Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (4): 781-794.DOI: 10.12211/2096-8280.2021-066
• Invited Review • Previous Articles Next Articles
Effects of mechanical signals on stem cell fate determination
SONG Chengzhi1, SUN Yang1, CAO Yi1,2
- 1.Department of physics,Nanjing University,Nanjing 210093,Jiangsu,China
2.Nanjing Laboratory of Solid State Microstructure,Nanjing University,Nanjing 210093,Jiangsu,China
-
Received:2021-06-09Revised:2021-10-30Online:2022-09-08Published:2022-08-31 -
Contact:CAO Yi
力信号在干细胞命运决定过程中的影响
宋成治1, 孙阳1, 曹毅1,2
- 1.南京大学物理学院,江苏 南京 210093
2.南京大学固体微结构物理国家重点实验室,江苏 南京 210093
-
通讯作者:曹毅 -
作者简介:宋成治 (1998—),男。研究方向为实验生物物理和理论生物物理。E-mail:857093988@qq.com曹毅 (1979—),男,教授,博士生导师。研究方向为单分子生物物理、蛋白质多肽生物材料、生物力学。E-mail:caoyi@nju.edu.cn -
基金资助:国家重点研发计划(2020YFA0908100)
CLC Number:
Cite this article
SONG Chengzhi, SUN Yang, CAO Yi. Effects of mechanical signals on stem cell fate determination[J]. Synthetic Biology Journal, 2022, 3(4): 781-794.
宋成治, 孙阳, 曹毅. 力信号在干细胞命运决定过程中的影响[J]. 合成生物学, 2022, 3(4): 781-794.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-066
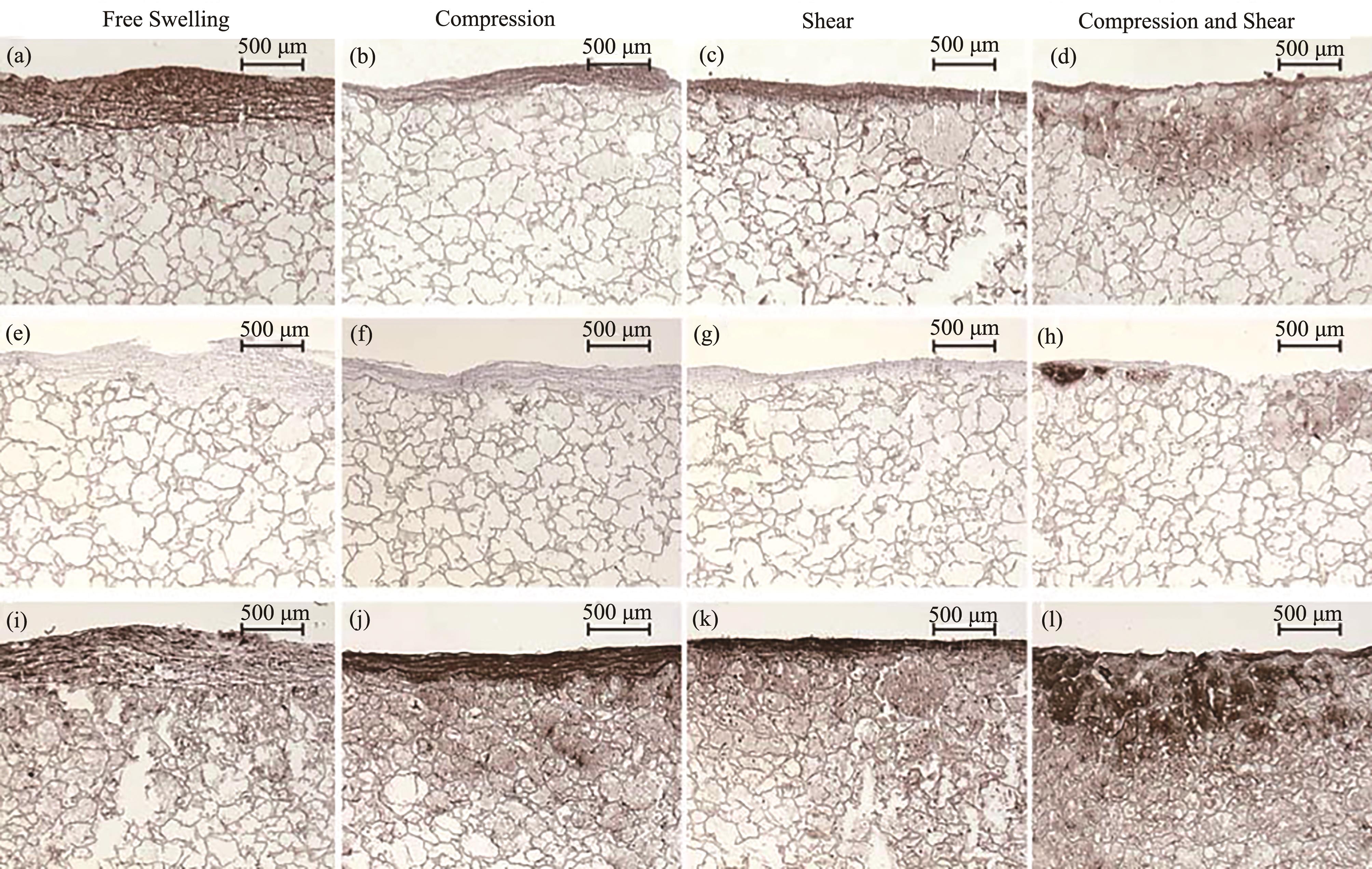
Fig. 2 Immunostaining for Col 1, Col 2, and Agg of MSC cryo-sections after different loading regimes[Agg(a~d), Col 2(e~h), Col 1(i~l). Agg and Col 2 are chondrogenic markers while Col 1 are osteogenic markers.]

Fig. 3 Schematic diagrams for mechanical signal transduction[External mechanical signal could be passed down to the nucleus either through a direct mechanical connection of ECM (extracellular matrix)-integrin-cytoskeleton or through some signaling molecules such as YAP/TAZ while cellular interaction is mediated through E-Cadherin (LINC: nucleoskeleton and cytoskeleton complex)]
| 1 | EVANS M J, KAUFMAN M H. Establishment in culture of pluripotential cells from mouse embryos[J]. Nature, 1981, 292(5819): 154-156. |
| 2 | DE LUCA M, AIUTI A, COSSU G, et al. Advances in stem cell research and therapeutic development[J]. Nature Cell Biology, 2019, 21(7): 801-811. |
| 3 | CHABANNON C, KUBALL J, BONDANZA A, et al. Hematopoietic stem cell transplantation in its 60 s: a platform for cellular therapies[J]. Science Translational Medicine, 2018, 10(436): eaap9630. |
| 4 | REN X, MOSER P T, GILPIN S E, et al. Engineering pulmonary vasculature in decellularized rat and human lungs[J]. Nature Biotechnology, 2015, 33(10): 1097-1102. |
| 5 | OTT H C, MATTHIESEN T S, GOH S K, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart[J]. Nature Medicine, 2008, 14(2): 213-221. |
| 6 | JIANG Y H, JAHAGIRDAR B N, REINHARDT R L, et al. Pluripotency of mesenchymal stem cells derived from adult marrow[J]. Nature, 2002, 418(6893): 41-49. |
| 7 | FU J P, WARMFLASH A, LUTOLF M P. Stem-cell-based embryo models for fundamental research and translation[J]. Nature Materials, 2021, 20(2): 132-144. |
| 8 | DISCHER D E, MOONEY D J, ZANDSTRA P W. Growth factors, matrices, and forces combine and control stem cells[J]. Science, 2009, 324(5935): 1673-1677. |
| 9 | CAMPBELL P A, PEREZ-IRATXETA C, ANDRADE-NAVARRO M A, et al. Oct4 targets regulatory nodes to modulate stem cell function[J]. PLoS One, 2007, 2(6): e553. |
| 10 | GRECO S J, LIU K, RAMESHWAR P. Functional similarities among genes regulated by OCT4 in human mesenchymal and embryonic stem cells[J]. Stem Cells, 2007, 25(12): 3143-3154. |
| 11 | ENGLER A J, SEN S, SWEENEY H L, et al. Matrix elasticity directs stem cell lineage specification[J]. Cell, 2006, 126(4): 677-689. |
| 12 | RHO J Y, ASHMAN R B, TURNER C H. Young's modulus of trabecular and cortical bone material: ultrasonic and microtensile measurements[J]. Journal of Biomechanics, 1993, 26(2): 111-119. |
| 13 | BANGASSER B L, SHAMSAN G A, CHAN C E, et al. Shifting the optimal stiffness for cell migration[J]. Nature Communications, 2017, 8: 15313. |
| 14 | VOGEL V, SHEETZ M. Local force and geometry sensing regulate cell functions[J]. Nature Reviews Molecular Cell Biology, 2006, 7(4): 265-275. |
| 15 | DISCHER D E, JANMEY P, WANG Y L. Tissue cells feel and respond to the stiffness of their substrate[J]. Science, 2005, 310(5751): 1139-1143. |
| 16 | ENGLER A, BACAKOVA L, NEWMAN C, et al. Substrate compliance versus ligand density in cell on gel responses[J]. Biophysical Journal, 2004, 86(1): 617-628. |
| 17 | PELHAM R J JR, WANG Y L. Cell locomotion and focal adhesions are regulated by substrate flexibility[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(25): 13661-13665. |
| 18 | WOLFENSON H, YANG B, SHEETZ M P. Steps in mechanotransduction pathways that control cell morphology[J]. Annual Review of Physiology, 2019, 81: 585-605. |
| 19 | SAHA K, KEUNG A J, IRWIN E F, et al. Substrate modulus directs neural stem cell behavior[J]. Biophysical Journal, 2008, 95(9): 4426-4438. |
| 20 | FU J P, WANG Y K, YANG M T, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates[J]. Nature Methods, 2010, 7(9): 733-736. |
| 21 | CHAUDHURI O, COOPER-WHITE J, JANMEY P A, et al. Effects of extracellular matrix viscoelasticity on cellular behaviour[J]. Nature, 2020, 584(7822): 535-546. |
| 22 | LEE H P, GU L, MOONEY D J, et al. Mechanical confinement regulates cartilage matrix formation by chondrocytes[J]. Nature Materials, 2017, 16(12): 1243-1251. |
| 23 | MADL C M, LESAVAGE B L, DEWI R E, et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling[J]. Nature Materials, 2017, 16(12): 1233-1242. |
| 24 | CAIAZZO M, OKAWA Y, RANGA A, et al. Defined three-dimensional microenvironments boost induction of pluripotency[J]. Nature Materials, 2016, 15(3): 344-352. |
| 25 | PAUL C D, MISTRIOTIS P, KONSTANTOPOULOS K. Cancer cell motility: lessons from migration in confined spaces[J]. Nature Reviews Cancer, 2017, 17(2): 131-140. |
| 26 | SCHULTZ K M, KYBURZ K A, ANSETH K S. Measuring dynamic cell-material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(29): E3757-E3764. |
| 27 | WISDOM K M, ADEBOWALE K, CHANG J L, et al. Matrix mechanical plasticity regulates cancer cell migration through confining microenvironments[J]. Nature Communications, 2018, 9: 4144. |
| 28 | CAMERON A R, FRITH J E, COOPER-WHITE J J. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype[J]. Biomaterials, 2011, 32(26): 5979-5993. |
| 29 | CHAUDHURI O, GU L, DARNELL M, et al. Substrate stress relaxation regulates cell spreading[J]. Nature Communications, 2015, 6: 6364. |
| 30 | CHAUDHURI O, GU L, KLUMPERS D, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity[J]. Nature Materials, 2016, 15(3): 326-334. |
| 31 | NAM S, GUPTA V K, LEE H P, et al. Cell cycle progression in confining microenvironments is regulated by a growth-responsive TRPV4-PI3K/Akt-p27 Kip1 signaling axis[J]. Science Advances, 2019, 5(8): eaaw6171. |
| 32 | NAM S, CHAUDHURI O. Mitotic cells generate protrusive extracellular forces to divide in three-dimensional microenvironments[J]. Nature Physics, 2018, 14(6): 621-628. |
| 33 | HUEBSCH N, LIPPENS E, LEE K, et al. Matrix elasticity of void-forming hydrogels controls transplanted-stem-cell-mediated bone formation[J]. Nature Materials, 2015, 14(12): 1269-1277. |
| 34 | HUEBSCH N, ARANY P R, MAO A S, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate[J]. Nature Materials, 2010, 9(6): 518-526. |
| 35 | BANGASSER B L, ROSENFELD S S, ODDE D J. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment[J]. Biophysical Journal, 2013, 105(3): 581-592. |
| 36 | ELOSEGUI-ARTOLA A, ORIA R, CHEN Y F, et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity[J]. Nature Cell Biology, 2016, 18(5): 540-548. |
| 37 | GONG Z, SZCZESNY S E, CALIARI S R, et al. Matching material and cellular timescales maximizes cell spreading on viscoelastic substrates[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): E2686-E2695. |
| 38 | UROZ M, GARCIA-PUIG A, TEKELI I, et al. Traction forces at the cytokinetic ring regulate cell division and polyploidy in the migrating zebrafish epicardium[J]. Nature Materials, 2019, 18(9): 1015-1023. |
| 39 | ATALA A, KIM W, PAIGE K T, et al. Endoscopic treatment of vesicoureteral reflux with a chondrocyte-alginate suspension[J]. The Journal of Urology, 1994, 152(2): 641-643. |
| 40 | CHHETRI D K, MENDELSOHN A H. Hyaluronic acid for the treatment of vocal fold scars[J]. Current Opinion in Otolaryngology & Head and Neck Surgery, 2010, 18(6): 498-502. |
| 41 | LIN X, LIU Y, BAI A B, et al. A viscoelastic adhesive epicardial patch for treating myocardial infarction[J]. Nature Biomedical Engineering, 2019, 3(8): 632-643. |
| 42 | RUVINOV E, COHEN S. Alginate biomaterial for the treatment of myocardial infarction: progress, translational strategies, and clinical outlook: from ocean algae to patient bedside[J]. Advanced Drug Delivery Reviews, 2016, 96: 54-76. |
| 43 | CUI Y D, HAMEED F M, YANG B, et al. Cyclic stretching of soft substrates induces spreading and growth[J]. Nature Communications, 2015, 6: 6333. |
| 44 | SAHA S, JI L, DE PABLO J J, et al. Inhibition of human embryonic stem cell differentiation by mechanical strain[J]. Journal of Cellular Physiology, 2006, 206(1): 126-137. |
| 45 | LI C W, LAU Y T, LAM K L, et al. Mechanically induced formation and maturation of 3D-matrix adhesions (3DMAs) in human mesenchymal stem cells[J]. Biomaterials, 2020, 258: 120292. |
| 46 | MAUCK R L, NICOLL S B, SEYHAN S L, et al. Synergistic action of growth factors and dynamic loading for articular cartilage tissue engineering[J]. Tissue Engineering, 2003, 9(4): 597-611. |
| 47 | SCHÄTTI O, GRAD S, GOLDHAHN J, et al. A combination of shear and dynamic compression leads to mechanically induced chondrogenesis of human mesenchymal stem cells[J]. European Cells & Materials, 2011, 22: 214-225. |
| 48 | HUANG A H, FARRELL M J, KIM M, et al. Long-term dynamic loading improves the mechanical properties of chondrogenic mesenchymal stem cell-laden hydrogel[J]. European Cells & Materials, 2010, 19: 72-85. |
| 49 | BAKER B M, SHAH R P, HUANG A H, et al. Dynamic tensile loading improves the functional properties of mesenchymal stem cell-laden nanofiber-based fibrocartilage[J]. Tissue Engineering Part A, 2011, 17(9/10): 1445-1455. |
| 50 | ISKRATSCH T, WOLFENSON H, SHEETZ M P. Appreciating force and shape-the rise of mechanotransduction in cell biology[J]. Nature Reviews Molecular Cell Biology, 2014, 15(12): 825-833. |
| 51 | SWAMINATHAN V, WATERMAN C M. The molecular clutch model for mechanotransduction evolves[J]. Nature Cell Biology, 2016, 18(5): 459-461. |
| 52 | MITCHISON T, KIRSCHNER M. Cytoskeletal dynamics and nerve growth[J]. Neuron, 1988, 1(9): 761-772. |
| 53 | VINING K H, MOONEY D J. Mechanical forces direct stem cell behaviour in development and regeneration[J]. Nature Reviews Molecular Cell Biology, 2017, 18(12): 728-742. |
| 54 | HAMADI A, BOUALI M, DONTENWILL M, et al. Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397[J]. Journal of Cell Science, 2005, 118(Pt 19): 4415-4425. |
| 55 | PRAGER-KHOUTORSKY M, LICHTENSTEIN A, KRISHNAN R, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing[J]. Nature Cell Biology, 2011, 13(12): 1457-1465. |
| 56 | TRICHET L, LE DIGABEL J, HAWKINS R J, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(18): 6933-6938. |
| 57 | VICENTE-MANZANARES M, ZARENO J, WHITMORE L, et al. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells (vol 176, pg 573, 2007)[J]. Journal of Cell Biology, 2007, 176(7): 1073. |
| 58 | WOLFENSON H, BERSHADSKY A, HENIS Y I, et al. Actomyosin-generated tension controls the molecular kinetics of focal adhesions[J]. Journal of Cell Science, 2011, 124(Pt 9): 1425-1432. |
| 59 | WENG S N, SHAO Y, CHEN W Q, et al. Mechanosensitive subcellular rheostasis drives emergent single-cell mechanical homeostasis[J]. Nature Materials, 2016, 15(9): 961-967. |
| 60 | CHOI C K, VICENTE-MANZANARES M, ZARENO J, et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner[J]. Nature Cell Biology, 2008, 10(9): 1039-1050. |
| 61 | YU C H, LAW J B K, SURYANA M, et al. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(51): 20585-20590. |
| 62 | SAXENA M, CHANGEDE R, HONE J, et al. Force-induced calpain cleavage of talin is critical for growth, adhesion development, and rigidity sensing[J]. Nano Letters, 2017, 17(12): 7242-7251. |
| 63 | SAXENA M, LIU S M, YANG B, et al. EGFR and HER2 activate rigidity sensing only on rigid matrices[J]. Nature Materials, 2017, 16(7): 775-781. |
| 64 | ROCA-CUSACHS P, DEL RIO A, PUKLIN-FAUCHER E, et al. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(15): E1361-E1370. |
| 65 | ZAMIR E, KATZ M, POSEN Y, et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts[J]. Nature Cell Biology, 2000, 2(4): 191-196. |
| 66 | ACHARYA B R, WU S K, LIEU Z Z, et al. Mammalian diaphanous 1 mediates a pathway for E-cadherin to stabilize epithelial barriers through junctional contractility[J]. Cell Reports, 2017, 18(12): 2854-2867. |
| 67 | BUCKLEY C D, TAN J Y, ANDERSON K L, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force[J]. Science, 2014, 346(6209): 1254211. |
| 68 | COLLINS C, DENISIN A K, PRUITT B L, et al. Changes in E-cadherin rigidity sensing regulate cell adhesion[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(29): E5835-E5844. |
| 69 | NEJSUM L N, NELSON W J. Epithelial cell surface polarity: The early steps[J]. Frontiers in Bioscience (Landmark Edition), 2009, 14(3): 1088-1098. |
| 70 | STRALE P O, DUCHESNE L, PEYRET G, et al. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity (vol 210, pg 333, 2015)[J]. Journal of Cell Biology, 2015, 210(6): 1033. |
| 71 | WU S K, BUDNAR S, YAP A S, et al. Pulsatile contractility of actomyosin networks organizes the cellular cortex at lateral cadherin junctions[J]. European Journal of Cell Biology, 2014, 93(10/11/12): 396-404. |
| 72 | WU Y, KANCHANAWONG P, ZAIDEL-BAR R. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions[J]. Developmental Cell, 2015, 32(2): 139-154. |
| 73 | REDDY P, LIU L, REN C, et al. Formation of E-cadherin-mediated cell-cell adhesion activates AKT and mitogen activated protein kinase via phosphatidylinositol 3 kinase and ligand-independent activation of epidermal growth factor receptor in ovarian cancer cells[J]. Molecular Endocrinology, 2005, 19(10): 2564-2578. |
| 74 | SWIFT J, IVANOVSKA I L, BUXBOIM A, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation[J]. Science, 2013, 341(6149): 1240104. |
| 75 | TAJIK A, ZHANG Y J, WEI F X, et al. Transcription upregulation via force-induced direct stretching of chromatin[J]. Nature Materials, 2016, 15(12): 1287-1296. |
| 76 | WOLFENSON H, MEACCI G, LIU S M, et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices[J]. Nature Cell Biology, 2016, 18(1): 33-42. |
| 77 | GIANNONE G, DUBIN-THALER B J, ROSSIER O, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation[J]. Cell, 2007, 128(3): 561-575. |
| 78 | NAUMANEN P, LAPPALAINEN P, HOTULAINEN P. Mechanisms of actin stress fibre assembly[J]. Journal of Microscopy, 2008, 231(3): 446-454. |
| 79 | IVANOVSKA J, MAHADEVAN V, SCHNEIDER-STOCK R. DAPK and cytoskeleton-associated functions[J]. Apoptosis, 2014, 19(2): 329-338. |
| 80 | BALESTRINI J L, CHAUDHRY S, SARRAZY V, et al. The mechanical memory of lung myofibroblasts[J]. Integrative Biology, 2012, 4(4): 410-421. |
| 81 | YANG C, TIBBITT M W, BASTA L, et al. Mechanical memory and dosing influence stem cell fate[J]. Nature Materials, 2014, 13(6): 645-652. |
| 82 | VOGEL V, SHEETZ M. Local force and geometry sensing regulate cell functions[J]. Nature Reviews Molecular Cell Biology, 2006, 7(4): 265-275. |
| 83 | GASPAR P, TAPON N. Sensing the local environment: actin architecture and Hippo signalling[J]. Current Opinion in Cell Biology, 2014, 31: 74-83. |
| 84 | PANCIERA T, AZZOLIN L, CORDENONSI M, et al. Mechanobiology of YAP and TAZ in physiology and disease[J]. Nature Reviews Molecular Cell Biology, 2017, 18(12): 758-770. |
| 85 | TOTARO A, PANCIERA T, PICCOLO S. YAP/TAZ upstream signals and downstream responses[J]. Nature Cell Biology, 2018, 20(8): 888-899. |
| 86 | MILLER C J, DAVIDSON L A. The interplay between cell signalling and mechanics in developmental processes[J]. Nature Reviews Genetics, 2013, 14(10): 733-744. |
| 87 | HENG B C, ZHANG X H, AUBEL D, et al. An overview of signaling pathways regulating YAP/TAZ activity[J]. Cellular and Molecular Life Sciences: CMLS, 2021, 78(2): 497-512. |
| 88 | POCATERRA A, ROMANI P, DUPONT S. YAP/TAZ functions and their regulation at a glance[J]. Journal of Cell Science, 2020, 133(2): jcs230425. |
| 89 | MEIER E, LAM M T. Role of mechanical stimulation in stem cell differentiation[J]. Biotechnology and Bioengineering, 2016, 3(3): 1069. |
| 90 | SAHA S, JI L, DE PABLO J J, et al. TGFbeta/Activin/Nodal pathway in inhibition of human embryonic stem cell differentiation by mechanical strain[J]. Biophysical Journal, 2008, 94(10): 4123-4133. |
| 91 | MAKITA R, UCHIJIMA Y, NISHIYAMA K, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ[J]. American Journal of Physiology Renal Physiology, 2008, 294(3): F542-F553. |
| 92 | MORIN-KENSICKI E M, BOONE B N, HOWELL M, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65[J]. Molecular and Cellular Biology, 2006, 26(1): 77-87. |
| 93 | ARAGONA M, PANCIERA T, MANFRIN A, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors[J]. Cell, 2013, 154(5): 1047-1059. |
| 94 | DUPONT S, MORSUT L, ARAGONA M, et al. Role of YAP/TAZ in mechanotransduction[J]. Nature, 2011, 474(7350): 179-183. |
| 95 | ROSENBLUH J, NIJHAWAN D, COX A G, et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis[J]. Cell, 2012, 151(7): 1457-1473. |
| 96 | SERO J E, BAKAL C. Multiparametric analysis of cell shape demonstrates that β-PIX directly couples YAP activation to extracellular matrix adhesion[J]. Cell Systems, 2017, 4(1): 84-96.e6. |
| 97 | ZHAO B, LI L, LU Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein[J]. Genes & Development, 2011, 25(1): 51-63. |
| 98 | MANA-CAPELLI S, PARAMASIVAM M, DUTTA S, et al. Angiomotins link F-actin architecture to Hippo pathway signaling[J]. Molecular Biology of the Cell, 2014, 25(10): 1676-1685. |
| 99 | KIRBY T J, LAMMERDING J. Emerging views of the nucleus as a cellular mechanosensor[J]. Nature Cell Biology, 2018, 20(4): 373-381. |
| 100 | PATHAK M M, NOURSE J L, TRAN T, et al. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(45): 16148-16153. |
| 101 | ZHAO B, LI L, WANG L, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis[J]. Genes & Development, 2012, 26(1): 54-68. |
| 102 | ELOSEGUI-ARTOLA A, ANDREU I, BEEDLE A E M, et al. Force triggers YAP nuclear entry by regulating transport across nuclear pores[J]. Cell, 2017, 171(6): 1397-1410.e14. |
| 103 | CONNELLY J T, GAUTROT J E, TRAPPMANN B, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions[J]. Nature Cell Biology, 2010, 12(7): 711-718. |
| 104 | VARTIAINEN M K, GUETTLER S, LARIJANI B, et al. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL[J]. Science, 2007, 316(5832): 1749-1752. |
| 105 | LEE J Y, CHANG J K, DOMINGUEZ A A, et al. YAP-independent mechanotransduction drives breast cancer progression[J]. Nature Communications, 2019, 10: 1848. |
| 106 | COSTE B, XIAO B L, SANTOS J S, et al. Piezo proteins are pore-forming subunits of mechanically activated channels[J]. Nature, 2012, 483(7388): 176-181. |
| 107 | LIN Y C, GUO Y R, MIYAGI A, et al. Force-induced conformational changes in PIEZO1[J]. Nature, 2019, 573(7773): 230-234. |
| 108 | 汝一雯, 张卫兵. 细胞力学信号转导中Piezo1机械敏感离子通道作用研究进展[J]. 中国细胞生物学学报, 2019, 41(8): 1622-1627. |
| RU Y W, ZHANG W B. Advances in the role of mechanosensitive ion channel Piezo1 in cellular mechanotransduction[J]. Chinese Journal of Cell Biology, 2019, 41(8): 1622-1627. | |
| 109 | TSE J R, ENGLER A J. Preparation of hydrogel substrates with tunable mechanical properties[J]. Current Protocols in Cell Biology, 2010, 47(1): 10.16.1-10.16.16. |
| 110 | JONKER A M, BODE S A, KUSTERS A H, et al. Soft PEG-hydrogels with independently tunable stiffness and RGDS-content for cell adhesion studies[J]. Macromolecular Bioscience, 2015, 15(10): 1338-1347. |
| 111 | DAVIRAN M, CATALANO J, SCHULTZ K M. Determining how human mesenchymal stem cells change their degradation strategy in response to microenvironmental stiffness[J]. Biomacromolecules, 2020, 21(8): 3056-3068. |
| 112 | TAN J L, TIEN J, PIRONE D M, et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(4): 1484-1489. |
| 113 | GHASSEMI S, MEACCI G, LIU S M, et al. Cells test substrate rigidity by local contractions on submicrometer pillars[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(14): 5328-5333. |
| 114 | TRICHET L, LE DIGABEL J, HAWKINS R J, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(18): 6933-6938. |
| 115 | SUN Y B, YONG K M A, VILLA-DIAZ L G, et al. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells[J]. Nature Materials, 2014, 13(6): 599-604. |
| 116 | BAI T, SUN F, ZHANG L, et al. Restraint of the differentiation of mesenchymal stem cells by a nonfouling zwitterionic hydrogel[J]. Angewandte Chemie International Edition, 2014, 53(47): 12729-12734. |
| 117 | HEO S J, SZCZESNY S E, KIM D H, et al. Expansion of mesenchymal stem cells on electrospun scaffolds maintains stemness, mechano-responsivity, and differentiation potential[J]. Journal of Orthopaedic Research, 2018, 36(2): 808-815. |
| 118 | JAIN N, IYER K V, KUMAR A, et al. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(28): 11349-11354. |
| 119 | MCWHORTER F Y, WANG T T, NGUYEN P, et al. Modulation of macrophage phenotype by cell shape[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(43): 17253-17258. |
| 120 | NEVES S R, TSOKAS P, SARKAR A, et al. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks[J]. Cell, 2008, 133(4): 666-680. |
| 121 | RANGAMANI P, LIPSHTAT A, AZELOGLU E U, et al. Decoding information in cell shape[J]. Cell, 2013, 154(6): 1356-1369. |
| 122 | ZHONG W L, ZHANG W G, WANG S Y, et al. Regulation of fibrochondrogenesis of mesenchymal stem cells in an integrated microfluidic platform embedded with biomimetic nanofibrous scaffolds[J]. PLoS One, 2013, 8(4): e61283. |
| 123 | SAUER F, OSWALD L, DE SCHELLENBERGER A A, et al. Collagen networks determine viscoelastic properties of connective tissues yet do not hinder diffusion of the aqueous solvent[J]. Soft Matter, 2019, 15(14): 3055-3064. |
| 124 | CHARRIER E E, POGODA K, WELLS R G, et al. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation[J]. Nature Communications, 2018, 9: 449. |
| 125 | YANG Z G, KOU S Z, WEI X, et al. Genetically programming stress-relaxation behavior in entirely protein-based molecular networks[J]. ACS Macro Letters, 2018, 7(12): 1468-1474. |
| 126 | BALIKOV D A, NEAL E H, LIPPMANN E S. Organotypic neurovascular models: past results and future directions[J]. Trends in Molecular Medicine, 2020, 26(3): 273-284. |
| 127 | BOEKHOVEN J, STUPP S I. 25th anniversary article: supramolecular materials for regenerative medicine[J]. Advanced Materials, 2014, 26(11): 1642-1659. |
| 128 | PRIOR N, INACIO P, HUCH M. Liver organoids: from basic research to therapeutic applications[J]. Gut, 2019, 68(12): 2228-2237. |
| 129 | SATO T, CLEVERS H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications[J]. Science, 2013, 340(6137): 1190-1194. |
| 130 | SHANSKY J, DEL TATTO M, CHROMIAK J, et al. A simplified method for tissue engineering skeletal muscle organoids in vitro [J]. In Vitro Cellular & Developmental Biology Animal, 1997, 33(9): 659-661. |
| 131 | BRAUNECKER W A, MATYJASZEWSKI K. Controlled/living radical polymerization: features, developments, and perspectives[J]. Progress in Polymer Science, 2007, 32(1): 93-146. |
| 132 | ONG L L, HANIKEL N, YAGHI O K, et al. Programmable self-assembly of three-dimensional nanostructures from 10 000 unique components[J]. Nature, 2017, 552(7683): 72-77. |
| 133 | ROSALES A M, ANSETH K S. The design of reversible hydrogels to capture extracellular matrix dynamics[J]. Nature Reviews Materials, 2016, 1: 15012. |
| 134 | WANG R, YANG Z G, LUO J R, et al. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(23): 5912-5917. |
| 135 | YANG Z G, YANG Y, WANG M, et al. Dynamically tunable, macroscopic molecular networks enabled by cellular synthesis of 4-arm star-like proteins[J]. Matter, 2020, 2(1): 233-249. |
| 136 | ZHANG X L, DONG C M, HUANG W Y, et al. Rational design of a photo-responsive UVR8-derived protein and a self-assembling peptide-protein conjugate for responsive hydrogel formation[J]. Nanoscale, 2015, 7(40): 16666-16670. |
| 137 | BAKER B M, CHEN C S. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues[J]. Journal of Cell Science, 2012, 125(Pt 13): 3015-3024. |
| 138 | CALIARI S R, VEGA S L, KWON M, et al. Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments[J]. Biomaterials, 2016, 103: 314-323. |
| 139 | GERECHT S, BURDICK J A, FERREIRA L S, et al. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(27): 11298-11303. |
| 140 | HARRIS A K, WILD P, STOPAK D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion[J]. Science, 1980, 208(4440): 177-179. |
| 141 | OLIVER T, DEMBO M, JACOBSON K. Measurement of traction forces in cells locomoting along a substratum[J]. Cell Motility and the Cytoskeleton, 1998, 39(4): 342-344. |
| 142 | DEL RIO A, PEREZ-JIMENEZ R, LIU R C, et al. Stretching single talin rod molecules activates vinculin binding[J]. Science, 2009, 323(5914): 638-641. |
| 143 | RIEF M, GAUTEL M, OESTERHELT F, et al. Reversible unfolding of individual titin immunoglobulin domains by AFM[J]. Science, 1997, 276(5315): 1109-1112. |
| 144 | GRASHOFF C, HOFFMAN B D, BRENNER M D, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics[J]. Nature, 2010, 466(7303): 263-266. |
| 145 | CHEN B C, LEGANT W R, WANG K, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution[J]. Science, 2014, 346(6208): 1257998. |
| 146 | GARAKANI K, SHAMS H, MOFRAD M R K. Mechanosensitive conformation of vinculin regulates its binding to MAPK1 [J]. Biophysical Journal, 2017, 112(9): 1885-1893. |
| [1] | CAO Rongkai, QIN Jianhua, WANG Yaqing. Advances in placenta-on-a-chip for reproductive medicine research [J]. Synthetic Biology Journal, 2024, 5(4): 831-850. |
| [2] | HU Bowen, TAN Jiaping, LIU Xiaodong. Advances in the development of human embryo models [J]. Synthetic Biology Journal, 2024, 5(4): 719-733. |
| [3] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [4] | HAN Yizhao, GUO Jia, SHAO Yue. Stem cell-based synthetic development: cellular components, embryonic models, and engineering approaches [J]. Synthetic Biology Journal, 2024, 5(4): 734-753. |
| [5] | LI Shikai, ZENG Dong′ao, DU Fangzhou, ZHANG Jingzhong, YU Shuang. The construction approaches and biomaterials for vascularized organoids [J]. Synthetic Biology Journal, 2024, 5(4): 851-866. |
| [6] | AI Zongyong, ZHANG Chengting, NIU Baohua, YIN Yu, YANG Jie, LI Tianqing. Early human embryo development and stem cells [J]. Synthetic Biology Journal, 2024, 5(4): 700-718. |
| [7] | ZHU Liyu, ZHAO Yulong, LI Wei, WANG Libin. Progress in mammalian chromosome engineering [J]. Synthetic Biology Journal, 2023, 4(2): 394-406. |
| [8] | ZHANG Can, SHI Liyang, DAI Jianwu. Cultured meat from biomaterials: challenges and prospects [J]. Synthetic Biology Journal, 2022, 3(4): 676-689. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||