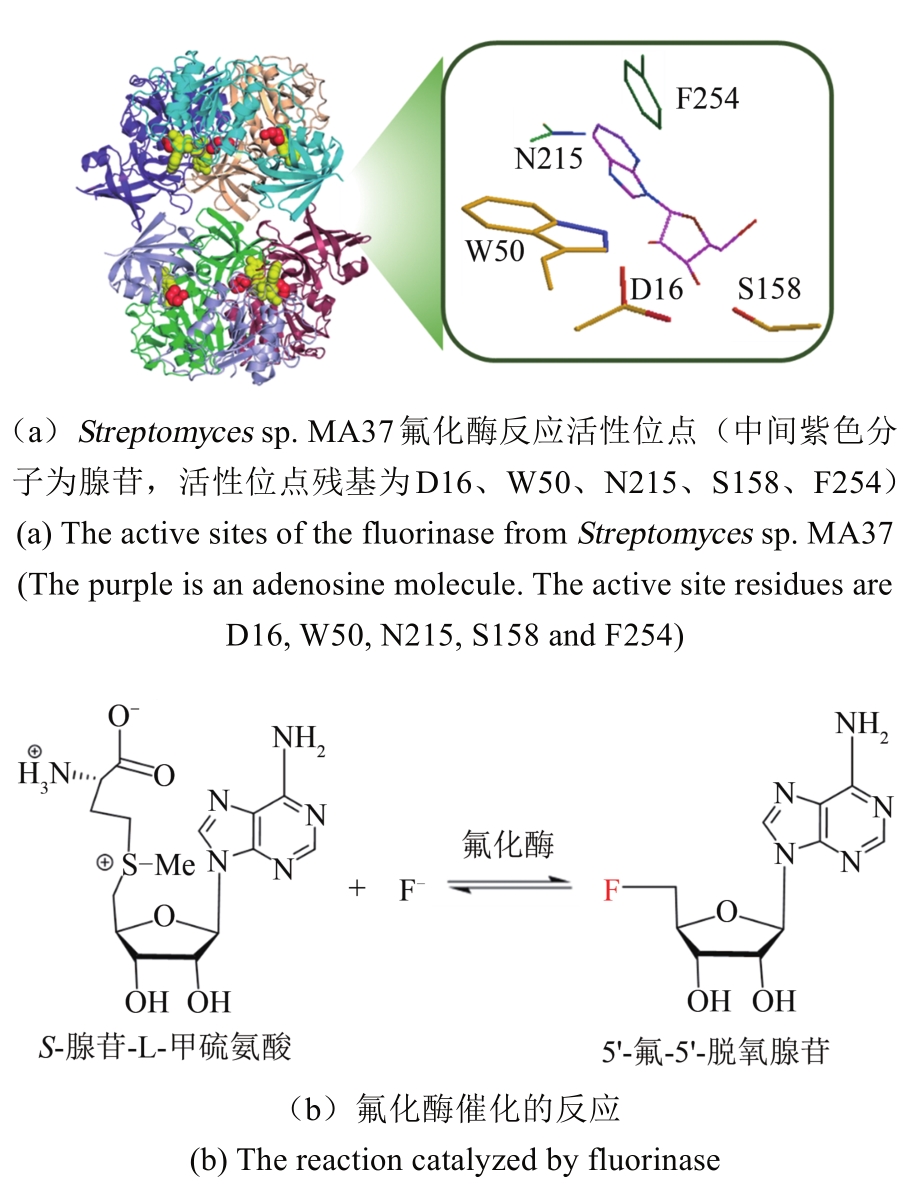
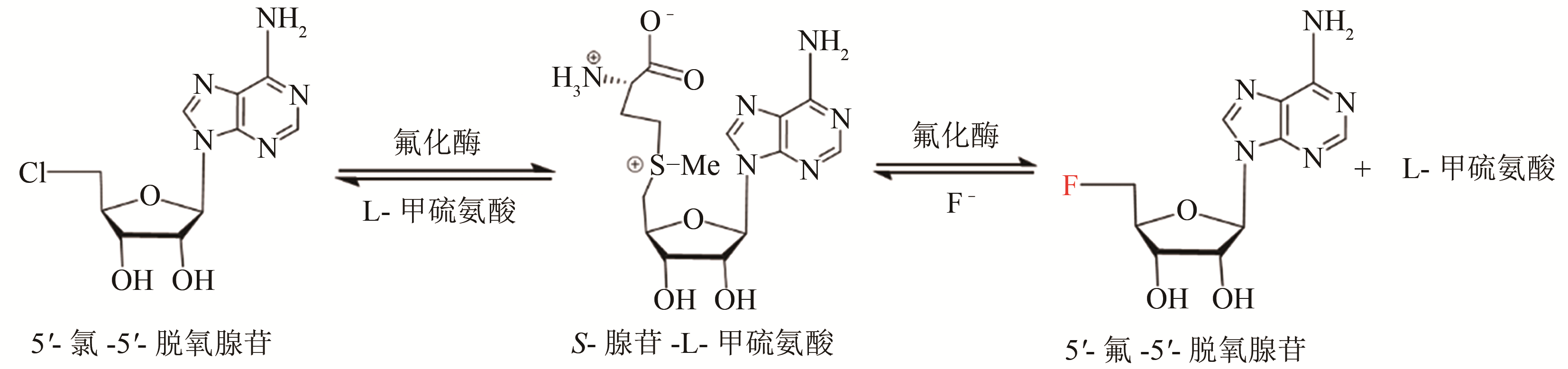
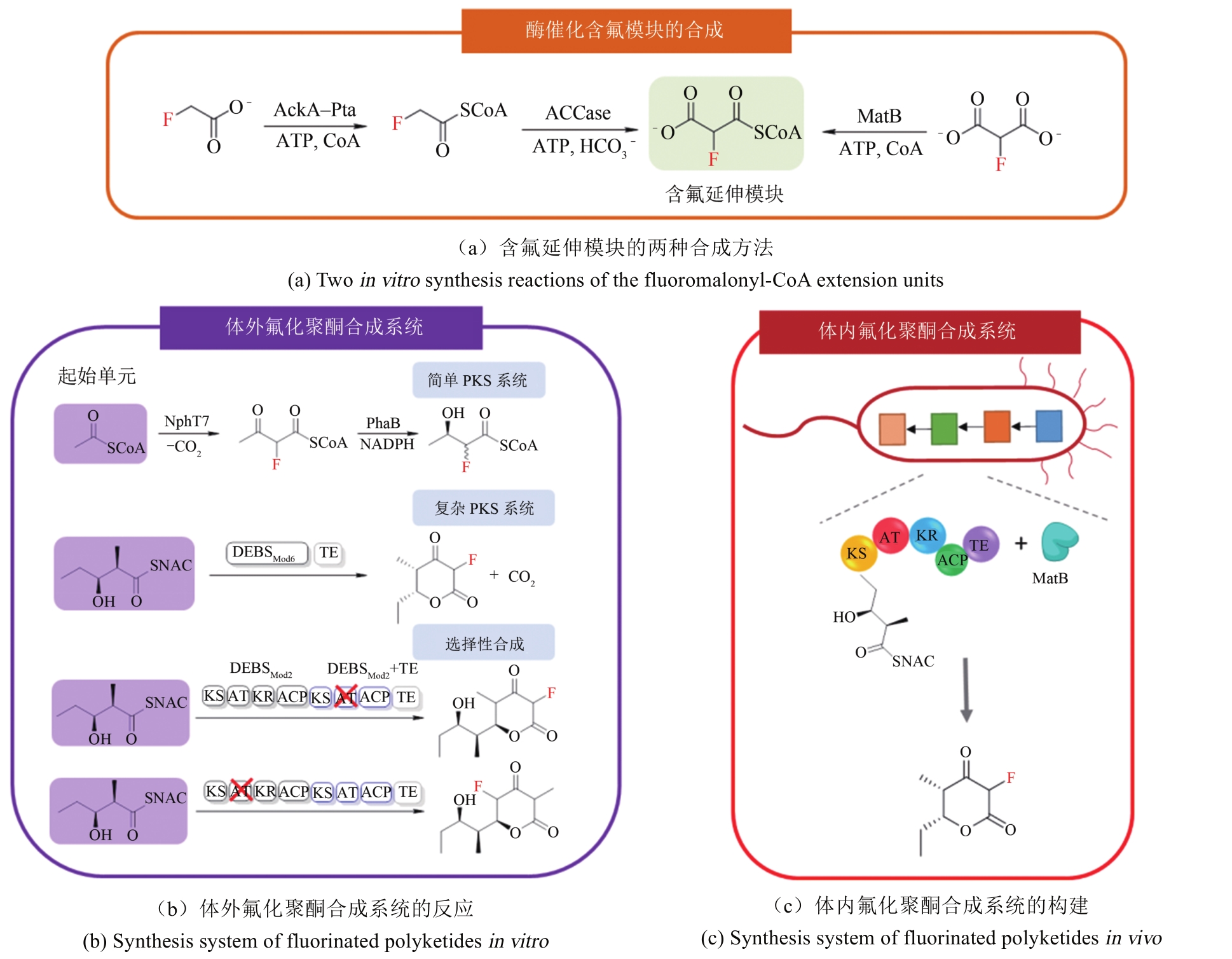
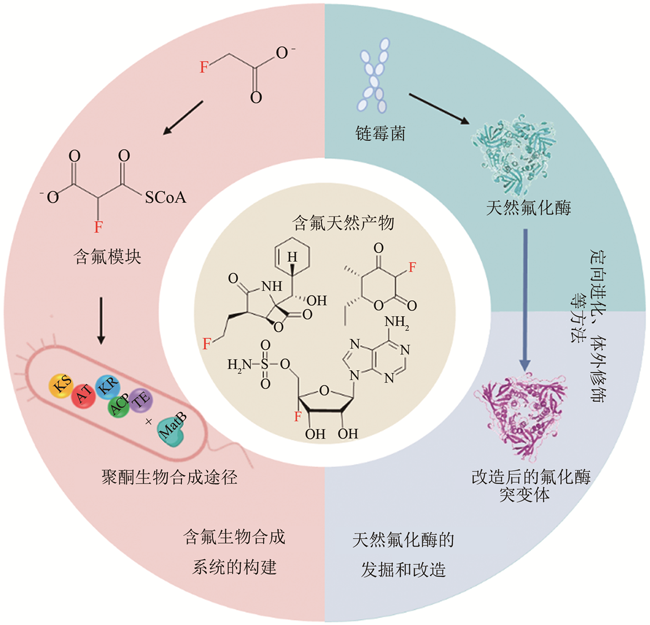
合成生物学 ›› 2020, Vol. 1 ›› Issue (3): 358-371.DOI: 10.12211/2096-8280.2020-003
合成生物学在含氟化合物生产中的应用
王高丽1, 金雪芮1, 罗云孜1,2
- 1.天津大学化工学院,系统生物工程教育部重点实验室,合成生物学前沿科学中心,天津 300072
2.天津大学化学化工协同创新中心,天津 300072
-
收稿日期:2020-02-27修回日期:2020-03-28出版日期:2020-06-30发布日期:2020-09-29 -
通讯作者:罗云孜 -
作者简介:王高丽(1996—),女,硕士研究生。E-mail:2019207301@tju.edu.cn
金雪芮(1997—),女,硕士研究生。E-mail:2019207303@tju.edu.cn
罗云孜(1985—),女,博士,教授,研究方向为合成生物学。E-mail:luoyunzi827@aliyun.com -
基金资助:天津市自然科学基金(19JCYBJC24200);国家重点研发计划 “合成生物学”重点专项(2018YFA0903300)
Applications of synthetic biology in the production of fluorinated compounds
WANG Gaoli1, JIN Xuerui1, LUO Yunzi1,2
- 1.Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education),School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
2.Collaborative Innovation Center of Chemical Science and Engineering (Tianjin),Tianjin University,Tianjin 300072,China
-
Received:2020-02-27Revised:2020-03-28Online:2020-06-30Published:2020-09-29 -
Contact:LUO Yunzi
摘要:
将氟原子引入有机化合物分子可以赋予化合物新的功能,使其具有更好的物理化学特性。含氟化合物常被应用于新药研发、医疗诊断等诸多方面,受到合成化学家的广泛关注。但利用有机化学方法合成氟化物往往难以实现氟原子的选择性引入,且含氟试剂成本高,产物分离纯化困难。近年来,合成生物学技术的发展为氟化物的生产提供了新思路。通过发掘链霉菌等微生物中天然的氟化酶,并通过定向进化、理性设计等方法对天然氟化酶进行优化改造,可催化合成特定的C—F键;通过将含氟模块引入天然产物生物合成途径,可合成新型复杂含氟天然产物,如氟化聚酮等。本文归纳总结了利用氟化酶和含氟模块构建氟化物生物合成系统的方法,讨论了合成生物学手段在含氟化合物生产中的重要应用,并从合成生物学的角度展望了优化改造氟化酶和构建新型含氟生物合成系统的未来发展方向。采用合成生物学新策略实现含氟产物的高效生物合成,将有望解决复杂手性含氟化合物的合成问题。
中图分类号:
引用本文
王高丽, 金雪芮, 罗云孜. 合成生物学在含氟化合物生产中的应用[J]. 合成生物学, 2020, 1(3): 358-371.
WANG Gaoli, JIN Xuerui, LUO Yunzi. Applications of synthetic biology in the production of fluorinated compounds[J]. Synthetic Biology Journal, 2020, 1(3): 358-371.
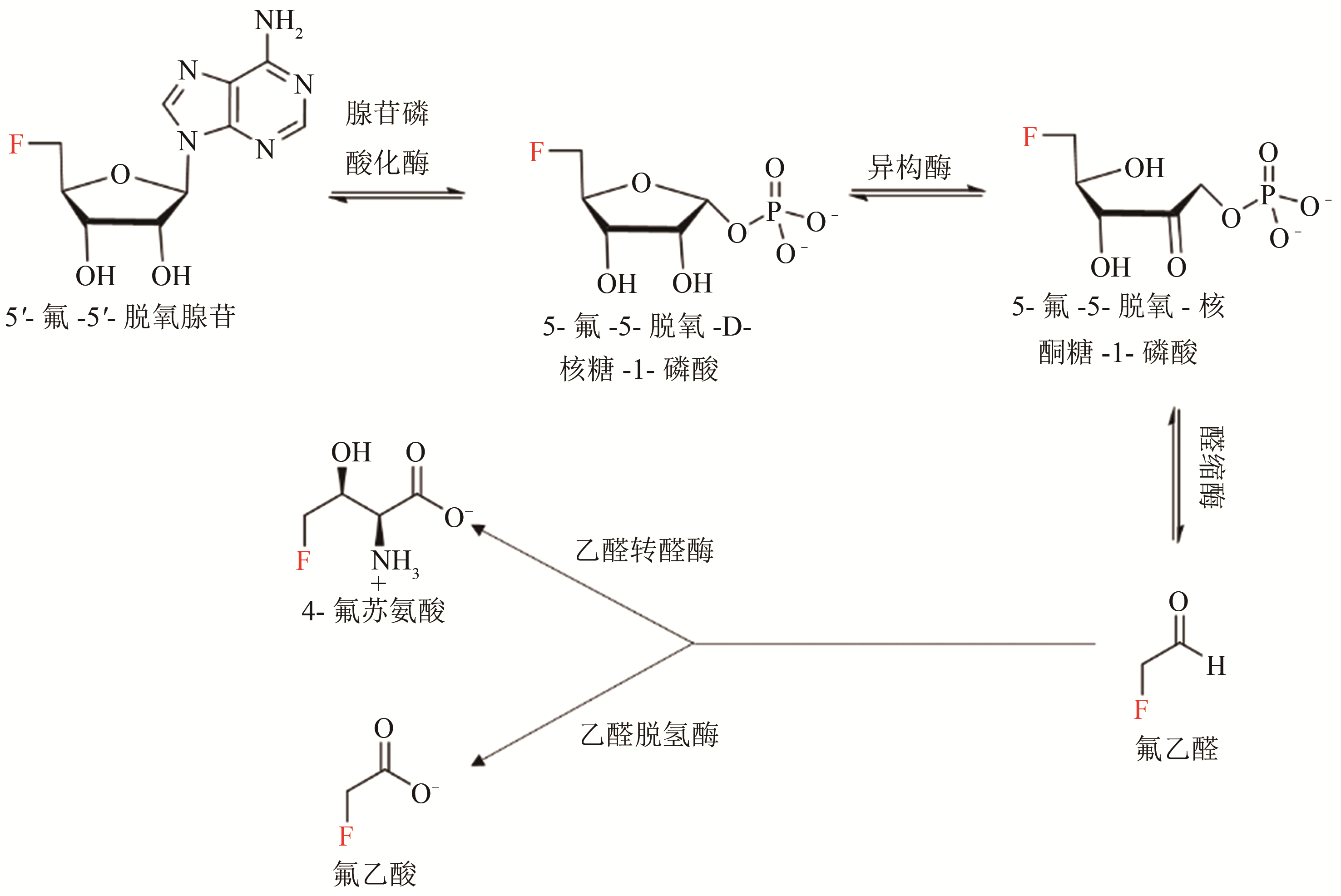
图3 S. cattleya中从5'-FDA到氟乙酸和4-氟苏氨酸的生物合成途径[30,36]
Fig. 3 Biosynthetic pathways for the production of fluoroacetate and 4-fluorthreonine from 5'-FDA in S. cattleya [30,36]
| 氟化酶来源 | KM(SAM)/ μmol·L-1 | 转化数kcat/min-1 | (kcat/KM)/×10-3 L·μmol-1·min-1 |
|---|---|---|---|
| S. cattleya[ | 29.2±2.41 | 0.083 | 2.84 |
| Streptomyces sp.MA37[ | 82.4±18.6 | 0.262 | 3.18 |
| N. brasiliensis[ | 27.8±4.23 | 0.122 | 4.40 |
| Actinoplanes sp. N902-109[ | 45.8±7.91 | 0.204 | 4.44 |
| Streptomyces xinghaiensis NRRL B24674[ | 7.04±0.94 | 0.277±0.007 | 39.5±1.51 |
表1 不同来源的氟化酶动力学参数汇总
Tab. 1 The summary for the kinetic parameters of fluorinase from different sources
| 氟化酶来源 | KM(SAM)/ μmol·L-1 | 转化数kcat/min-1 | (kcat/KM)/×10-3 L·μmol-1·min-1 |
|---|---|---|---|
| S. cattleya[ | 29.2±2.41 | 0.083 | 2.84 |
| Streptomyces sp.MA37[ | 82.4±18.6 | 0.262 | 3.18 |
| N. brasiliensis[ | 27.8±4.23 | 0.122 | 4.40 |
| Actinoplanes sp. N902-109[ | 45.8±7.91 | 0.204 | 4.44 |
| Streptomyces xinghaiensis NRRL B24674[ | 7.04±0.94 | 0.277±0.007 | 39.5±1.51 |
| 1 | GRIBBLE G W. Occurrence of halogenated alkaloids[J]. The Alkaloids: Chemistry and Biology, 2012, 71:1-165. |
| 2 | O’HAHAN D, PERRY R, LOCK J M, et al. Identification of exceptionally high levels of monofluoroacetate in Dichapetalum braunii from Southeastern Tanzania[J]. Phytochemistry, 1993, 33: 1043–1046. |
| 3 | PAGUIGAN N D, HUNITI M H A, RAJA H A, et al. Chemoselective fluorination and chemoinformatic analysis of griseofulvin: natural vs fluorinated fungal metabolites[J]. Bioorganic and Medicinal Chemistry, 2017, 25(20): 5238-5246. |
| 4 | BOHM H J, BANNER D, BENDELS S, et al. Fluorine in medicinal chemistry[J]. ChemBioChem, 2004, 5(5):637-643. |
| 5 | LIN X X, RONG F, FU D G, et al. Enhanced photocatalytic activity of fluorine doped TiO2 by loaded with Ag for degradation of organic pollutants[J]. Powder Technology, 2012, 219:173-178. |
| 6 | LIANG J, LUO Y Z, ZHAO H M. Synthetic biology: putting synthesis into biology[J]. Wiley Interdisciplinary Reviews Systems Biology & Medicine, 2011, 3(1):7-20. |
| 7 | LUO Y Z, COBB E R, ZHAO H M, Recent advances in natural product discovery [J]. Current Opinion in Biotechnology, 2014, 30: 230-237. |
| 8 | ZHAO Q Y, WANG L P, LUO Y Z. Recent advances in natural products exploitation in Streptomyces via synthetic biology[J]. Engineering in Life Sciences, 2019, 19(6): 452-462. |
| 9 | 王丽苹, 罗云孜. 合成生物学在天然产物研究中的应用[J]. 生物技术通报, 2017, 33(1): 35-47. |
| WANG L P, LUO Y Z. Applications of synthetic biology in the research of natural product[J]. Biotechnology Bulletin, 2017, 33(1): 35-47. | |
| 10 | LUO Y Z, LI B Z, LIU D, et al. Engineered biosynthesis of natural products in heterologous hosts[J]. Chemical Society Reviews, 2015, 44(15): 5265-5290. |
| 11 | PALAZZOTTO E, TONG Y J, LEE S Y, et al. Synthetic biology and metabolic engineering of actinomycetes for natural product discovery[J]. Biotechnology Advances, 2019, 37(6):107366. |
| 12 | TILBURG A Y V, CAO H J, MEULEN S B V D, et al. Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories[J]. Current Opinion in Biotechnology, 2019, 59: 1-7. |
| 13 | FREY R, HAYASHI T, BULLER R M. Directed evolution of carbon-hydrogen bond activating enzymes[J]. Current Opinion in Biotechnology, 2019, 60: 29-38. |
| 14 | LI F Z, ZHANG X, RENATA H. Enzymatic C—H functionalizations for natural product synthesis[J]. Current Opinion in Chemical Biology, 2019, 49:25-32. |
| 15 | OELSNITZ S D, ELLINGTON A. Continuous directed evolution for strain and protein engineering[J]. Current Opinion in Biotechnology, 2018, 53:158-163. |
| 16 | BELSARE K D, HORN T, RUFF A J, et al. Directed evolution of P450cin for mediated electron transfer[J]. Protein Engineering Design and Selection, 2017, 30(2):119-127. |
| 17 | BALE J B, GONEN S, LIU Y, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes[J]. Science, 2016, 353(6297):389-394. |
| 18 | BAKER D. Computationally designed protein activation[J]. National Science Review, 2019, 6(4): 609-610. |
| 19 | WU Q, PENG Z L, ANISHCHENKO I, et al. Protein contact prediction using metagenome sequence data and residual neural networks[J]. Bioinformatics, 2020, 36(1):41-48. |
| 20 | CONG Q, ANISHCHENKO I, OVCHINNIKOV S, et al. Protein interaction networks revealed by proteome coevolution[J]. Science, 2019, 365(6449):185-189. |
| 21 | NIELAEN J. Cell factory engineering for improved production of natural products[J]. Natural Product Reports, 2019, 36(9):1233-1236. |
| 22 | CHEVRETTE M G, GARCIA K G, MOJJCA N S, et al. Evolutionary dynamics of natural product biosynthesis in bacteria[J]. Natural Product Reports, 2020, 37: 566-599. |
| 23 | JENSEN P E, SCHARFF L B. Engineering of plastids to optimize the production of high-value metabolites and proteins[J]. Current Opinion in Biotechnology, 2019, 59:8-15. |
| 24 | WANG W, LI S, LI Z, et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces [J]. Nature Biotechnology, 2020, 38: 76-83. |
| 25 | SHAO Z, RAO G, LI C, et al. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold[J]. ACS Synthetic Biology, 2013, 2(11):662-669. |
| 26 | LI L, JIANG W, LU Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products[J]. Biotechnology Advances, 2017, 35(8):936-949. |
| 27 | SPRAKER J E, LUU G T, SANCHEZ L M. Imaging mass spectrometry for natural products discovery: a review of ionization methods[J]. Natural Product Reports,2019,2: |
| 28 | LUO Y Z, HUANG H, LIANG J, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster[J]. Nature Communications, 2013, 4:2894. |
| 29 | CABRERA V L, LOPEZ R O, GONZALEZ R E, et al. Complete genome sequence of Nocardia brasiliensis HUJEG-1[J]. Journal of Bacteriology, 2012, 194(10): 2761-2762. |
| 30 | SUN H H, YEO W L, LIM Y H, et al. Directed evolution of a fluorinase for improved fluorination efficiency with a non-native substrate[J]. Angewandte Chemie-International Edition, 2016, 128(46):14489-14492. |
| 31 | WALKER M C, THURONYI B. W, CHARKOUDIAN L K, et al . Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways[J]. Science, 2013, 341(6150): 1089-1094. |
| 32 | FRALRY A E, SHERMAN D H. Halogenase engineering and its utility in medicinal chemistry[J]. Bioorganic and Medicinal Chemistry Letters, 2018, 11: 1992-1999. |
| 33 | SANADA M, MIYANO T, IWADARE S W, et al. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya [J]. The Journal of Antibiotics, 1986, 39: 259-265. |
| 34 | ZHAO C H, LI P, DENG Z X, et al. Insights into fluorometabolite biosynthesis in Streptomyces cattleya DSM46488 through genome sequence and knockout mutants[J]. Bioorganic Chemistry, 2012, 44: 1-7. |
| 35 | O' HAGAN D, SCHAFFRATH C, COBB S L, et al. Biochemistry: biosynthesis of an organofluorine molecule [J]. Nature, 2002, 416(6878): 279-279. |
| 36 | DENG H, MA L, BANDARANAYAKA N, et al. Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining[J]. ChemBioChem, 2014, 15(3): 364-368. |
| 37 | WANG Y Y, DENG Z X, QU X D. Characterization of a SAM-dependent fluorinase from a latent biosynthetic pathway for fluoroacetate and 4-fluorothreonine formation in Nocardia brasiliensis [J]. F1000Research, 2014, 3: 61. |
| 38 | HUANG S, MA L, TONG M H, et al. Fluoroacetate biosynthesis from the marine-derived bacterium Streptomyces xinghaiensis NRRL B-24674[J]. Organic & Biomolecular Chemistry,2014,12(27):4828-4831. |
| 39 | MA L, LI Y F, MENG L P, et al. Biological fluorination from the sea: discovery of a SAM-dependent nucleophilic fluorinating enzyme from the marine-derived bacterium Streptomyces xinghaiensis NRRL B24674[J]. RSC Advances, 2016, 6: 27047-27051. |
| 40 | MEKLAT A, BOURAS N, ZITOUNI A, et al. Actinopolyspora mzabensis sp. nov., a halophilic actinomycete isolated from an Algerian Saharan soil[J]. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(10):3787-3792. |
| 41 | SOOKLAL S A, KONING C D, DEAN B, et al. Identification and characterisation of a fluorinase from Actinopolyspora mzabensis [J]. Protein Expression and Purification, 2020, 166: 105508. |
| 42 | DONG C J, HUANG F L, DENG H, et al. Crystal structure and mechanism of a bacterial fluorinating enzyme[J]. Nature, 2004, 427(6974): 561-565. |
| 43 | PITEL S B R, ZHAO H M. Recent advances in biocatalysis by directed enzyme evolution[J]. Comb. Chem. High Throughput Screen., 2006, 9:247-257. |
| 44 | YEO W L, CHEW X, SMITH D J, et al. Probing the molecular determinants of fluorinase specificity[J]. Chemical Communication, 2017, 53(17):2559-2562. |
| 45 | DENG H, COBB S L, MCEWAN A R, et al. McEwan the fluorinase from Streptomyces cattleya is also a chlorinase[J]. Angewandte Chemie International Edition, 2006, 45(5):759-762. |
| 46 | LOWE P T, COBB S L, O'HAGAN D. An enzymatic Finkelstein reaction: fluorinase catalyses direct halogen exchange[J]. Organic and Biomolecular Chemistry, 2019, 17(32): 7493-7496. |
| 47 | SERGEEV M E, MORGIA F, JAVED M R, et al. Enzymatic radiofluorination: fluorinase accepts methylaza-analog of SAM as substrate for FDA synthesis[J]. Journal of Molecular Catalysis B: Enzymatic, 2013, 97:74-79. |
| 48 | SUN H H, ZHAO H M, ANG E L. A coupled chlorinase-fluorinase system with a high efficiency of trans-halogenation and a shared substrate tolerance[J]. Chemical Communications, 2018, 54(68): 9458-9461. |
| 49 | ZHAO W X, DU G C, LIU S. An efficient thermostabilization strategy based on self-assembling amphipathic peptides for fusion tags[J]. Enzyme and Microbial Technology, 2018,121:68-77. |
| 50 | TU C H, ZHOU J, PENG L. Self-assembled nano-aggregates of fluorinases demonstrate enhanced enzymatic activity, thermostability and reusability[J]. Biomaterials Science, 2019: 2047-4849. |
| 51 | O' HAGAN D. Fluorine in health care: organofluorine containing blockbuster drugs[J]. J. Fluor. Chem., 2010, 131: 1071-1081. |
| 52 | 刘栓栓,王晶,许斌,等.近5年美国FDA批准上市的含氟药物研究进展[J].药学进展,2016,40(10):783-794. |
| LIU S S, WANG J, XU B, et al. Recent advances in R&D of fluorinated drugs approved by FDA in the past five years[J]. Progress in Pharmaceutical Sciences, 2016, 40(10):783-794 | |
| 53 | 赵方诺.氟原子在药物设计中的主要应用以及引入方法[J].国际公关,2019(7):247-248. |
| ZHAO F N. The main applications and introduction methods of fluorine atom in drug design[J]. PR Magazine, 2019(7):247-248. | |
| 54 | 张霁,金传飞,张英俊.含氟药物研究进展和芳(杂)环氟化及N(n=1,2,3)氟甲基化新方法[J].有机化学,2014,34(4):662-680. |
| ZHANG J, JIN C F, ZHANG Y J. Recent advances in research and development of fluorinated drugs and new methods for fluorination, mono-, di-and tri-fluoromethylation[J]. Chinese Journal of Organic Chemistry, 2014, 34(4):662-680. | |
| 55 | 弓添添,肖美娟,陈樱,等.克唑替尼药物关键手性中间体合成进展[J].浙江化工,2018,49(10):10-14. |
| GONG T T, XIAO M J, CHEN Y, et al. Research progress in synthesis of a key chiral intermediate of drug crizotinib[J]. Zhejiang Chemical Industry, 2018, 49(10):10-14. | |
| 56 | 孙冰,赵会,王玉军,等.马来酸阿法替尼的合成工艺研究[J].中国药物化学杂志,2019,29(1):44-48. |
| SUN B, ZHAO H, WANG Y J, et al. Study on synthetic process of afatinib dimaleate[J]. Chinese Journal of Medicinal Chemistry, 2019, 29(1):44-48. | |
| 57 | ODAR C, WINKLER M, WILTSCHI B. Fluoro amino acids: a rarity in nature, yet a prospect for protein engineering [J]. Biotechnology Journal, 2015, 10(3):427-446. |
| 58 | CHAN K K J, O'HAGAN D. The rare fluorinated natural products and biotechnological prospects for fluorine enzymology[J]. Methods in Enzymology, 2012, 516: 219-233. |
| 59 | ZHU X M, HACKL S, THAKER M N, et al. Biosynthesis of the fluorinated natural product nucleocidin in Streptomyces calvus is dependent on the bldA-specified Leu-tRNAUUA molecule[J]. ChemBioChem, 2015,16(17): 2498-2506. |
| 60 | WEISSLEDER R, MAHMOOD U. Molecular imaging[J]. Radiology, 2001, 219 (2): 316-333 |
| 61 | JACOBSON O, KIESEWETTER D O, CHEN X Y. Fluorine-18 radiochemistry, labeling strategies and synthetic routes[J]. Bioconjugate Chemistry, 2015, 26(1):1-18. |
| 62 | THOMPSON S, FLEMING I N, O' HAGAN D, et al. Enzymatic transhalogenation of dendritic RGD peptide constructs with the fluorinase[J]. Organic and Biomolecular Chemistry, 2016, 14(11): 3120-3129. |
| 63 | THOMPSON S, ONEGA M, ASHWORTH S, et al. A two-step fluorinase enzyme mediated 18F labelling of an RGD peptide for positron emission tomography[J]. Chemical Communications, 2015, 51(70): 13542-13545. |
| 64 | ZHANG Q Z, DALL'ANGELO S, FLEMING I N, et al. Last-step enzymatic [18F]-fluorination of cysteine-tethered RGD peptides using modified barbas linkers[J]. Chemistry, 2016, 22(31): 10998-11004. |
| 65 | LOWE P T, DALL'ANGELO S, KRIEGER T M, et al. A new class of fluorinated A2A adenosine receptor agonist with application to last-step enzymatic [18F]fluorination for PET imaging[J]. ChemBioChem, 2017, 18(21): 2156-2164. |
| 66 | LOWE P T, DALL'ANGELO S, DEVINE A, et al. Enzymatic fluorination of biotin and tetrazine conjugates for pretargeting approaches to positron emission tomography imaging[J]. ChemBioChem, 2019, 19(18): 1969-1978. |
| 67 | LOWE P T, DALL'ANGELO S, FLEMING I N, et al. Enzymatic radiosynthesis of a 18F-Glu-Ureido-Lys ligand for the prostate-specific membrane antigen (PSMA) [J]. Organic and Biomolecular Chemistry, 2019, 17(6): 1480-1486. |
| 68 | EUSTAQUIO A S, O'HAGAN D, MOORE B S. Engineering fuorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide[J]. Journal of Natural Products, 2010, 73: 378-382. |
| 69 | LUO Y Z, LEE J K, ZHAO H M. Challenges and opportunities in synthetic biology for chemical engineers[J]. Chemical Engineering Science. 2013, 103: 115-119. |
| 70 | WEISSMAN K J. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology[J]. Natural Product Reports, 2016, 33(2): 203-230 |
| 71 | WU L R, MAGLABGIT F, DENG H. Fluorine biocatalysis[J]. Current Opinion in Chemical Biology, 2020, 55:119-126 |
| 72 | KLOPEIRS S, KOOPMANS K R M, GARCIA E S, et al. Biosynthesis with fluorine[J]. ChemBioChem, 2014, 15(4): 495-497. |
| 73 | THURONYI B. W, CHANG M C Y. Synthetic biology approaches to fluorinated polyketides[J]. Accounts of Chemical Research, 2015, 48(3): 584-592. |
| 74 | THURONYI B. W, PRIVALSKY T M, CHANG, M C Y. Engineered fluorine metabolism and fluoropolymer production in living cell[J]. Angewandte Chemie-International Edition, 2017, 56(44): 13637-13640. |
| 75 | AD O, THURONYI B. W, CHANG M C Y. Elucidating the mechanism of fluorinated extender unit loading for improved production of fluorine-containing polyketides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(5): E660-E668. |
| 76 | JOSE R C, RAJA H A, GRAF T N. Biosynthesis of fluorinated peptaibols using a site-directed building block incorporation approach[J]. Journal of Natural Products, 2017, 80(6): 1883-1892. |
| 77 | FANG J, HAIT D, GORDON M H, et al. Chemoenzymatic platform for synthesis of chiral organofluorines based on type II aldolases[J]. Angewandte Chemie-International Edition, 2019, 131(34): 11967-11971. |
| 78 | ZHANG J, HUANG X Y, ZHANG R K, et al. Enantiodivergent α‑amino C—H fluoroalkylation catalyzed by engineered cytochrome P450s[J]. Journal of the American Chemical Society, 2019, 141: 9798-9802. |
| 79 | GILLIS E P, EASTMAN K J, HILL M D, et al. Applications of fluorine in medicinal chemistry [J]. Journal of Medicinal Chemistry, 2015, 58: 8315-8359. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 仲泉周, 单依怡, 裴清云, 金艳芸, 王艺涵, 孟璐远, 王歆韵, 张雨鑫, 刘坤媛, 王慧中, 冯尚国. 生物合成法生产α-熊果苷的研究进展[J]. 合成生物学, 2025, 6(1): 118-135. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [11] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [12] | 李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517. |
| [13] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [14] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [15] | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||