Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (3): 548-560.DOI: 10.12211/2096-8280.2023-090
• Invited Review • Previous Articles Next Articles
Bacterial inter-PKS hybrids and the biosynthetic algorithm of polyketides
ZHANG Rui, JIN Wenzheng, CHEN Yijun
- Laboratory of Chemical Biology,School of Life Science and Technology,China Pharmaceutical University,Nanjing 211198,Jiangsu,China
-
Received:2023-11-28Revised:2024-03-04Online:2024-07-12Published:2024-06-30 -
Contact:CHEN Yijun
细菌聚酮合酶间的杂合方式及聚酮化合物生物合成逻辑
张瑞, 金文铮, 陈依军
- 中国药科大学生命科学与技术学院化学生物学教研室,江苏 南京 211198
-
通讯作者:陈依军 -
作者简介:张瑞 (1999—),女,硕士研究生。研究方向为聚酮化合物的生物合成。E-mail:zhangrui19990303@163.com陈依军 (1962—),男,教授。研究方向为药物合成生物学。E-mail:yjchen@cpu.edu.cn
CLC Number:
Cite this article
ZHANG Rui, JIN Wenzheng, CHEN Yijun. Bacterial inter-PKS hybrids and the biosynthetic algorithm of polyketides[J]. Synthetic Biology Journal, 2024, 5(3): 548-560.
张瑞, 金文铮, 陈依军. 细菌聚酮合酶间的杂合方式及聚酮化合物生物合成逻辑[J]. 合成生物学, 2024, 5(3): 548-560.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-090
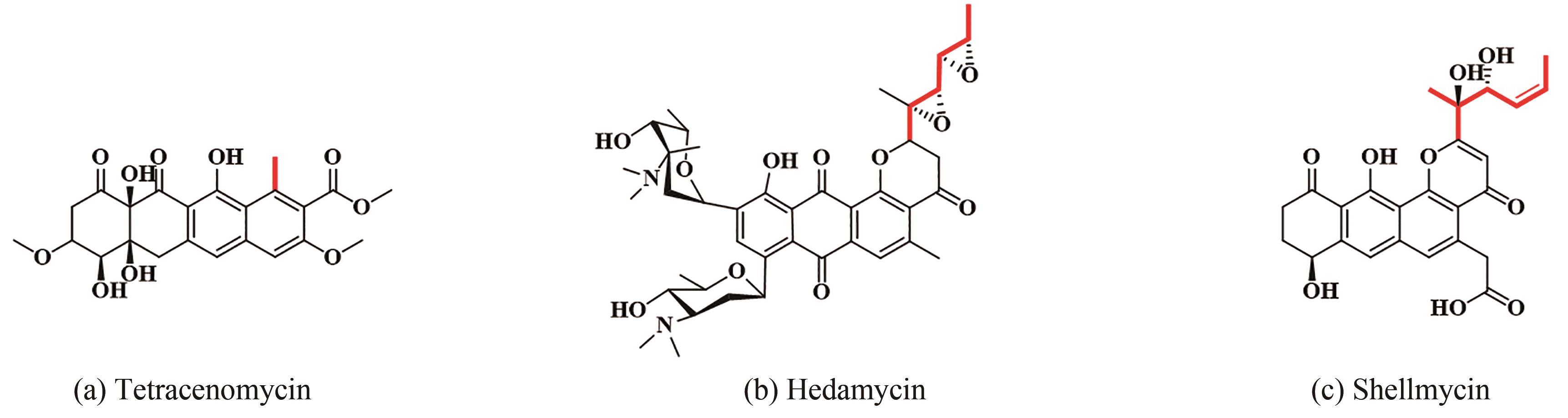
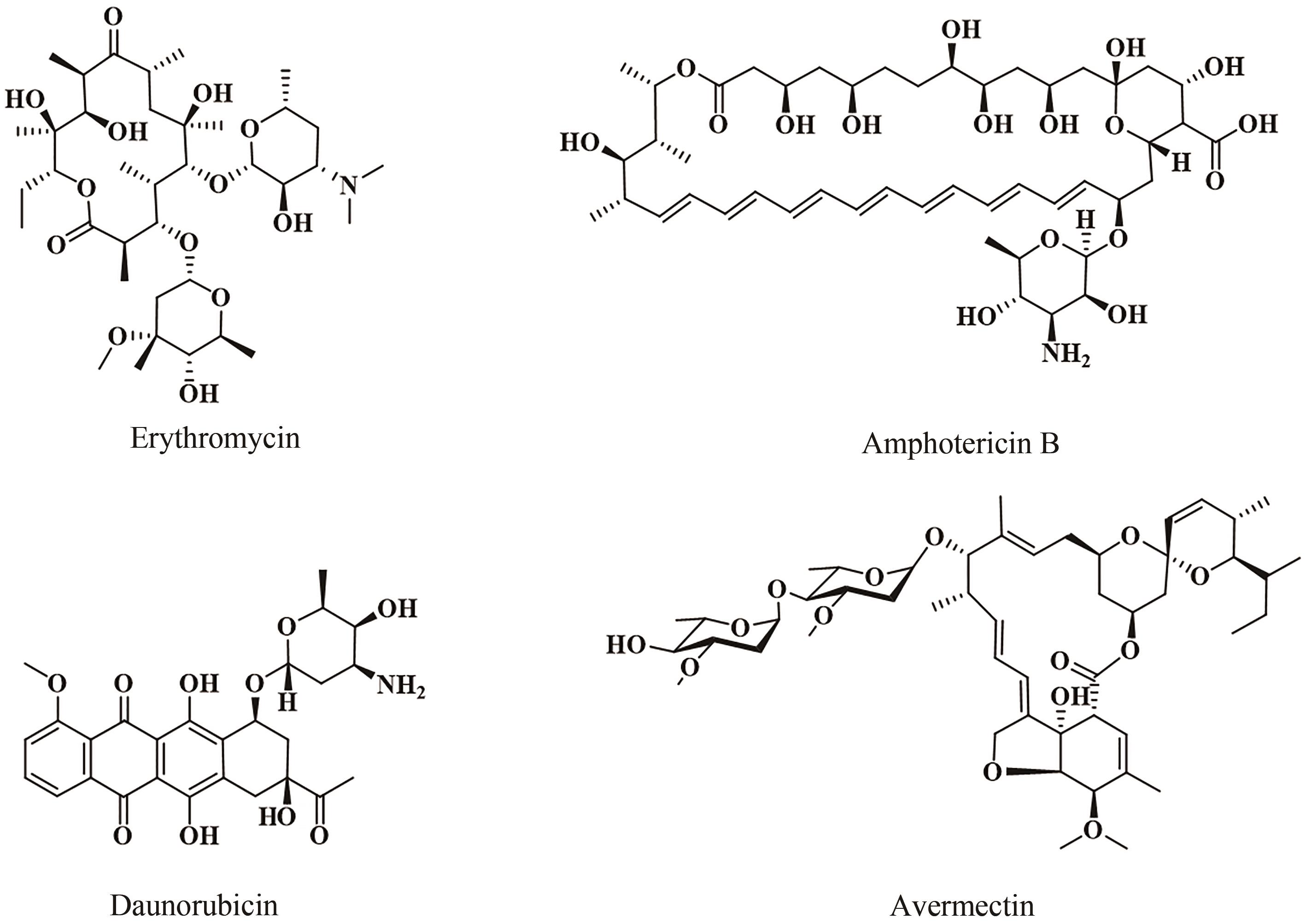
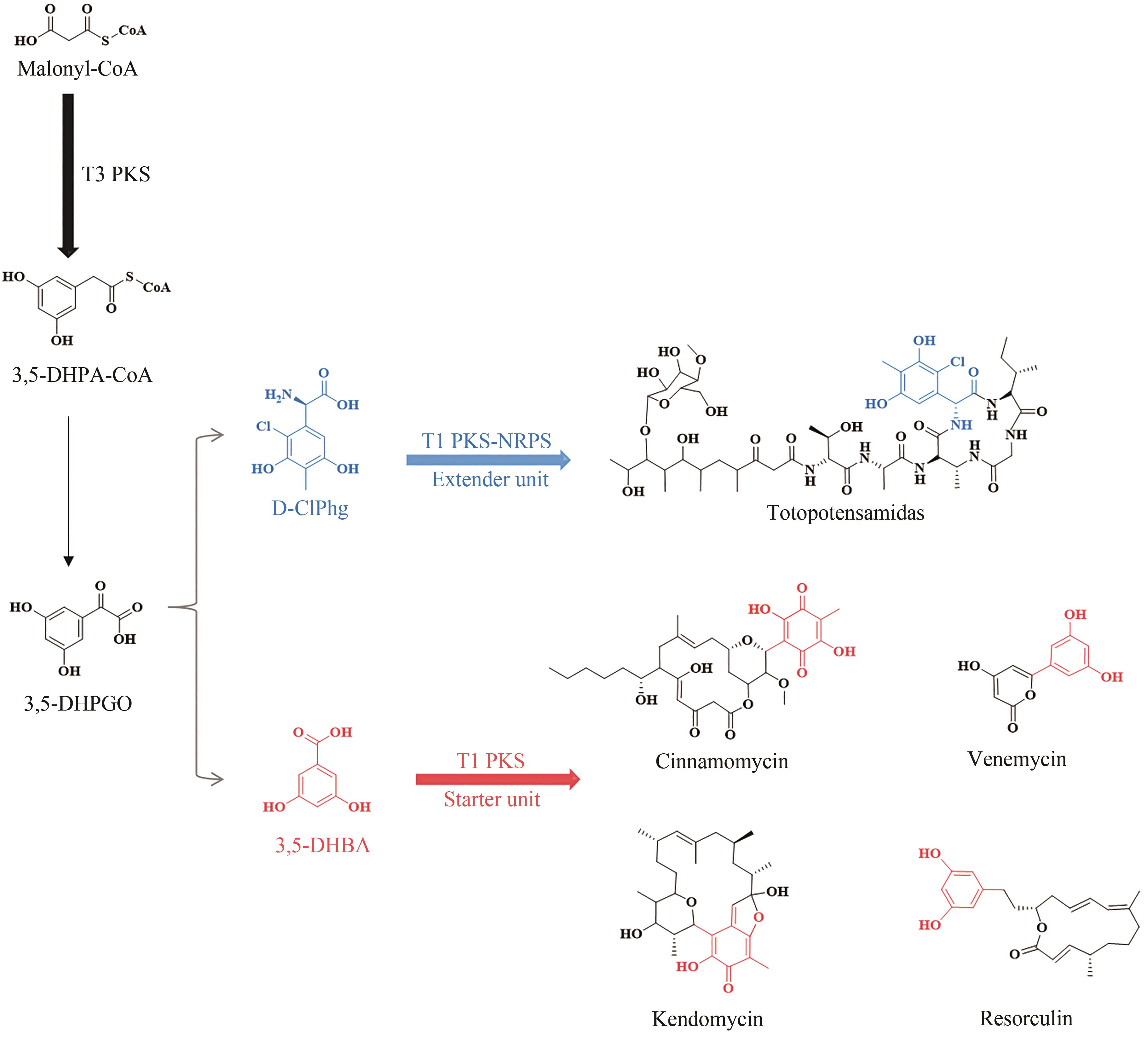
Fig. 5 Structures of products synthesized under the catalysis of type Ⅱ PKS and type Ⅰ/Ⅱ PKS hybrids(The moieties from starter units in polyketidess are highlighted in red.)
| 1 | TOOPAANG W, BUNNAK W, SRISUKSAM C, et al. Microbial polyketides and their roles in insect virulence: from genomics to biological functions[J]. Natural Product Reports, 2022, 39(11): 2008-2029. |
| 2 | HUR G H, VICKERY C R, BURKART M D. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology[J]. Natural Product Reports, 2012, 29(10): 1074-1098. |
| 3 | AVALOS M, GARBEVA P, VADER L, et al. Biosynthesis, evolution and ecology of microbial terpenoids[J]. Natural Product Reports, 2022, 39(2): 249-272. |
| 4 | MONTALBÁN-LÓPEZ M, SCOTT T A, RAMESH S, et al. New developments in RiPP discovery, enzymology and engineering[J]. Natural Product Reports, 2021, 38(1): 130-239. |
| 5 | LI S S, YANG B W, TAN G Y, et al. Polyketide pesticides from actinomycetes[J]. Current Opinion in Biotechnology, 2021, 69: 299-307. |
| 6 | GRAHAM E M. Erythromycin[J]. Obstetrics and Gynecology Clinics of North America, 1992, 19(3): 539-549. |
| 7 | HAMILL R J. Amphotericin B formulations: a comparative review of efficacy and toxicity[J]. Drugs, 2013, 73(9): 919-934. |
| 8 | POURMADADI M, GHAEMI A, SHAMSABADIPOUR A, et al. Nanoparticles loaded with Daunorubicin as an advanced tool for cancer therapy[J]. European Journal of Medicinal Chemistry, 2023, 258: 115547. |
| 9 | IKEDA H, OMURA S. Avermectin biosynthesis[J]. Chemical Reviews, 1997, 97(7): 2591-2610. |
| 10 | HOPWOOD D A. Genetic contributions to understanding polyketide synthases[J]. Chemical Reviews, 1997, 97(7): 2465-2498. |
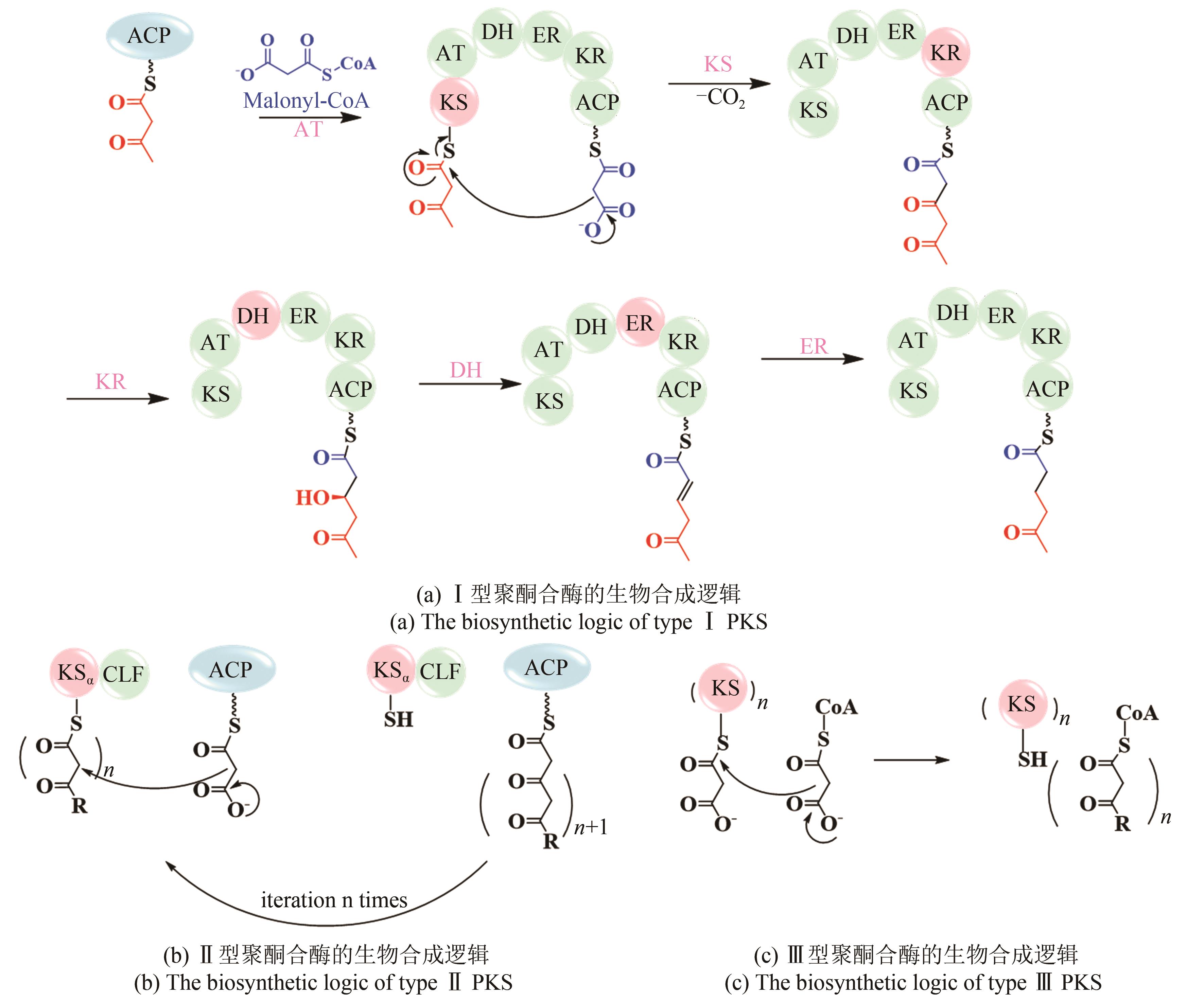
| 11 | SHEN B. Polyketide biosynthesis beyond the type Ⅰ, Ⅱ and Ⅲ polyketide synthase paradigms[J]. Current Opinion in Chemical Biology, 2003, 7(2): 285-295. |
| 12 | KEATINGE-CLAY A T. The structures of type Ⅰ polyketide synthases[J]. Natural Product Reports, 2012, 29(10): 1050-1073. |
| 13 | HERTWECK C. The biosynthetic logic of polyketide diversity[J]. Angewandte Chemie International Edition, 2009, 48(26): 4688-4716. |
| 14 | KEATINGE-CLAY A T. Stereocontrol within polyketide assembly lines[J]. Natural Product Reports, 2016, 33(2): 141-149. |
| 15 | CHEN H T, DU L C. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria[J]. Applied Microbiology and Biotechnology, 2016, 100(2): 541-557. |
| 16 | SONG L J, JENNER M, MASSCHELEIN J, et al. Discovery and biosynthesis of gladiolin: a Burkholderia gladioli antibiotic with promising activity against Mycobacterium tuberculosis [J]. Journal of the American Chemical Society, 2017, 139(23): 7974-7981. |
| 17 | DAS A, KHOSLA C. Biosynthesis of aromatic polyketides in bacteria[J]. Accounts of Chemical Research, 2009, 42(5): 631-639. |
| 18 | DREIER J, KHOSLA C. Mechanistic analysis of a type Ⅱ polyketide synthase. Role of conserved residues in the beta-ketoacyl synthase-chain length factor heterodimer[J]. Biochemistry, 2000, 39(8): 2088-2095. |
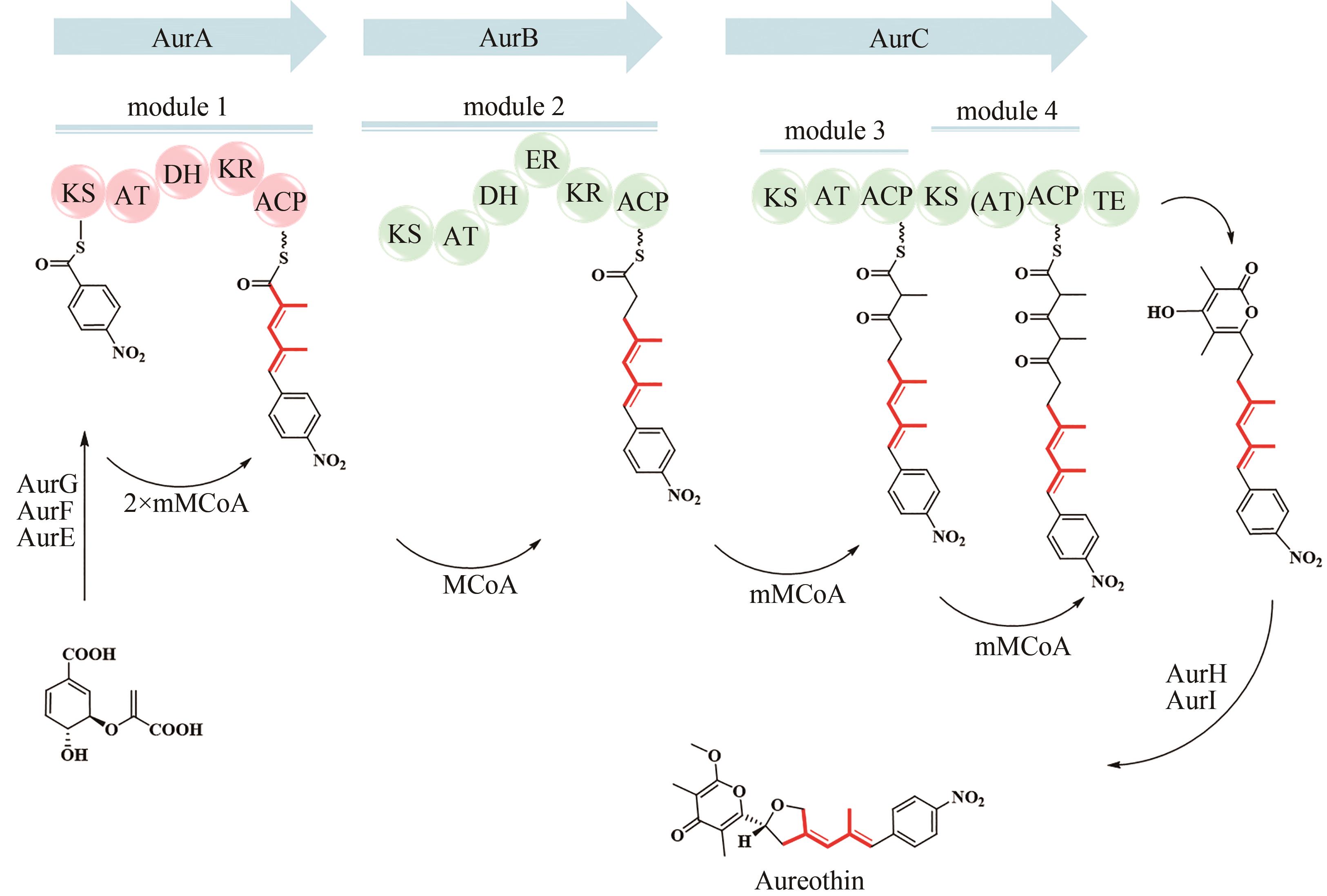
| 19 | HERTWECK C, LUZHETSKYY A, REBETS Y, et al. Type Ⅱ polyketide synthases: gaining a deeper insight into enzymatic teamwork[J]. Natural Product Reports, 2007, 24(1): 162-190. |
| 20 | XIE S L, ZHANG L H. Type Ⅱ polyketide synthases: a bioinformatics-driven approach[J]. ChemBioChem, 2023, 24(9): e202200775. |
| 21 | YU D Y, XU F C, ZENG J, et al. Type Ⅲ polyketide synthases in natural product biosynthesis[J]. IUBMB Life, 2012, 64(4): 285-295. |
| 22 | LI Y Y, MÜLLER R. Non-modular polyketide synthases in myxobacteria[J]. Phytochemistry, 2009, 70(15-16): 1850-1857. |
| 23 | MIYANAGA A, KUDO F, EGUCHI T. Protein-protein interactions in polyketide synthase-nonribosomal peptide synthetase hybrid assembly lines[J]. Natural Product Reports, 2018, 35(11): 1185-1209. |
| 24 | JENKE-KODAMA H, DITTMANN E. Bioinformatic perspectives on NRPS/PKS megasynthases: advances and challenges[J]. Natural Product Reports, 2009, 26(7): 874-883. |
| 25 | SCHWECKE T, APARICIO J F, MOLNÁR I, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin[J]. Proceedings of the National Academy of Sciences of the United States of America, 1995, 92(17): 7839-7843. |
| 26 | CHEN H, O’CONNOR S, CANE D E, et al. Epothilone biosynthesis: assembly of the methylthiazolylcarboxy starter unit on the EpoB subunit[J]. Chemistry & Biology, 2001, 8(9): 899-912. |
| 27 | DU L, SÁNCHEZ C, CHEN M, et al. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase[J]. Chemistry & Biology, 2000, 7(8): 623-642. |
| 28 | KOZAKAI R, ONO T, HOSHINO S, et al. Acyltransferase that catalyses the condensation of polyketide and peptide moieties of goadvionin hybrid lipopeptides[J]. Nature Chemistry, 2020, 12(9): 869-877. |
| 29 | HUANG Y, HOEFGEN S, VALIANTE V. Biosynthesis of fungal drimane-type sesquiterpene esters[J]. Angewandte Chemie International Edition, 2021, 60(44): 23763-23770. |
| 30 | DONADIO S, STAVER M J, MCALPINE J B, et al. Modular organization of genes required for complex polyketide biosynthesis[J]. Science, 1991, 252(5006): 675-679. |
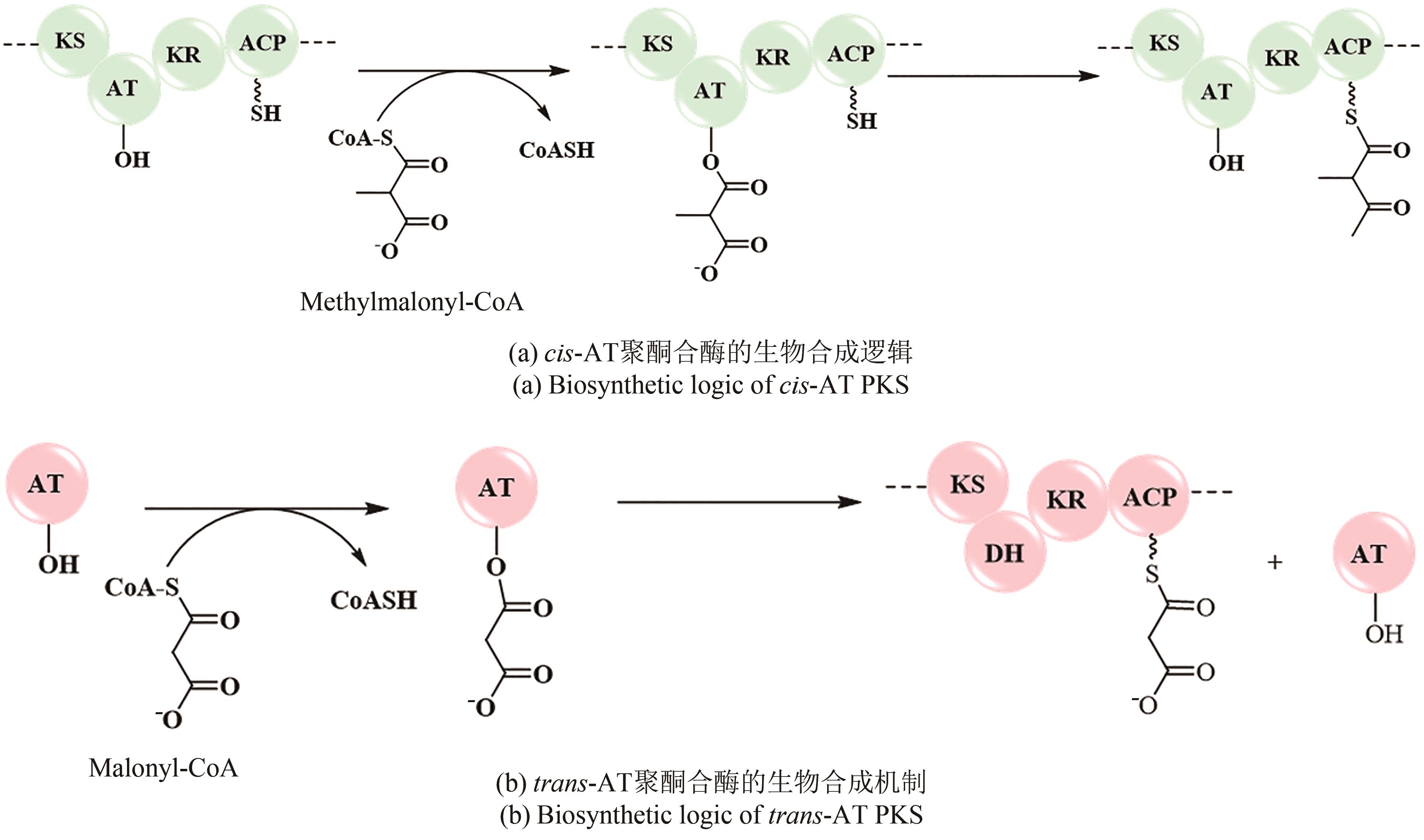
| 31 | OMURA S, IKEDA H, ISHIKAWA J, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(21): 12215-12220. |
| 32 | STAUNTON J, WEISSMAN K J. Polyketide biosynthesis: a millennium review[J]. Natural Product Reports, 2001, 18(4): 380-416. |
| 33 | HELFRICH E J N, REITER S, PIEL J. Recent advances in genome-based polyketide discovery[J]. Current Opinion in Biotechnology, 2014, 29: 107-115. |
| 34 | WINTER J M, BEHNKEN S, HERTWECK C. Genomics-inspired discovery of natural products[J]. Current Opinion in Chemical Biology, 2011, 15(1): 22-31. |
| 35 | NIEHS S P, KUMPFMÜLLER J, DOSE B, et al. Insect-associated bacteria assemble the antifungal butenolide gladiofungin by non-canonical polyketide chain termination[J]. Angewandte Chemie International Edition, 2020, 59(51): 23122-23126. |
| 36 | ZAZOPOULOS E, HUANG K X, STAFFA A, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways[J]. Nature Biotechnology, 2003, 21(2): 187-190. |
| 37 | PENG H Y, ISHIDA K, HERTWECK C. Loss of single-domain function in a modular assembly line alters the size and shape of a complex polyketide[J]. Angewandte Chemie International Edition, 2019, 58(50): 18252-18256. |
| 38 | ZHANG J J, TANG X Y, HUAN T, et al. Pass-back chain extension expands multimodular assembly line biosynthesis[J]. Nature Chemical Biology, 2020, 16(1): 42-49. |
| 39 | HIRATA Y, NAKATA H, YAMADA K, et al. The structure of aureothin, a nitro compound obtained from Streptomyces thioluteus [J]. Tetrahedron, 1961, 14(3-4): 252-274. |
| 40 | HE J, HERTWECK C. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster[J]. Chemistry & Biology, 2003, 10(12): 1225-1232. |
| 41 | HE J, HERTWECK C. Functional analysis of the aureothin iterative type Ⅰ polyketide synthase[J]. ChemBioChem, 2005, 6(5): 908-912. |
| 42 | BUSCH B, UEBERSCHAAR N, SUGIMOTO Y, et al. Interchenar retrotransfer of aureothin intermediates in an iterative polyketide synthase module[J]. Journal of the American Chemical Society, 2012, 134(30): 12382-12385. |
| 43 | ARAI M, HAMANO K. Isolation of three main components. F3, F4 and F5, from azalomycin F-complex[J]. The Journal of Antibiotics, 1970, 23(3): 107-112. |
| 44 | YUAN G J, LIN H P, WANG C, et al. 1H and 13C assignments of two new macrocyclic lactones isolated from Streptomyces sp. 211726 and revised assignments of azalomycins F3a, F4a and F5a [J]. Magnetic Resonance in Chemistry, 2011, 49(1): 30-37. |
| 45 | ZHAI G F, WANG W Y, XU W, et al. Cross-module enoylreduction in the Azalomycin F polyketide synthase[J]. Angewandte Chemie International Edition, 2020, 59(50): 22738-22742. |
| 46 | XU W, ZHAI G F, LIU Y Z, et al. An iterative module in the Azalomycin F polyketide synthase contains a switchable enoylreductase domain[J]. Angewandte Chemie International Edition, 2017, 56(20): 5503-5506. |
| 47 | CHEN A Y, SCHNARR N A, KIM C Y, et al. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase[J]. Journal of the American Chemical Society, 2006, 128(9): 3067-3074. |
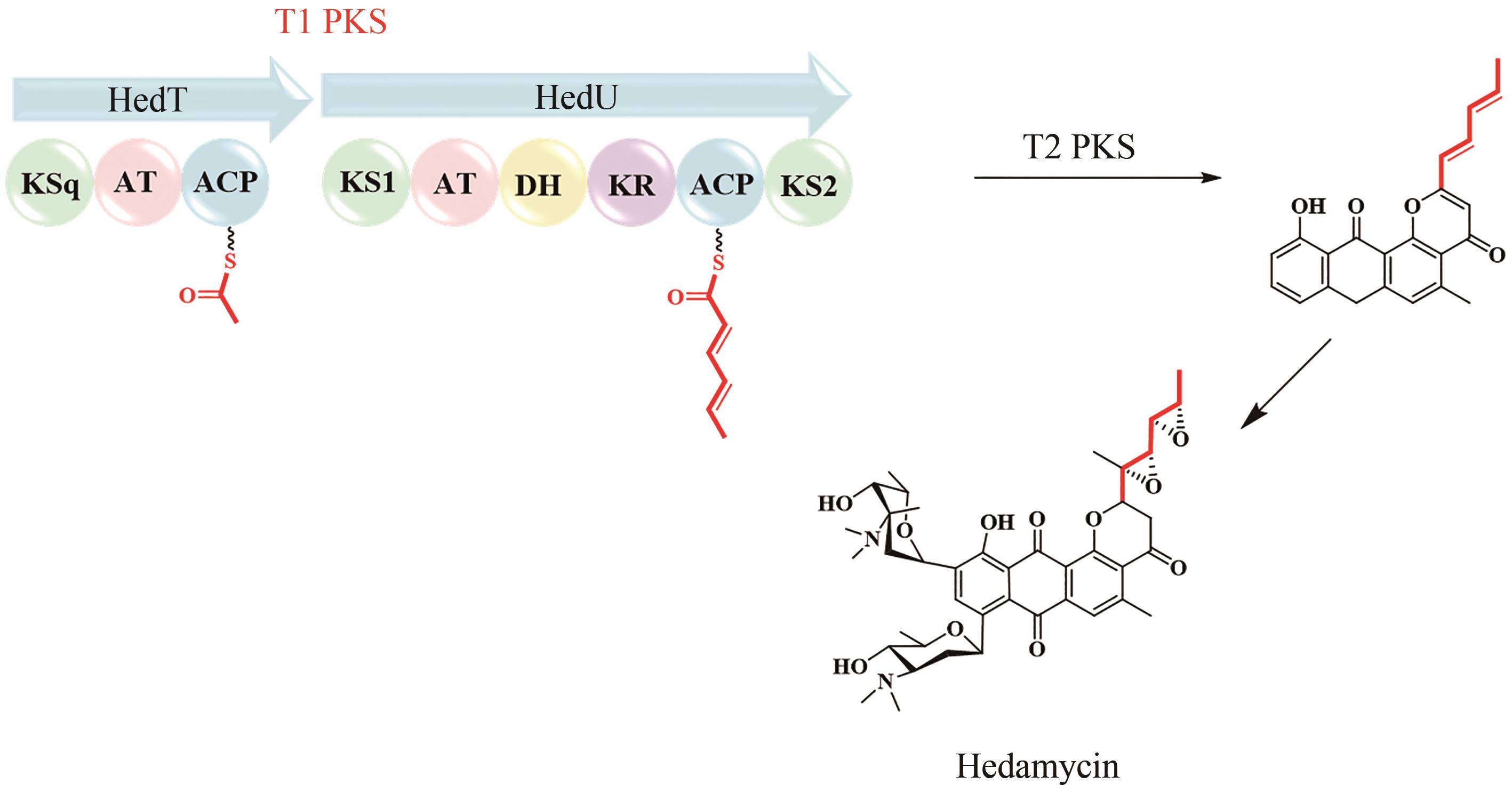
| 48 | KAPUR S, CHEN A Y, CANE D E, et al. Molecular recognition between ketosynthase and acyl carrier protein domains of the 6-deoxyerythronolide B synthase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(51): 22066-22071. |
| 49 | TAN F H, PUTOCZKI T L, STYLLI S S, et al. Ponatinib: a novel multi-tyrosine kinase inhibitor against human malignancies[J]. OncoTargets and Therapy, 2019, 12: 635-645. |
| 50 | SUGIMOTO Y, ISHIDA K, TRAITCHEVA N, et al. Freedom and constraint in engineered noncolinear polyketide assembly lines[J]. Chemistry & Biology, 2015, 22(2): 229-240. |
| 51 | FISCH K M. Biosynthesis of natural products by microbial iterative hybrid PKS-NRPS[J]. RSC Advances, 2013, 3(40): 18228-18247. |
| 52 | CHENG Y Q, TANG G L, SHEN B. Type Ⅰ polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(6): 3149-3154. |
| 53 | DODGE G J, MALONEY F P, SMITH J L. Protein-protein interactions in “cis-AT” polyketide synthases[J]. Natural Product Reports, 2018, 35(10): 1082-1096. |
| 54 | KOSOL S, JENNER M, LEWANDOWSKI J R, et al. Protein-protein interactions in trans-AT polyketide synthases[J]. Natural Product Reports, 2018, 35(10): 1097-1109. |
| 55 | WOLF H, CHINALI G, PARMEGGIANI A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu[J]. Proceedings of the National Academy of Sciences of the United States of America, 1974, 71(12): 4910-4914. |
| 56 | WEBER T, LAIPLE K J, PROSS E K, et al. Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety[J]. Chemistry & Biology, 2008, 15(2): 175-188. |
| 57 | ROBERTSEN H L, MUSIOL-KROLL E M, DING L, et al. Filling the gaps in the kirromycin biosynthesis: deciphering the role of genes involved in ethylmalonyl-CoA supply and tailoring reactions[J]. Scientific Reports, 2018, 8(1): 3230. |
| 58 | MUSIOL E M, HÄRTNER T, KULIK A, et al. Supramolecular templating in kirromycin biosynthesis: the acyltransferase KirCⅡ loads ethylmalonyl-CoA extender onto a specific ACP of the trans-AT PKS[J]. Chemistry & Biology, 2011, 18(4): 438-444. |
| 59 | MUSIOL E M, GREULE A, HÄRTNER T, et al. The AT2 domain of KirCⅠ loads malonyl extender units to the ACPs of the kirromycin PKS[J]. ChemBioChem, 2013, 14(11): 1343-1352. |
| 60 | JENKE-KODAMA H, BÖRNER T, DITTMANN E. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis [J]. PLoS Computational Biology, 2006, 2(10): 1210-1218.e132. |
| 61 | NGUYEN T, ISHIDA K, JENKE-KODAMA H, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection[J]. Nature Biotechnology, 2008, 26(2): 225-233. |
| 62 | PIEL J. Biosynthesis of polyketides by trans-AT polyketide synthases[J]. Natural Product Reports, 2010, 27(7): 996-1047. |
| 63 | YEO W L, HENG E, TAN L, et al. Biosynthetic engineering of the antifungal, anti-MRSA auroramycin[J]. Microbial Cell Factories, 2020, 19(1): 3. |
| 64 | DUNN B J, WATTS K R, ROBBINS T, et al. Comparative analysis of the substrate specificity of trans-versus cis-acyltransferases of assembly line polyketide synthases[J]. Biochemistry, 2014, 53(23): 3796-3806. |
| 65 | SIRIRUNGRUANG S, AD O, PRIVALSKY T M, et al. Engineering site-selective incorporation of fluorine into polyketides[J]. Nature Chemical Biology, 2022, 18(8): 886-893. |
| 66 | SHEN B, NAKAYAMA H, HUTCHINSON C R. Isolation and structural elucidation of tetracenomycin F2 and tetracenomycin F1: early intermediates in the biosynthesis of tetracenomycin C in Streptomyces glaucescens [J]. Journal of Natural Products, 1993, 56(8): 1288-1293. |
| 67 | KOTOWSKA M, PAWLIK K. Roles of type Ⅱ thioesterases and their application for secondary metabolite yield improvement[J]. Applied Microbiology and Biotechnology, 2014, 98(18): 7735-7746. |
| 68 | CHEN A, RE R N, BURKART M D. Type Ⅱ fatty acid and polyketide synthases: deciphering protein-protein and protein-substrate interactions[J]. Natural Product Reports, 2018, 35(10): 1029-1045. |
| 69 | DAS A, KHOSLA C. In vivo and in vitro analysis of the hedamycin polyketide synthase[J]. Chemistry & Biology, 2009, 16(11): 1197-1207. |
| 70 | HAN Y, WANG Y, YANG Y H, et al. Shellmycin A-D, novel bioactive tetrahydroanthra-γ-pyrone antibiotics from marine Streptomyces sp. Shell-016[J]. Marine Drugs, 2020, 18(1): 58. |
| 71 | BILILIGN T, HYUN C G, WILLIAMS J S, et al. The hedamycin locus implicates a novel aromatic PKS priming mechanism[J]. Chemistry & Biology, 2004, 11(7): 959-969. |
| 72 | ABE I. Biosynthesis of medicinally important plant metabolites by unusual type Ⅲ polyketide synthases[J]. Journal of Natural Medicines, 2020, 74(4): 639-646. |
| 73 | MURRAY L A M, MCKINNIE S M K, MOORE B S, et al. Meroterpenoid natural products from Streptomyces bacteria- the evolution of chemoenzymatic syntheses[J]. Natural Product Reports, 2020, 37(10): 1334-1366. |
| 74 | AUSTIN M B, NOEL J P. The chalcone synthase superfamily of type Ⅲ polyketide synthases[J]. Natural Product Reports, 2003, 20(1): 79-110. |
| 75 | CHEN H, TSENG C C, HUBBARD B K, et al. Glycopeptide antibiotic biosynthesis: enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(26): 14901-14906. |
| 76 | WENZEL S C, BODE H B, KOCHEMS I, et al. A type Ⅰ/type Ⅲ polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin[J]. ChemBioChem, 2008, 9(16): 2711-2721. |
| 77 | SONG R T, SHI H X, ZHU J, et al. A single-component flavoenzyme catalyzed regioselective halogenation of pyrone in the biosynthesis of venemycins[J]. ACS Chemical Biology, 2019, 14(12): 2533-2537. |
| 78 | LACEY H J, CHEN R, VUONG D, et al. Resorculins: hybrid polyketide macrolides from Streptomyces sp. MST-91080[J]. Organic & Biomolecular Chemistry, 2023, 21(12): 2531-2538. |
| 79 | ZHANG B, JIN W Z, ZHANG Y Y, et al. A type Ⅰ/type Ⅲ PKS hybrid generates cinnamomycin A-D[J]. Organic Letters, 2023, 25(15): 2560-2564. |
| 80 | CORTES J, HAYDOCK S F, ROBERTS G A, et al. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea [J]. Nature, 1990, 348(6297): 176-178. |
| 81 | GOMES E S, SCHUCH V, DE MACEDO LEMOS E G. Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics[J]. Brazilian Journal of Microbiology, 2013, 44(4): 1007-1034. |
| 82 | ZHANG J Y, SUN Y, WANG Y J, et al. Genome mining of novel rubiginones from Streptomyces sp. CB02414 and characterization of the post-PKS modification steps in rubiginone biosynthesis[J]. Microbial Cell Factories, 2021, 20(1): 192. |
| 83 | BAGDE S R, MATHEWS I I, FROMME J C, et al. Modular polyketide synthase contains two reaction chambers that operate asynchronously[J]. Science, 2021, 374(6568): 723-729. |
| 84 | RISDIAN C, MOZEF T, WINK J. Biosynthesis of polyketides in Streptomyces [J]. Microorganisms, 2019, 7(5): 124. |
| 85 | PRUSOV E V. Total synthesis of antibiotics: recent achievements, limitations, and perspectives[J]. Applied Microbiology and Biotechnology, 2013, 97(7): 2773-2795. |
| 86 | ZHANG S W, XIE Q, SUN C L, et al. Cytotoxic kendomycins containing the carbacylic Ansa scaffold from the marine-derived Verrucosispora sp. SCSIO 07399[J]. Journal of Natural Products, 2019, 82(12): 3366-3371. |
| [1] | XIE Xiangqian, GUO Wen, WANG Huan, LI Jin. Biosynthesis and chemical synthesis of ribosomally synthesized and post-translationally modified peptides containing aminovinyl cysteine [J]. Synthetic Biology Journal, 2024, 5(5): 981-996. |
| [2] | FENG Jin, PAN Haixue, TANG Gongli. Research advances in biosynthesis of natural product drugs within the past decade [J]. Synthetic Biology Journal, 2024, 5(3): 408-446. |
| [3] | Huang XIE, Yilei ZHENG, Yiting SU, Jingyi RUAN, Yongquan LI. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [4] | YU Xuchang, WU Hui, LI Lei. Library construction and targeted BGC screening for more efficient discovery of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 492-506. |
| [5] | Jun ZHANG, Shixue JIN, Qian YUN, Xudong QU. Biosynthesis of the unnatural extender units with polyketides and their structural modifications for applications in medicines [J]. Synthetic Biology Journal, 2024, 5(3): 561-570. |
| [6] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [7] | LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids [J]. Synthetic Biology Journal, 2024, 5(3): 507-526. |
| [8] | Xinjie SHI, Yiling DU. Research advances in the biosynthesis of nonribosomal peptides within the bisintercalator family as anticancer drugs [J]. Synthetic Biology Journal, 2024, 5(3): 593-611. |
| [9] | SONG Yongxiang, ZHANG Xiufeng, LI Yanqin, XIAO Hua, YAN Yan. Resistance-gene directed discovery of bioactive natural products [J]. Synthetic Biology Journal, 2024, 5(3): 474-491. |
| [10] | Zhen HUI, Xiaoyu TANG. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| [11] | Zhehui HU, Juan XU, Guangkai BIAN. Application of automated high-throughput technology in natural product biosynthesis [J]. Synthetic Biology Journal, 2023, 4(5): 932-946. |
| [12] | Fanzhong ZHANG, Changjun XIANG, Lihan ZHANG. Advances and applications of evolutionary analysis and big-data guided bioinformatics in natural product research [J]. Synthetic Biology Journal, 2023, 4(4): 629-650. |
| [13] | Jingwei LYU, Zixin DENG, Qi ZHANG, Wei DING. Identification of RiPPs precursor peptides and cleavage sites based on deep learning [J]. Synthetic Biology Journal, 2022, 3(6): 1262-1276. |
| [14] | Jiayu DONG, Min LI, Zonghua XIAO, Ming HU, Yudai MATSUDA, Weiguang WANG. Recent advances in heterologous production of natural products using Aspergillus oryzae [J]. Synthetic Biology Journal, 2022, 3(6): 1126-1149. |
| [15] | Faguang ZHANG, Ge QU, Zhoutong SUN, Jun′an MA. From chemical synthesis to biosynthesis: trends toward total synthesis of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 674-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||