Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (1): 88-106.DOI: 10.12211/2096-8280.2023-074
• Invited Review • Previous Articles Next Articles
Advances in applications of deep learning for predicting sequence-based protein interactions
ZHU Jingyong1,2,3, LI Junxiang3,4, LI Xuhui3,5, ZHANG Jin2, WU Wenjing2
- 1.College of Life Sciences and Medicine,Zhejiang Sci-tech University,Hangzhou 310018,Zhejiang,China
2.College of Biological Chemical Sciences and Engineering,Jiaxing University,Jiaxing 314000,Zhejiang,China
3.Agecode R&D Center,Yangtze Delta Region Institute of Tsinghua University,Jiaxing 341001,Zhejiang,China
4.Harvest Biotech. Co. ,Ltd. ,Jiaxing 341001,Zhejiang,China
5.Zhejiang Provincial Key Laboratory of Applied Enzymology,Yangtze Delta Region Institute of Tsinghua University,Jiaxing 314006,Zhejiang,China
-
Received:2023-10-24Revised:2023-11-28Online:2024-03-20Published:2024-02-29 -
Contact:ZHANG Jin
深度学习在基于序列的蛋白质互作预测中的应用进展
朱景勇1,2,3, 李钧翔3,4, 李旭辉3,5, 张瑾2, 毋文静2
- 1.浙江理工大学生命科学与医药学院,浙江 杭州 310018
2.嘉兴学院生物与化学工程学院,浙江 嘉兴 314000
3.浙江清华长三角研究院,衰老科学创新研发中心,浙江 嘉兴 341001
4.禾美生物科技(浙江)有限公司,浙江 嘉兴 341001
5.浙江清华长三角研究院,浙江省应用酶学重点实验室,浙江 嘉兴 314006
-
通讯作者:张瑾 -
作者简介:朱景勇 (1998—),男,硕士研究生。研究方向为深度学习预测蛋白质互作。 E-mail:jingyongzhu2016@163.com张瑾 (1976—),男,教授,硕士生导师。研究方向为代谢疾病的致病机理及药物靶点发掘。 E-mail:zhangjin7688@163.com -
基金资助:国家自然科学基金(32172708);浙江省自然科学基金重点项目(LZ23C170002)
CLC Number:
Cite this article
ZHU Jingyong, LI Junxiang, LI Xuhui, ZHANG Jin, WU Wenjing. Advances in applications of deep learning for predicting sequence-based protein interactions[J]. Synthetic Biology Journal, 2024, 5(1): 88-106.
朱景勇, 李钧翔, 李旭辉, 张瑾, 毋文静. 深度学习在基于序列的蛋白质互作预测中的应用进展[J]. 合成生物学, 2024, 5(1): 88-106.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-074
| 数据库名称Database name | 简介 Description | 链接 URL | 最近更新 Last update | 参考文献Reference |
|---|---|---|---|---|
| BioGRID | 以蛋白质为核心的互作数据库 | https://thebiogrid.org | 2023 | [ |
| DIP | 通过实验验证和文献确认的PPI互作信息 | https://dip.doe-mbi.ucla.edu/dip | 2020 | [ |
| HIPPIE | 人工整合的超过60 000条PPI互作数据 | https://cbdm.uni-mainz.de/hippie | 2023 | [ |
| HPRD | 人类PPI数据库,包括41 327对互作信息 | https://hprd.org | 2010 | [ |
| HVIDB | HVIDB重点介绍了48 643个经过实验验证的人与病毒PPI | http://zzdlab.com/hvidb/ | 2020 | [ |
| Intact | 从22 954份文献中提取1 194 594份互作数据信息 | https://www.ebi.ac.uk/intact/home | 2022 | [ |
| Negatome | 通过整理文献和分析蛋白质复合物的三维结构得到的非相互作用信息 | http://mips.helmholtz-muenchen.de/proj/ppi/negatome | 2014 | [ |
| STRING | 蛋白质互作数据库,涉及14 094种生物的67 592 464个蛋白质 | https://cn.string-db.org | 2023 | [ |
Table 1 Public databases and basic information
| 数据库名称Database name | 简介 Description | 链接 URL | 最近更新 Last update | 参考文献Reference |
|---|---|---|---|---|
| BioGRID | 以蛋白质为核心的互作数据库 | https://thebiogrid.org | 2023 | [ |
| DIP | 通过实验验证和文献确认的PPI互作信息 | https://dip.doe-mbi.ucla.edu/dip | 2020 | [ |
| HIPPIE | 人工整合的超过60 000条PPI互作数据 | https://cbdm.uni-mainz.de/hippie | 2023 | [ |
| HPRD | 人类PPI数据库,包括41 327对互作信息 | https://hprd.org | 2010 | [ |
| HVIDB | HVIDB重点介绍了48 643个经过实验验证的人与病毒PPI | http://zzdlab.com/hvidb/ | 2020 | [ |
| Intact | 从22 954份文献中提取1 194 594份互作数据信息 | https://www.ebi.ac.uk/intact/home | 2022 | [ |
| Negatome | 通过整理文献和分析蛋白质复合物的三维结构得到的非相互作用信息 | http://mips.helmholtz-muenchen.de/proj/ppi/negatome | 2014 | [ |
| STRING | 蛋白质互作数据库,涉及14 094种生物的67 592 464个蛋白质 | https://cn.string-db.org | 2023 | [ |
类型 Type | 编码方法 Encoding method | 形式 Form | 向量长度 Vector length | 参考文献 Reference |
|---|---|---|---|---|
| 基于序列成分 | 二肽组成 dipeptide composition | 一维向量 | 400 | [ |
| 氨基酸组成 amino acid composition | 一维向量 | 20 | [ | |
| 伪氨基酸组成 pseudo-amino acid composition | 一维向量 | 20+L,L为最大滞后值 | [ | |
| 联合三元组 conjoint triad | 一维向量 | 343 | [ | |
| 基于自相关 | 自协方差 auto covariance | 一维向量 | L×理化性质个数,L为最大滞后值 | [ |
| 交叉协方差 cross covariance | 一维向量 | L×N×(N-1),L为最大滞后值,N为理化性质个数 | [ | |
| 自交叉协方差 auto-cross covariance | 一维向量 | L×N×N,L为最大滞后值,N为理化性质个数 | [ | |
| Moran自相关 Moran autocorrelation | 一维向量 | L×理化性质个数,L为最大滞后值 | [ | |
| Geary自相关 Geary autocorrelation | 一维向量 | L×理化性质个数,L为最大滞后值 | [ | |
| 物理化学距离变换 physicochemical distance transformation | 一维向量 | 531×β,β为最大间隔距离 | [ | |
| 基于进化信息 | 位置特异性得分矩阵PSSM | 二维矩阵 | 20×N,N为序列长度 | [ |
| ACC-PSSM | 一维向量 | 400×L,L为最大滞后值 | [ | |
| Top-n-grams | 一维向量 | L×N,L为序列长度,N为阈值 | [ | |
| BLOSUM62 | 二维矩阵 | 20×N,N为序列长度 | [ | |
| 伪位置特异性得分矩阵Pseudo-PSSM | 一维向量 | 40 | [ |
Table 2 Protein encoding methods in sequence-based PPI prediction models
类型 Type | 编码方法 Encoding method | 形式 Form | 向量长度 Vector length | 参考文献 Reference |
|---|---|---|---|---|
| 基于序列成分 | 二肽组成 dipeptide composition | 一维向量 | 400 | [ |
| 氨基酸组成 amino acid composition | 一维向量 | 20 | [ | |
| 伪氨基酸组成 pseudo-amino acid composition | 一维向量 | 20+L,L为最大滞后值 | [ | |
| 联合三元组 conjoint triad | 一维向量 | 343 | [ | |
| 基于自相关 | 自协方差 auto covariance | 一维向量 | L×理化性质个数,L为最大滞后值 | [ |
| 交叉协方差 cross covariance | 一维向量 | L×N×(N-1),L为最大滞后值,N为理化性质个数 | [ | |
| 自交叉协方差 auto-cross covariance | 一维向量 | L×N×N,L为最大滞后值,N为理化性质个数 | [ | |
| Moran自相关 Moran autocorrelation | 一维向量 | L×理化性质个数,L为最大滞后值 | [ | |
| Geary自相关 Geary autocorrelation | 一维向量 | L×理化性质个数,L为最大滞后值 | [ | |
| 物理化学距离变换 physicochemical distance transformation | 一维向量 | 531×β,β为最大间隔距离 | [ | |
| 基于进化信息 | 位置特异性得分矩阵PSSM | 二维矩阵 | 20×N,N为序列长度 | [ |
| ACC-PSSM | 一维向量 | 400×L,L为最大滞后值 | [ | |
| Top-n-grams | 一维向量 | L×N,L为序列长度,N为阈值 | [ | |
| BLOSUM62 | 二维矩阵 | 20×N,N为序列长度 | [ | |
| 伪位置特异性得分矩阵Pseudo-PSSM | 一维向量 | 40 | [ |
类型 Type | 发表时间 Published time | 模型名称 Model name | 算法框架 Algorithm framework | 预测类型 Prediction type | 参考文献 Reference |
|---|---|---|---|---|---|
| 基于DNN | 2018 | — | DNN | PPI | [ |
| 2019 | EnsDNN | DNN | PPI | [ | |
| 2019 | — | DNN | PPI | [ | |
| 2019 | — | DNN | PPI | [ | |
| 2019 | DeepFEPPI | DNN | PPI | [ | |
| 2019 | DNN-PPI | DNN | PPI | [ | |
| 2020 | — | DNN | PPI | [ | |
| 2020 | — | DNN | PPI | [ | |
| 2022 | DNN-XGB | DNN,XGB | PPI | [ | |
| 2022 | DWPPI | DNN | PPI | [ | |
| 2022 | — | DNN | PPI | [ | |
| 2022 | CT-DNN | DNN | PPI | [ | |
| 2022 | — | DNN | PPI | [ | |
| 基于CNN | 2018 | DPPI | CNN | PPI | [ |
| 2019 | — | CNN-RF | PPI | [ | |
| 2019 | — | CNN | PPI | [ | |
| 2020 | Visual | CNN | PPIsite | [ | |
| 2020 | DeepPPISP | TextCNN | PPIsite | [ | |
| 2020 | EnAmDNN | CNN | PPI | [ | |
| 2020 | — | CNN | PPIsite | [ | |
| 2021 | TransPPI | CNN | PPI | [ | |
| 2021 | — | CNN | PPI | [ | |
| 2021 | D-script | CNN | PPI | [ | |
| 2021 | CAMP | CNN | PepPI | [ | |
| 2021 | DeepViral | CNN | PPI | [ | |
| 2022 | DeepTrio | CNN | PPI | [ | |
| 2023 | EResCNN | ResCNN+RF | PPI | [ | |
| 2023 | ProtInteract | CNN | PPI | [ | |
| 基于RNN及变体 | 2019 | — | RNN | PPI | [ |
| 2019 | DLPred | LSTM | PPIsite | [ | |
| 2019 | — | LSTM | PPI | [ | |
| 2020 | — | LSTM | PPI | [ | |
| 2021 | — | GRNN | PPI | [ | |
| 2021 | LSTM-PHV | LSTM | PPI | [ | |
| 2023 | — | RNN | PPI | [ | |
| 基于注意力机制和Transformer | 2021 | HANPPIS | Stratified attention | PPIsite | [ |
| 2022 | Cross-attention PHV | Cross-attention | PPI | [ | |
| 2022 | ADH-PPI | Self-attention | PPI | [ | |
| 2022 | SDNN | Self-attention | PPI | [ | |
| 2023 | EnsemPPIS | Transformer | PPIsite | [ | |
| 基于混合网络 | 2018 | DNN-PPI | CNN+LSTM | PPI | [ |
| 2019 | PIPR | RNN+CNN | PPI | [ | |
| 2019 | IPPI | DNN+LSTM | PPI | [ | |
| 2021 | DELPHI | RNN+CNN | PPIsite | [ | |
| 2021 | OR-RCNN | RNN+CNN | PPI | [ | |
| 2023 | MM-StackEns | SNN+GNN | PPI | [ |
Table 3 Computational models developed within the past 5 years
类型 Type | 发表时间 Published time | 模型名称 Model name | 算法框架 Algorithm framework | 预测类型 Prediction type | 参考文献 Reference |
|---|---|---|---|---|---|
| 基于DNN | 2018 | — | DNN | PPI | [ |
| 2019 | EnsDNN | DNN | PPI | [ | |
| 2019 | — | DNN | PPI | [ | |
| 2019 | — | DNN | PPI | [ | |
| 2019 | DeepFEPPI | DNN | PPI | [ | |
| 2019 | DNN-PPI | DNN | PPI | [ | |
| 2020 | — | DNN | PPI | [ | |
| 2020 | — | DNN | PPI | [ | |
| 2022 | DNN-XGB | DNN,XGB | PPI | [ | |
| 2022 | DWPPI | DNN | PPI | [ | |
| 2022 | — | DNN | PPI | [ | |
| 2022 | CT-DNN | DNN | PPI | [ | |
| 2022 | — | DNN | PPI | [ | |
| 基于CNN | 2018 | DPPI | CNN | PPI | [ |
| 2019 | — | CNN-RF | PPI | [ | |
| 2019 | — | CNN | PPI | [ | |
| 2020 | Visual | CNN | PPIsite | [ | |
| 2020 | DeepPPISP | TextCNN | PPIsite | [ | |
| 2020 | EnAmDNN | CNN | PPI | [ | |
| 2020 | — | CNN | PPIsite | [ | |
| 2021 | TransPPI | CNN | PPI | [ | |
| 2021 | — | CNN | PPI | [ | |
| 2021 | D-script | CNN | PPI | [ | |
| 2021 | CAMP | CNN | PepPI | [ | |
| 2021 | DeepViral | CNN | PPI | [ | |
| 2022 | DeepTrio | CNN | PPI | [ | |
| 2023 | EResCNN | ResCNN+RF | PPI | [ | |
| 2023 | ProtInteract | CNN | PPI | [ | |
| 基于RNN及变体 | 2019 | — | RNN | PPI | [ |
| 2019 | DLPred | LSTM | PPIsite | [ | |
| 2019 | — | LSTM | PPI | [ | |
| 2020 | — | LSTM | PPI | [ | |
| 2021 | — | GRNN | PPI | [ | |
| 2021 | LSTM-PHV | LSTM | PPI | [ | |
| 2023 | — | RNN | PPI | [ | |
| 基于注意力机制和Transformer | 2021 | HANPPIS | Stratified attention | PPIsite | [ |
| 2022 | Cross-attention PHV | Cross-attention | PPI | [ | |
| 2022 | ADH-PPI | Self-attention | PPI | [ | |
| 2022 | SDNN | Self-attention | PPI | [ | |
| 2023 | EnsemPPIS | Transformer | PPIsite | [ | |
| 基于混合网络 | 2018 | DNN-PPI | CNN+LSTM | PPI | [ |
| 2019 | PIPR | RNN+CNN | PPI | [ | |
| 2019 | IPPI | DNN+LSTM | PPI | [ | |
| 2021 | DELPHI | RNN+CNN | PPIsite | [ | |
| 2021 | OR-RCNN | RNN+CNN | PPI | [ | |
| 2023 | MM-StackEns | SNN+GNN | PPI | [ |
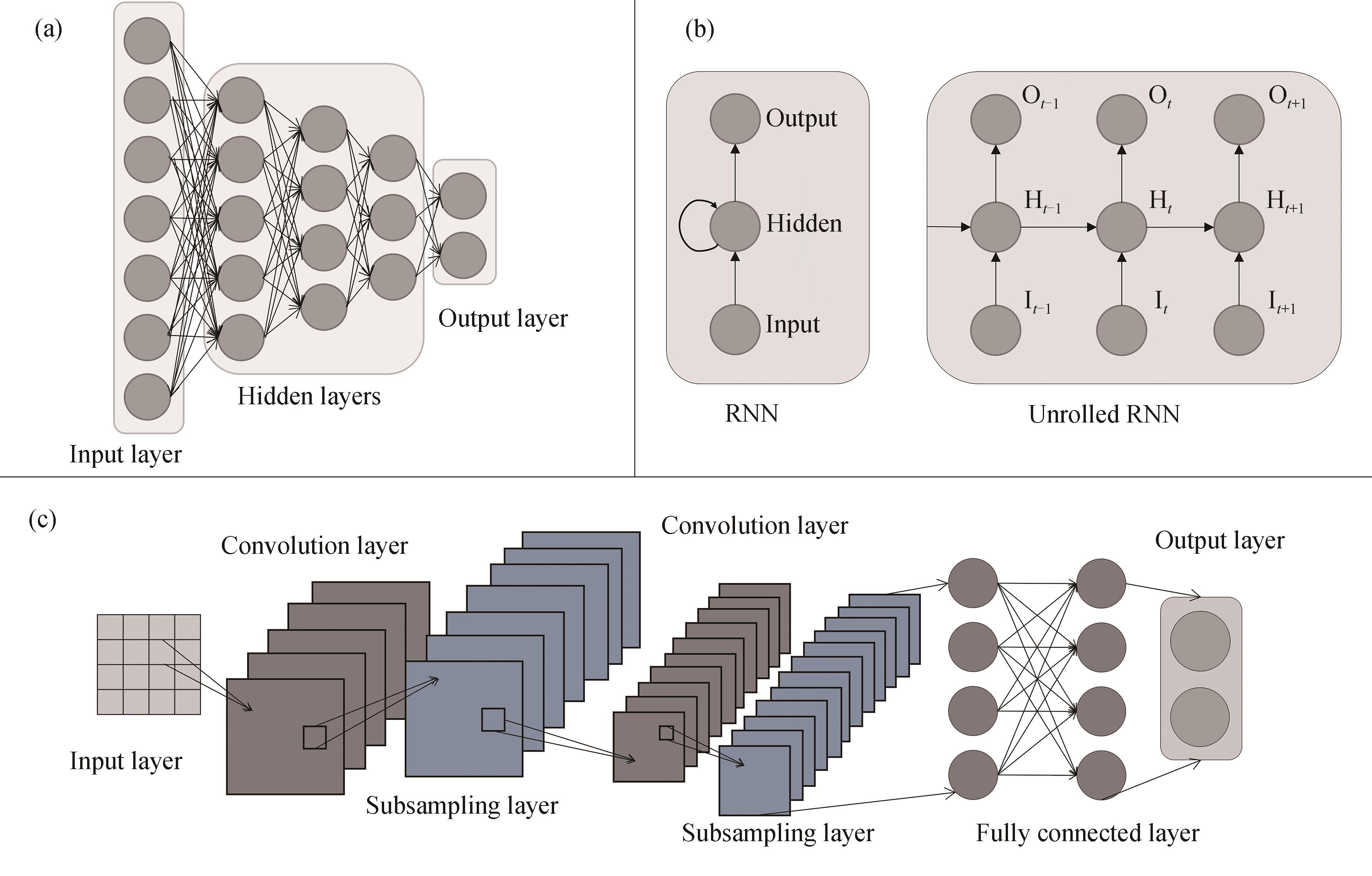
Fig. 3 Basic network structure of DNN, RNN, and CNN(a) DNN contains an input layer, multiple hidden layers, and an output layer; (b) RNN consists of an input unit (Input), a hidden unit (Hidden), and an output unit (Output) to unfold the RNN in time, with the subscript of the symbols representing the time, indicating that H t receives the inputs from I t and H t-1, and then propagates the results of the computation to O t and H t+1; (c) CNNs include an input layer, multiple convolutional and pooling layers, and fully connected and output layers
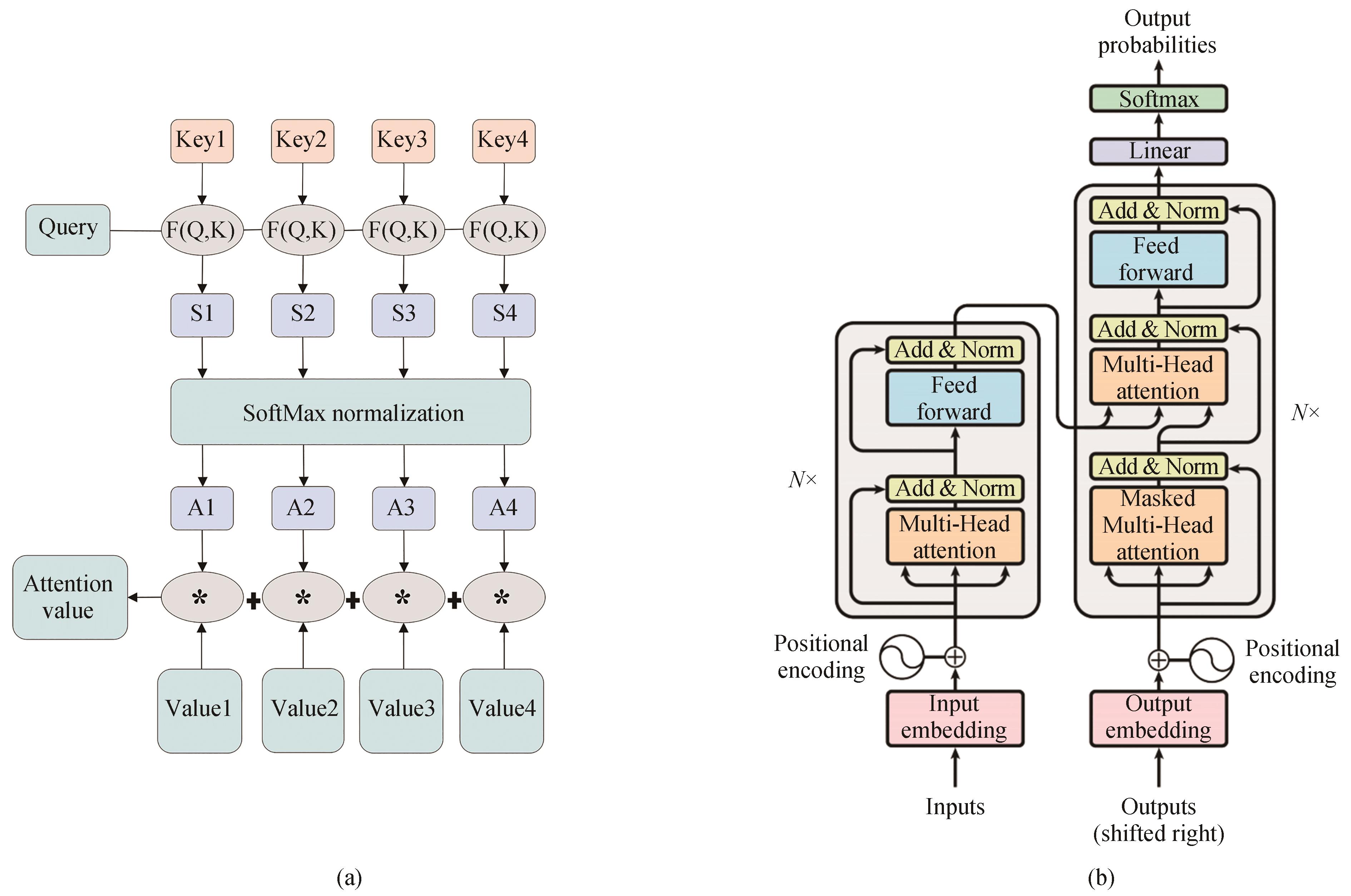
Fig. 4 Basic structure of the basic structure of Attention mechanism and Transformer(a) Providing a detailed depiction of the core operations in the attention mechanism, such as Query, Key, and corresponding value for each Key. The similarity scores (S1 to S4) between each Query and Key are first computed using the function F (Q, K), which are then normalized through the Softmax function to derive weights for each Key (A1 to A4). Every Key is associated with a "value" (Value 1 to Value 4). The final attention value is derived by summing up the product of each value and its respective weight. (b) Adapted from reference [72], showcasing the fundamental architecture of the transformer model that is split into encoder and decoder components. The encoder on the left initially receives inputs and pre-processes them through "input embedding" and "positional encoding", and then repeatedly passes them through structural units containing the "multi-head attention" mechanism and feed-forward neural networks. The objective is to transform the input sequence into a context-rich continuous vector representation. On the right, the decoder is responsible for producing an output sequence based on the context provided by the encoder. Its initial input is labeled as "Output", ensuring that the output at the current position in the decoding process only depends on prior information. The decoder also undergoes multiple rounds of "multi-head attention" and feed-forward network processing, ultimately yielding an output probability distribution via linear transformation and a Softmax layer, representing a probability for the potential output
预测结果 Prediction result | 英文名称 English name | 描述 Description |
|---|---|---|
| 真阳性 | true positive, TP | 对阳性样本预测为阳性 |
| 假阳性 | false positive, FP | 对阴性样本预测为阳性 |
| 真阴性 | true negative, TN | 对阴性样本预测为阴性 |
| 假阴性 | false negative, FN | 对阳性样本预测为阴性 |
Table 4 Four basic prediction results and their specific meanings
预测结果 Prediction result | 英文名称 English name | 描述 Description |
|---|---|---|
| 真阳性 | true positive, TP | 对阳性样本预测为阳性 |
| 假阳性 | false positive, FP | 对阴性样本预测为阳性 |
| 真阴性 | true negative, TN | 对阴性样本预测为阴性 |
| 假阴性 | false negative, FN | 对阳性样本预测为阴性 |
评估指标 Evaluation index | 计算公式 Calculation formula |
|---|---|
| 准确率(accuracy) | |
| 精准率(precision) | |
| 敏感性(sensitivity) | |
| 特异性(specificity) | |
| F1值(F1 score) | |
| 马修斯相关系数(Matthews correlation coefficient) |
Table 5 Six evaluation metrics and their calculation formulas
评估指标 Evaluation index | 计算公式 Calculation formula |
|---|---|
| 准确率(accuracy) | |
| 精准率(precision) | |
| 敏感性(sensitivity) | |
| 特异性(specificity) | |
| F1值(F1 score) | |
| 马修斯相关系数(Matthews correlation coefficient) |
| 1 | HOSSEINI S, ILIE L. PITHIA: protein interaction site prediction using multiple sequence alignments and attention[J]. International Journal of Molecular Sciences, 2022, 23(21): 12814. |
| 2 | FIELDS S, STERNGLANZ R. The two-hybrid system: an assay for protein-protein interactions[J]. Trends in Genetics, 1994, 10(8): 286-292. |
| 3 | RIGAUT G, SHEVCHENKO A, RUTZ B, et al. A generic protein purification method for protein complex characterization and proteome exploration[J]. Nature Biotechnology, 1999, 17(10): 1030-1032. |
| 4 | ZHU H, BILGIN M, BANGHAM R, et al. Global analysis of protein activities using proteome chips[J]. Science, 2001, 293(5537): 2101-2105. |
| 5 | ZHAN X K, XIAO M, YOU Z H, et al. Predicting protein-protein interactions based on ensemble learning-based model from protein sequence[J]. Biology, 2022, 11(7): 995. |
| 6 | LEE H, DENG M H, SUN F Z, et al. An integrated approach to the prediction of domain-domain interactions[J]. BMC Bioinformatics, 2006, 7(1): 269. |
| 7 | HSIN LIU C, LI K C, YUAN S. Human protein-protein interaction prediction by a novel sequence-based co-evolution method: co-evolutionary divergence[J]. Bioinformatics, 2013, 29(1): 92-98. |
| 8 | KOVÁCS I A, LUCK K, SPIROHN K, et al. Network-based prediction of protein interactions[J]. Nature Communications, 2019, 10: 1240. |
| 9 | SMITH G R, STERNBERG M J E. Prediction of protein-protein interactions by docking methods[J]. Current Opinion in Structural Biology, 2002, 12(1): 28-35. |
| 10 | ZHOU J, CUI G Q, HU S D, et al. Graph neural networks: a review of methods and applications[J]. AI open, 2020, 1: 57-81. |
| 11 | PENG X Q, WANG J X, PENG W, et al. Protein-protein interactions: detection, reliability assessment and applications[J]. Briefings in Bioinformatics, 2017, 18(5): 798-819. |
| 12 | NORTHEY T C, BAREŠIĆ A, MARTIN A C R. IntPred: a structure-based predictor of protein-protein interaction sites[J]. Bioinformatics, 2018, 34(2): 223-229. |
| 13 | BARANWAL M, MAGNER A, SALDINGER J, et al. Struct2Graph: a graph attention network for structure based predictions of protein-protein interactions[J]. BMC Bioinformatics, 2022, 23(1): 370. |
| 14 | LEE A C L, HARRIS J L, KHANNA K K, et al. A comprehensive review on current advances in peptide drug development and design[J]. International Journal of Molecular Sciences, 2019, 20(10): 2383. |
| 15 | WEI L Y, ZOU Q A. Recent progress in machine learning-based methods for protein fold recognition[J]. International Journal of Molecular Sciences, 2016, 17(12): 2118. |
| 16 | SHEN J W, ZHANG J, LUO X M, et al. Predicting protein-protein interactions based only on sequences information[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(11): 4337-4341. |
| 17 | ZHOU C, YU H A, DING Y J, et al. Multi-scale encoding of amino acid sequences for predicting protein interactions using gradient boosting decision tree[J]. PLoS One, 2017, 12(8): e0181426. |
| 18 | LIN X T, CHEN X W. Heterogeneous data integration by tree-augmented naïve Bayes for protein-protein interactions prediction[J]. Proteomics, 2013, 13(2): 261-268. |
| 19 | LI J Q, YOU Z H, LI X, et al. PSPEL: In silico prediction of self-interacting proteins from amino acids sequences using ensemble learning[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2017, 14(5): 1165-1172. |
| 20 | BAI Q F, MA J, LIU S, et al. WADDAICA: a webserver for aiding protein drug design by artificial intelligence and classical algorithm[J]. Computational and Structural Biotechnology Journal, 2021, 19: 3573-3579. |
| 21 | CHAI J Y, ZENG H, LI A M, et al. Deep learning in computer vision: a critical review of emerging techniques and application scenarios[J]. Machine Learning with Applications, 2021, 6: 100134. |
| 22 | LOPEZ M M, KALITA J. Deep learning applied to NLP[EB/OL]. arXiv, 2017: 1703.03091[2023-10-01]. . |
| 23 | TANG B H, PAN Z X, YIN K, et al. Recent advances of deep learning in bioinformatics and computational biology[J]. Frontiers in Genetics, 2019, 10: 214. |
| 24 | CONSORTIUM T U. UniProt: a hub for protein information[J]. Nucleic Acids Research, 2015, 43(D1): D204-D212. |
| 25 | STARK C, BREITKREUTZ B J, REGULY T, et al. BioGRID: a general repository for interaction datasets[J]. Nucleic Acids Research, 2006, 34(): D535-D539. |
| 26 | KESHAVA PRASAD T S, GOEL R, KANDASAMY K, et al. Human protein reference database—2009 update[J]. Nucleic Acids Research, 2009, 37(): D767-D772. |
| 27 | XENARIOS I, SALWÍNSKI L, DUAN X J, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions[J]. Nucleic Acids Research, 2002, 30(1): 303-305. |
| 28 | KANDEL D, MATIAS Y, UNGER R, et al. Shuffling biological sequences[J]. Discrete Applied Mathematics, 1996, 71(1/2/3): 171-185. |
| 29 | BLOHM P, FRISHMAN G, SMIALOWSKI P, et al. Negatome 2.0: a database of non-interacting proteins derived by literature mining, manual annotation and protein structure analysis[J]. Nucleic Acids Research, 2014, 42(D1): D396-D400. |
| 30 | ALANIS-LOBATO G, ANDRADE-NAVARRO M A, SCHAEFER M H. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks[J]. Nucleic Acids Research, 2017, 45(D1): D408-D414. |
| 31 | YANG X D, LIAN X Y, FU C, et al. HVIDB: a comprehensive database for human-virus protein-protein interactions[J]. Briefings in Bioinformatics, 2021, 22(2): 832-844. |
| 32 | KERRIEN S, ARANDA B, BREUZA L, et al. The IntAct molecular interaction database in 2012[J]. Nucleic Acids Research, 2012, 40(D1): D841-D846. |
| 33 | SZKLARCZYK D, GABLE A L, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Research, 2019, 47(D1): D607-D613. |
| 34 | HE J A, WU Y L, PU X M, et al. A transfer-learning-based deep convolutional neural network for predicting leukemia-related phosphorylation sites from protein primary sequences[J]. International Journal of Molecular Sciences, 2022, 23(3): 1741. |
| 35 | LIU B, LIU F L, WANG X L, et al. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences[J]. Nucleic Acids Research, 2015, 43(W1): W65-W71. |
| 36 | BHASIN M, RAGHAVA G P S. Classification of nuclear receptors based on amino acid composition and dipeptide composition[J]. The Journal of Biological Chemistry, 2004, 279(22): 23262-23266. |
| 37 | ZHANG C T, CHOU K C. An optimization approach to predicting protein structural class from amino acid composition[J]. Protein Science, 1992, 1(3): 401-408. |
| 38 | CHOU K C. Prediction of protein cellular attributes using pseudo-amino acid composition[J]. Proteins: Structure, Function, and Bioinformatics, 2001, 43(3): 246-255. |
| 39 | GUO Y Z, YU L Z, WEN Z N, et al. Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences[J]. Nucleic Acids Research, 2008, 36(9): 3025-3030. |
| 40 | XIAO X, WANG P, CHOU K C. iNR-PhysChem: a sequence-based predictor for identifying nuclear receptors and their subfamilies via physical-chemical property matrix[J]. PLoS One, 2012, 7(2): e30869. |
| 41 | DONG Q W, ZHOU S G, GUAN J H. A new taxonomy-based protein fold recognition approach based on autocross-covariance transformation[J]. Bioinformatics, 2009, 25(20): 2655-2662. |
| 42 | FENG Z P, ZHANG C T. Prediction of membrane protein types based on the hydrophobic index of amino acids[J]. Journal of Protein Chemistry, 2000, 19(4): 269-275. |
| 43 | SOKAL R R, THOMSON B A. Population structure inferred by local spatial autocorrelation: an example from an Amerindian tribal population[J]. American Journal of Physical Anthropology, 2006, 129(1): 121-131. |
| 44 | LIU B, WANG X L, CHEN Q C, et al. Using amino acid physicochemical distance transformation for fast protein remote homology detection[J]. PLoS One, 2012, 7(9): e46633. |
| 45 | ZAHIRI J, YAGHOUBI O, MOHAMMAD-NOORI M, et al. PPIevo: protein-protein interaction prediction from PSSM based evolutionary information[J]. Genomics, 2013, 102(4): 237-242. |
| 46 | LIU B, WU H, CHOU K C. Pse-in-One 2.0: an improved package of web servers for generating various modes of pseudo components of DNA, RNA, and protein sequences [J]. Natural Science, 2017, 9(4): 67. |
| 47 | LIU B, WANG X L, LIN L, et al. A discriminative method for protein remote homology detection and fold recognition combining Top-n-grams and latent semantic analysis[J]. BMC Bioinformatics, 2008, 9: 510. |
| 48 | LI A, WANG L R, SHI Y Z, et al. Phosphorylation site prediction with a modified k-nearest neighbor algorithm and BLOSUM62 matrix[C/OL]//2005 IEEE Engineering in Medicine and Biology 27th Annual Conference. January 17-18, 2006, Shanghai. IEEE, 2006: 6075-6078 [2023-10-01]. . |
| 49 | SHEN H B, CHOU K C. Nuc-PLoc: a new web-server for predicting protein subnuclear localization by fusing PseAA composition and PsePSSM[J]. Protein Engineering, Design & Selection, 2007, 20(11): 561-567. |
| 50 | YANG K K, WU Z, BEDBROOK C N, et al. Learned protein embeddings for machine learning[J]. Bioinformatics, 2018, 34(15): 2642-2648. |
| 51 | DUNKER A K, LAWSON J D, BROWN C J, et al. Intrinsically disordered protein[J]. Journal of Molecular Graphics and Modelling, 2001, 19(1): 26-59. |
| 52 | FONG J H, SHOEMAKER B A, GARBUZYNSKIY S O, et al. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis[J]. PLoS Computational Biology, 2009, 5(3): e1000316. |
| 53 | TOMPA P, FUXREITER M, OLDFIELD C J, et al. Close encounters of the third kind: disordered domains and the interactions of proteins[J]. BioEssays, 2009, 31(3): 328-335. |
| 54 | WEATHERITT R J, GIBSON T J. Linear motifs: lost in (pre)translation[J]. Trends in Biochemical Sciences, 2012, 37(8): 333-341. |
| 55 | DYSON H J, WRIGHT P E. Coupling of folding and binding for unstructured proteins[J]. Current Opinion in Structural Biology, 2002, 12(1): 54-60. |
| 56 | DUNKER A K, CORTESE M S, ROMERO P, et al. Flexible nets: the roles of intrinsic disorder in protein interaction networks[J]. The FEBS Journal, 2005, 272(20): 5129-5148. |
| 57 | LEI Y P, LI S Y, LIU Z Y, et al. A deep-learning framework for multi-level peptide-protein interaction prediction[J]. Nature Communications, 2021, 12: 5465. |
| 58 | MÉSZÁROS B, ERDŐS G, DOSZTÁNYI Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding[J]. Nucleic Acids Research, 2018, 46(W1): W329-W337. |
| 59 | BASU S, GSPONER J, KURGAN L. DEPICTER2: a comprehensive webserver for intrinsic disorder and disorder function prediction[J]. Nucleic Acids Research, 2023, 51(W1): W141-W147. |
| 60 | DEL CONTE A, BOUHRAOUA A, MEHDIABADI M, et al. CAID prediction portal: a comprehensive service for predicting intrinsic disorder and binding regions in proteins[J]. Nucleic Acids Research, 2023, 51(W1): W62-W69. |
| 61 | HANSON J, PALIWAL K K, LITFIN T, et al. SPOT-Disorder2: improved protein intrinsic disorder prediction by ensembled deep learning[J]. Genomics, Proteomics & Bioinformatics, 2019, 17(6): 645-656. |
| 62 | HU J, SHEN L, SUN G. Squeeze-and-excitation networks[C/OL]//2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. June 18-23, 2018, Salt Lake City, UT, USA. IEEE, 2018: 7132-7141 [2023-10-01]. . |
| 63 | GRAVES A. Long short-term memory[M/OL]// Supervised sequence labelling with recurrent neural networks. Berlin, Heidelberg: Springer, 2012: 37-45 [2023-10-01]. . |
| 64 | KAWASHIMA S, POKAROWSKI P, POKAROWSKA M, et al. AAindex: amino acid index database, progress report 2008[J]. Nucleic Acids Research, 2008, 36(): D202-D205. |
| 65 | ALTSCHUL S F, MADDEN T L, SCHÄFFER A A, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389-3402. |
| 66 | EDDY S R. Where did the BLOSUM62 alignment score matrix come from?[J]. Nature Biotechnology, 2004, 22(8): 1035-1036. |
| 67 | CHOU K C, SHEN H B. MemType-2L: a Web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM[J]. Biochemical and Biophysical Research Communications, 2007, 360(2): 339-345. |
| 68 | SUN T L, ZHOU B, LAI L H, et al. Sequence-based prediction of protein protein interaction using a deep-learning algorithm[J]. BMC Bioinformatics, 2017, 18(1): 1-8. |
| 69 | MIIKKULAINEN R, LIANG J, MEYERSON E, et al. Evolving deep neural networks[M/OL]//Artificial intelligence in the age of neural networks and brain computing. Amsterdam: Elsevier. 2019: 293-312 [2023-10-01]. . |
| 70 | GU J X, WANG Z H, KUEN J, et al. Recent advances in convolutional neural networks[J]. Pattern Recognition, 2018, 77: 354-377. |
| 71 | SHERSTINSKY A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network[J]. Physica D: Nonlinear Phenomena, 2020, 404: 132306. |
| 72 | VASWANI A, SHAZEER N, PARMAR N, et al. Attention is all you need[C/OL]//Proceedings of the 31st International Conference on Neural Information Processing Systems. December 4-9, 2017, Long Beach, California, USA. New York: ACM, 2017: 6000-6010 [2023-10-01]. . |
| 73 | GUI Y M, WANG R J, WEI Y Y, et al. Construction of protein-protein interactions model by deep neural networks[C/OL]//Proceedings of the 2018 International Workshop on Bioinformatics, Biochemistry, Biomedical Sciences (BBBS 2018). April 14-15, 2018. Hangzhou City, China. Paris, France: Atlantis Press, 2018: 221-229 [2023-10-01]. . |
| 74 | ZHANG L, YU G X, XIA D W, et al. Protein-protein interactions prediction based on ensemble deep neural networks[J]. Neurocomputing, 2019, 324: 10-19. |
| 75 | WANG X E, WU Y J, WANG R J, et al. A novel matrix of sequence descriptors for predicting protein-protein interactions from amino acid sequences[J]. PLoS One, 2019, 14(6): e0217312. |
| 76 | WANG X, WANG R J, WEI Y Y, et al. A novel conjoint triad auto covariance (CTAC) coding method for predicting protein-protein interaction based on amino acid sequence[J]. Mathematical Biosciences, 2019, 313: 41-47. |
| 77 | YAO Y, DU X Q, DIAO Y Y, et al. An integration of deep learning with feature embedding for protein-protein interaction prediction[J]. PeerJ, 2019, 7: e7126. |
| 78 | GUI Y M, WANG R J, WEI Y Y, et al. DNN-PPI: a large-scale prediction of protein-protein interactions based on deep neural networks[J]. Journal of Biological Systems, 2019, 27(1): 1-18. |
| 79 | HANGGARA F S, ANAM K. Sequence-based protein-protein interaction prediction using greedy layer-wise training of deep neural networks[C/OL]//AIP Conference Proceedings. Novosibirsk, Russia: AIP Publishing, 2020, 2278(1): 020050 [2023-10-01]. . |
| 80 | GUI Y M, WANG R J, WANG X E, et al. Using deep neural networks to improve the performance of protein-protein interactions prediction[J]. International Journal of Pattern Recognition and Artificial Intelligence, 2020, 34(13): 2052012. |
| 81 | MAHAPATRA S, GUPTA V R, SAHU S S, et al. Deep neural network and extreme gradient boosting based hybrid classifier for improved prediction of protein-protein interaction[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, 19(1): 155-165. |
| 82 | PAN J, YOU Z H, LI L P, et al. DWPPI: a deep learning approach for predicting protein-protein interactions in plants based on multi-source information with a large-scale biological network[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 807522. |
| 83 | KHANH LE N Q, KHA Q H. Prediction of protein-protein interactions through deep learning based on sequence feature extraction and interaction network[C/OL]//2022 IEEE Biomedical Circuits and Systems Conference (BioCAS). October 13-15, 2022, Taipei, China. IEEE, 2022: 539-543 [2023-10-01]. . |
| 84 | WANG J H, WANG X D, CHEN W T. Prediction of protein interactions based on CT-DNN[C/OL]//Proceedings of the 2022 9th International Conference on Biomedical and Bioinformatics Engineering. November 10-13, 2022, Kyoto, Japan. New York: ACM, 2022: 81-87 [2023-10-01]. . |
| 85 | MEWARA B, LALWANI S. Strengthening auto-feature engineering of deep learning architecture in protein-protein interaction prediction[C/OL]//SHARMA H, SHRIVASTAVA V, KUMARI BHARTI K, et al. Communication and intelligent systems: Proceedings of ICCIS 2021. Singapore: Springer, 2022: 1205-1216 [2023-10-01]. . |
| 86 | HASHEMIFAR S, NEYSHABUR B, KHAN A A, et al. Predicting protein-protein interactions through sequence-based deep learning[J]. Bioinformatics, 2018, 34(17): i802-i810. |
| 87 | WANG L, WANG H F, LIU S R, et al. Predicting protein-protein interactions from matrix-based protein sequence using convolution neural network and feature-selective rotation forest[J]. Scientific Reports, 2019, 9: 9848. |
| 88 | WANG Y B, YOU Z H, YANG S, et al. A high efficient biological language model for predicting protein-protein interactions[J]. Cells, 2019, 8(2): 122. |
| 89 | WARDAH W, DEHZANGI A, TAHERZADEH G, et al. Predicting protein-peptide binding sites with a deep convolutional neural network[J]. Journal of Theoretical Biology, 2020, 496: 110278. |
| 90 | ZENG M, ZHANG F H, WU F X, et al. Protein-protein interaction site prediction through combining local and global features with deep neural networks[J]. Bioinformatics, 2020, 36(4): 1114-1120. |
| 91 | LI F F, ZHU F, LING X H, et al. Protein interaction network reconstruction through ensemble deep learning with attention mechanism[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 390. |
| 92 | XIE Z Y, DENG X Y, SHU K X. Prediction of protein-protein interaction sites using convolutional neural network and improved data sets[J]. International Journal of Molecular Sciences, 2020, 21(2): 467. |
| 93 | YANG X D, YANG S P, LIAN X Y, et al. Transfer learning via multi-scale convolutional neural layers for human-virus protein-protein interaction prediction[J]. Bioinformatics, 2021, 37(24): 4771-4778. |
| 94 | WANG Y, LI Z C, ZHANG Y F, et al. Performance improvement for a 2D convolutional neural network by using SSC encoding on protein-protein interaction tasks[J]. BMC Bioinformatics, 2021, 22(1): 184. |
| 95 | SLEDZIESKI S, SINGH R, COWEN L, et al. D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions[J]. Cell Systems, 2021, 12(10): 969-982.e6. |
| 96 | LIU-WEI W, KAFKAS Ş, CHEN J, et al. DeepViral: prediction of novel virus-host interactions from protein sequences and infectious disease phenotypes[J]. Bioinformatics, 2021, 37(17): 2722-2729. |
| 97 | HU X T, FENG C, ZHOU Y C, et al. DeepTrio: a ternary prediction system for protein-protein interaction using mask multiple parallel convolutional neural networks[J]. Bioinformatics, 2022, 38(3): 694-702. |
| 98 | GAO H L, CHEN C, LI S Y, et al. Prediction of protein-protein interactions based on ensemble residual convolutional neural network[J]. Computers in Biology and Medicine, 2023, 152: 106471. |
| 99 | SOLEYMANI F, PAQUET E, VIKTOR H L, et al. ProtInteract: a deep learning framework for predicting protein-protein interactions[J]. Computational and Structural Biotechnology Journal, 2023, 21: 1324-1348. |
| 100 | GONZALEZ-LOPEZ F, MORALES-CORDOVILLA J A, VILLEGAS-MORCILLO A, et al. End-to-end prediction of protein-protein interaction based on embedding and recurrent neural networks[C/OL]//2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). December 3-6, 2018, Madrid, Spain. IEEE, 2019: 2344-2350 [2023-10-01]. . |
| 101 | ZHANG B Z, LI J Y, QUAN L J, et al. Sequence-based prediction of protein-protein interaction sites by simplified long short-term memory network[J]. Neurocomputing, 2019, 357: 86-100. |
| 102 | ALAKUS T B, TURKOGLU I. Prediction of protein-protein interactions with LSTM deep learning model[C/OL]//2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT). October 11-13, 2019, Ankara, Turkey. IEEE, 2019: 1-5 [2023-10-01]. . |
| 103 | YANG L, HAN Y K, ZHANG H X, et al. Prediction of protein-protein interactions with local weight-sharing mechanism in deep learning[J]. BioMed Research International, 2020, 2020: 5072520. |
| 104 | XU H X, XU D, ZHANG N Q, et al. Protein-protein interaction prediction based on spectral radius and general regression neural network[J]. Journal of Proteome Research, 2021, 20(3): 1657-1665. |
| 105 | TSUKIYAMA S, HASAN M M, FUJII S, et al. LSTM-PHV: prediction of human-virus protein-protein interactions by LSTM with word2vec[J]. Briefings in Bioinformatics, 2021, 22(6): bbab228. |
| 106 | MEWARA B, LALWANI S. Sequence-based prediction of protein-protein interaction using auto-feature engineering of RNN-based model[J]. Research on Biomedical Engineering, 2023, 39(1): 259-272. |
| 107 | TANG M L, WU L X, YU X Y, et al. Prediction of protein-protein interaction sites based on stratified attentional mechanisms[J]. Frontiers in Genetics, 2021, 12: 784863. |
| 108 | TSUKIYAMA S, KURATA H. Cross-attention PHV: prediction of human and virus protein-protein interactions using cross-attention-based neural networks[J]. Computational and Structural Biotechnology Journal, 2022, 20: 5564-5573. |
| 109 | ASIM M N, IBRAHIM M ALI, MALIK M I, et al. ADH-PPI: an attention-based deep hybrid model for protein-protein interaction prediction[J]. iScience, 2022, 25(10): 105169. |
| 110 | LI X, HAN P F, WANG G, et al. SDNN-PPI: self-attention with deep neural network effect on protein-protein interaction prediction[J]. BMC Genomics, 2022, 23(1): 474. |
| 111 | MOU M J, PAN Z Q, ZHOU Z M, et al. A transformer-based ensemble framework for the prediction of protein-protein interaction sites[J]. Research, 2023, 6: 0240. |
| 112 | LI H, GONG X J, YU H A, et al. Deep neural network based predictions of protein interactions using primary sequences[J]. Molecules, 2018, 23(8): 1923. |
| 113 | CHEN M H, JU C J T, ZHOU G Y, et al. Multifaceted protein-protein interaction prediction based on Siamese residual RCNN[J]. Bioinformatics, 2019, 35(14): i305-i314. |
| 114 | GUO Y, CHEN X. A deep learning framework for improving protein interaction prediction using sequence properties[EB/OL]. bioRxiv, 2019: 843755[2023-10-01]. . |
| 115 | LI Y W, GOLDING G B, ILIE L. DELPHI: accurate deep ensemble model for protein interaction sites prediction[J]. Bioinformatics, 2021, 37(7): 896-904. |
| 116 | XU W X, GAO Y Y, WANG Y, et al. Protein-protein interaction prediction based on ordinal regression and recurrent convolutional neural networks[J]. BMC Bioinformatics, 2021, 22(): 485. |
| 117 | ALBU A I, BOCICOR M I, CZIBULA G. MM-StackEns: a new deep multimodal stacked generalization approach for protein-protein interaction prediction[J]. Computers in Biology and Medicine, 2023, 153: 106526. |
| 118 | DU X Q, SUN S W, HU C L, et al. DeepPPI: boosting prediction of protein-protein interactions with deep neural networks[J]. Journal of Chemical Information and Modeling, 2017, 57(6): 1499-1510. |
| 119 | YANG L, XIA J F, GUI J E. Prediction of protein-protein interactions from protein sequence using local descriptors[J]. Protein & Peptide Letters, 2010, 17(9): 1085-1090. |
| 120 | CHUNG J, GULCEHRE C, CHO K, et al. Empirical evaluation of gated recurrent neural networks on sequence modeling[EB/OL]. arXiv, 2014: 1412.3555[2023-10-01]. . |
| 121 | NAMBIAR A, LIU S, HEFLIN M, et al. Transformer neural networks for protein family and interaction prediction tasks[J]. Journal of Computational Biology, 2023, 30(1): 95-111. |
| 122 | JAISWAL A, BABU A R, ZADEH M Z, et al. A survey on contrastive self-supervised learning[J]. Technologies, 2020, 9(1): 2. |
| 123 | HU X, CHU L Y, PEI J, et al. Model complexity of deep learning: a survey[J]. Knowledge and Information Systems, 2021, 63(10): 2585-2619. |
| 124 | OUYANG L, WU J, JIANG X, et al. Training language models to follow instructions with human feedback[C/OL]//Advances in Neural Information Processing Systems 35 (NeurIPS 2022), 2022, 35: 27730-27744 [2023-10-01]. . |
| 125 | MIKOLOV T, CHEN K, CORRADO G, et al. Efficient estimation of word representations in vector space[EB/OL]. arXiv, 2013: 1301.3781[2023-10-01]. . |
| 126 | ELNAGGAR A, HEINZINGER M, DALLAGO C, et al. ProtTrans: toward understanding the language of life through self-supervised learning[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 44(10): 7112-7127. |
| 127 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| [1] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [2] | LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids [J]. Synthetic Biology Journal, 2024, 5(3): 507-526. |
| [3] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [4] | Sheng WANG, Zechen WANG, Weihua CHEN, Ke CHEN, Xiangda PENG, Fafen OU, Liangzhen ZHENG, Jinyuan SUN, Tao SHEN, Guoping ZHAO. Design of synthetic biology components based on artificial intelligence and computational biology [J]. Synthetic Biology Journal, 2023, 4(3): 422-443. |
| [5] | Liqi KANG, Pan TAN, Liang HONG. Enzyme engineering in the age of artificial intelligence [J]. Synthetic Biology Journal, 2023, 4(3): 524-534. |
| [6] | Qiaozhen MENG, Fei GUO. Applications of foldability in intelligent enzyme engineering and design: take AlphaFold2 for example [J]. Synthetic Biology Journal, 2023, 4(3): 571-589. |
| [7] | Yiming TANG, Yifei YAO, Zhongyuan YANG, Yun ZHOU, Zichao WANG, Guanghong WEI. Pathological aggregation and liquid-liquid phase separation of proteins associated with neurodegenerative diseases [J]. Synthetic Biology Journal, 2023, 4(3): 590-610. |
| [8] | Zhihang CHEN, Menglin JI, Yifei QI. Research progress of artificial intelligence in desiging protein structures [J]. Synthetic Biology Journal, 2023, 4(3): 464-487. |
| [9] | Qilong LAI, Shuai YAO, Yuguo ZHA, Hong BAI, Kang NING. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials [J]. Synthetic Biology Journal, 2023, 4(3): 611-627. |
| [10] | Yidong SONG, Qianmu YUAN, Yuedong YANG. Application of deep learning in protein function prediction [J]. Synthetic Biology Journal, 2023, 4(3): 488-506. |
| [11] | Jingwei LYU, Zixin DENG, Qi ZHANG, Wei DING. Identification of RiPPs precursor peptides and cleavage sites based on deep learning [J]. Synthetic Biology Journal, 2022, 3(6): 1262-1276. |
| [12] | Jiahao BIAN, Guangyu YANG. Artificial intelligence-assisted protein engineering [J]. Synthetic Biology Journal, 2022, 3(3): 429-444. |
| [13] | Ye WANG, Haochen WANG, Minghao YAN, Guanhua HU, Xiaowo WANG. Design of biomolecular sequences by artificial intelligence [J]. Synthetic Biology Journal, 2021, 2(1): 1-14. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||