-37. |
| [5] | zhiqiang ZHANG, Yang ZHANG, Weibao QIU, Hairong ZHENG. Progress and prospect of ultrasonic liquid transfer and low-volume liquid transfer technology [J]. Synthetic Biology Journal, 2023, 4(5): 916-931. |
| [6] | Can ZHANG, Liyang SHI, Jianwu DAI. Cultured meat from biomaterials: challenges and prospects [J]. Synthetic Biology Journal, 2022, 3(4): 676-689. |
| [7] | Liangzhi XIE. Contributions to the booming biologics industrialization —a dedicated pioneer: Professor. Daniel I. C. Wang [J]. Synthetic Biology Journal, 2021, 2(4): 482-496. |
| [8] | Jiangnan CHEN, Xiaoning CHEN, Xinyi LIU, Wei WAN, Yixin ZHANG, Zihao ZHANG, Yifei ZHENG, Taoran ZHENG, Xuan WANG, Ziyu WANG, Xu YAN, Xu ZHANG, Fuqing WU, Guoqiang CHEN. Engineering Halomonas spp. for next generation industrial biotechnology (NGIB) [J]. Synthetic Biology Journal, 2020, 1(5): 516-527. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||