合成生物学 ›› 2020, Vol. 1 ›› Issue (6): 621-634.DOI: 10.12211/2096-8280.2020-012
合成微生物群落研究进展
曲泽鹏, 陈沫先, 曹朝辉, 左文龙, 陈业, 戴磊
- 中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东 深圳 518055
-
收稿日期:2020-02-09修回日期:2020-06-02出版日期:2020-12-31发布日期:2021-01-15 -
通讯作者:戴磊 -
作者简介:曲泽鹏(1993—),男,硕士,研究助理。研究方向为合成微生物组学。E-mail:zp.qu@siat.ac.cn
戴磊(1987—),男,博士,研究员。研究方向为合成微生物组学。E-mail:lei.dai@siat.ac.cn -
基金资助:国家自然科学基金(31971513);深圳合成生物学创新研究院科研基金(ZTXM20190003)
Research advances in synthetic microbial communities
QU Zepeng, CHEN Moxian, CAO Zhaohui, ZUO Wenlong, CHEN Ye, DAI Lei
- Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
-
Received:2020-02-09Revised:2020-06-02Online:2020-12-31Published:2021-01-15 -
Contact:DAI Lei
摘要:
合成微生物群落属于合成生物学和微生物组学的交叉领域,是新兴的研究方向。合成微生物群落是人工合成的多个物种共培养的微生物体系,具有组成明确、可操控性高等特点,在研究微生物组的功能和生态机制方面有着明显的优势。本文将从以下几个方面介绍合成微生物群落的研究进展:①微生物群落生态学,包括微生物之间的相互作用、宿主和其他环境因素对群落结构的影响;②合成微生物群落的研究方法,围绕设计-构建-测试-学习循环;③合成微生物群落在多个领域的应用,包括人体疾病治疗、植物抗逆、工业生产、环境修复等多个领域。最后,提出了合成微生物群落领域有待解决的重要问题,包括如何构建可控和稳定的微生物互作网络、如何表征和控制微生物群落的空间结构以及如何精准地控制微生物群落的功能。
中图分类号:
引用本文
曲泽鹏, 陈沫先, 曹朝辉, 左文龙, 陈业, 戴磊. 合成微生物群落研究进展[J]. 合成生物学, 2020, 1(6): 621-634.
QU Zepeng, CHEN Moxian, CAO Zhaohui, ZUO Wenlong, CHEN Ye, DAI Lei. Research advances in synthetic microbial communities[J]. Synthetic Biology Journal, 2020, 1(6): 621-634.
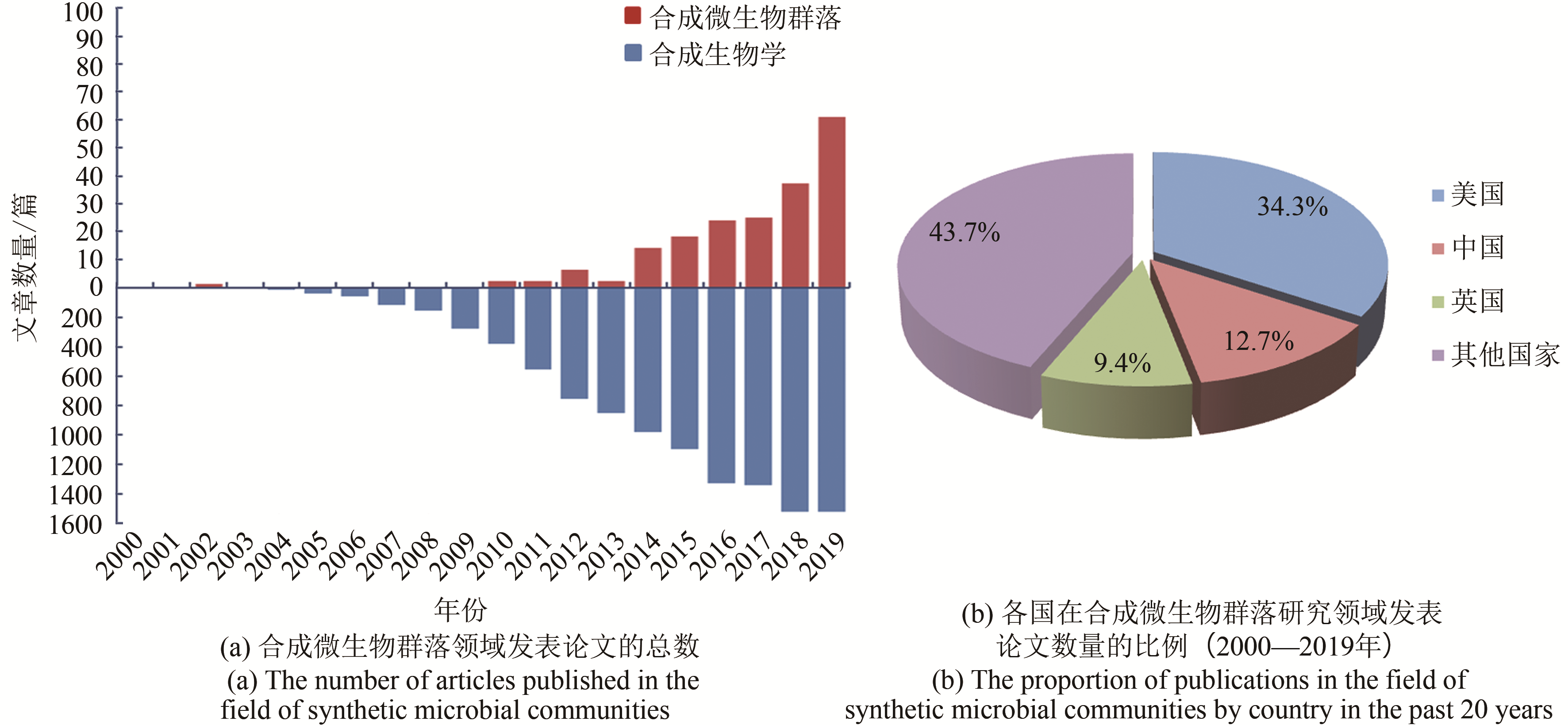
图1 合成微生物群落领域的研究趋势(基于Web of Science数据库,以“synthetic microbial communities/consortia,synthetic microbiota/microbiome,microbiome engineering”为关键词,检索2000—2019年合成微生物群落领域的论文发表情况。合成生物学领域的论文检索以“synthetic biology”为关键词)
Fig. 1 Research trends in synthetic microbial communities(Based on the Web of Science database, search the publications in the past 20 years, in the field of synthetic microbial communities with‘synthetic microbial communities/consortia, synthetic microbiota/microbiome, microbiome engineering’ and in the field of synthetic biology with ‘synthetic biology’)
| 相互作用类型 | 研究案例 |
|---|---|
| 互利互惠(+/+) | Shou等[ |
| Summers等[ | |
| 偏利共生(+/0) | Freilich等[ |
| Kedia等[ | |
| Lu等[ | |
| 寄生或捕食(+/-) | Contreras-Moreno等[ |
| Zhang等[ | |
| 竞争(-/-) | Gadaga等[ |
| Gause[ |
表1 微生物相互作用类型的研究案例
Tab. 1 Examples of different types of microbial interactions
| 相互作用类型 | 研究案例 |
|---|---|
| 互利互惠(+/+) | Shou等[ |
| Summers等[ | |
| 偏利共生(+/0) | Freilich等[ |
| Kedia等[ | |
| Lu等[ | |
| 寄生或捕食(+/-) | Contreras-Moreno等[ |
| Zhang等[ | |
| 竞争(-/-) | Gadaga等[ |
| Gause[ |
| 1 | GROSSKOPF T, SOYER O S. Synthetic microbial communities[J]. Current Opinion in Microbiology, 2014, 18:72-77. |
| 2 | KAREN D R, MASSIMO M, PIETER V A, et al. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities[J]. Environmental Microbiology, 2014, 16(6):1472-1481. |
| 3 | BERNSTEIN H C, CARLSON R P. Microbial consortia engineering for cellular factories: in vitro to in silico systems[J]. Computational and Structural Biotechnology Journal, 2012, 3(4):e201210017. |
| 4 | FALKOWSKI P G, FENCHEL T, DELONG E F. The microbial engines that drive Earth's biogeochemical cycles[J]. Science, 2008, 320(5879):1034-1039. |
| 5 | LEY R E, TURNBAUGH P J, KLEIN S, et al. Microbial ecology - human gut microbes associated with obesity[J]. Nature, 2006, 444(7122):1022-1023. |
| 6 | LUKENS J R, GURUNG P, VOGEL P, et al. Dietary modulation of the microbiome affects autoinflammatory disease[J]. Nature, 2014, 516(7530):246-249. |
| 7 | BERG G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture[J]. Applied Microbiology and Biotechnology, 2009, 84(1):11-18. |
| 8 | PATLE S, LAL B. Ethanol production from hydrolysed agricultural wastes using mixed culture of Zymomonas mobilis and Candida tropicalis [J]. Biotechnology Letters, 2007, 29(12):1839-1843. |
| 9 | PAWELCZYK S, ABRAHAM W R, HARMS H, et al. Community-based degradation of 4-chorosalicylate tracked on the single cell level[J]. Journal of Microbiological Methods, 2008, 75(1):117-126. |
| 10 | RAES J, BORK P. Systems microbiology - timeline - molecular eco-systems biology: towards an understanding of community function[J]. Nature Reviews Microbiology, 2008, 6(9):693-699. |
| 11 | PROSSER J I, BOHANNAN B J M, CURTIS T P, et al. The role of ecological theory in microbial ecology[J]. Nature Reviews Microbiology, 2007, 5(5):384-392. |
| 12 | VENTURELLI O S, CARR A V, FISHER G, et al. Deciphering microbial interactions in synthetic human gut microbiome communities[J]. Molecular Systems Biology, 2018, 14(6):e8157. |
| 13 | LIU F, MAO J W, KONG W T, et al. Interaction variability shapes succession of synthetic microbial ecosystems[J]. Nature Communications, 2020, 11(1):e309. |
| 14 | COYTE K Z, SCHLUTER J, FOSTER K R. The ecology of the microbiome: networks, competition, and stability[J]. Science, 2015, 350(6261):663-666. |
| 15 | BANERJEE S, SCHLAEPPI K, VAN DER H M G. A. Keystone taxa as drivers of microbiome structure and functioning[J]. Nature Reviews Microbiology, 2018, 16(9):567-576. |
| 16 | SONG H, DING M Z, JIA X Q, et al. Synthetic microbial consortia: from systematic analysis to construction and applications[J]. Chemical Society Reviews, 2014, 43(20):6954-6981. |
| 17 | FAUST K, RAES J. Microbial interactions: from networks to models[J]. Nature Reviews Microbiology, 2012, 10(8):538-550. |
| 18 | SHOU W Y, RAM S, VILAR J M G. Synthetic cooperation in engineered yeast populations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6):1877-1882. |
| 19 | SUMMERS Z M, FOGARTY H E, LEANG C, et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria[J]. Science, 2010, 330(6009):1413-1415. |
| 20 | FREILICH S, ZARECKI R, EILAM O, et al. Competitive and cooperative metabolic interactions in bacterial communities[J]. Nature Communications, 2011, 2:e589. |
| 21 | KEDIA G, WANG R, PATEL H, et al. Use of mixed cultures for the fermentation of cereal-based substrates with potential probiotic properties[J]. Process Biochemistry, 2007, 42(1):65-70. |
| 22 | LU Y Y, PUTRA S D, LIU S Q. A novel non-dairy beverage from durian pulp fermented with selected probiotics and yeast[J]. International Journal of Food Microbiology, 2018, 265:1-8. |
| 23 | FRANCISCO J C, JOSE M, NATALIA I J, et al. Copper and melanin play a role in Myxococcus xanthus predation on Sinorhizobium meliloti[J]. Frontiers in Microbiology, 2020, 11:e94. |
| 24 | ZHANG W C, WANG Y, LU H N, et al. Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the Single-Cell Level[J]. Applied and Environmental Microbiology, 2020, 86(3):e02286-19. |
| 25 | GADAGA T H, MUTUKUMIRA A N, NARVHUS J A. The growth and interaction of yeasts and lactic acid bacteria isolated from zimbabwean naturally fermented milk in UHT milk[J]. International Journal of Food Microbiology, 2001, 68(1):21-32. |
| 26 | GAUSE G F. The struggle for existence[J]. Williams &Wilkins, 1935, 16(4): 656-657. |
| 27 | NIU B, PAULSON J N, ZHENG X, et al. Simplified and representative bacterial community of maize roots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(12):2450-2459. |
| 28 | WINTERMUTE E H, SILVER P A. Dynamics in the mixed microbial concourse[J]. Genes & Development, 2010, 24(23):2603-2614. |
| 29 | BALAGADDE F K, SONG H, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem[J]. Molecular Systems Biology, 2008, 4:e187. |
| 30 | HU B, DU J, ZOU R Y, et al. An environment-sensitive synthetic microbial ecosystem[J]. PLoS One, 2010, 5(5):e10619. |
| 31 | HONG S H, HEGDE M, KIM J, et al. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device[J]. Nature Communications, 2012, 3:e613. |
| 32 | CHEN Y, KIM J K, HIRNING A J, et al. Emergent genetic oscillations in a synthetic microbial consortium[J]. Science, 2015, 349(6251):986-989. |
| 33 | KONG W T, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14(8):821-829. |
| 34 | KERNER A, PARK J, WILLIAMS A, et al. A programmable Escherichia coli consortium via tunable symbiosis[J]. PLoS One, 2012, 7(3):e34032. |
| 35 | KUBO I, HOSODA K, SUZUKI S, et al. Construction of bacteria-eukaryote synthetic mutualism[J]. Biosystems, 2013, 113(2):66-71. |
| 36 | HUANG A C, JIANG T, LIU Y X, et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota[J]. Science, 2019, 364(6440):eaau6389. |
| 37 | TEIXEIRA P, COLAIANNI N R, FITZPATRICK C R, et al. Beyond pathogens: microbiota interactions with the plant immune system[J]. Current Opinion in Microbiology, 2019, 49:7-17. |
| 38 | RAWLS J F, MAHOWALD M A, LEY R E, et al. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection[J]. Cell, 2006, 127(2):423-433. |
| 39 | ROESELERS G, MITTGE E K, STEPHENS W Z, et al. Evidence for a core gut microbiota in the zebrafish[J]. ISME Journal, 2011, 5(10):1595-1608. |
| 40 | RATZKE C, GORE J. Modifying and reacting to the environmental pH can drive bacterial interactions[J]. PLoS Biology, 2018, 16(3):e2004248. |
| 41 | FOERSTNER K U, VON M C, BORK P. Comparative analysis of environmental sequences: potential and challenges[J]. Philosophical Transactions of the Royal Society B-Biological Sciences, 2006, 361(1467):519-523. |
| 42 | RAMETTE A. Multivariate analyses in microbial ecology[J]. FEMS Microbiology Ecology, 2007, 62(2):142-160. |
| 43 | BERTIN P N, MEDIGUE C, NORMAND P. Advances in environmental genomics: towards an integrated view of micro-organisms and ecosystems[J]. Microbiology-(UK), 2008, 154:347-359. |
| 44 | SHONG J, JIMENEZ D M R, COLLINS C H. Towards synthetic microbial consortia for bioprocessing[J]. Current Opinion in Microbiology, 2012, 23(5):798-802. |
| 45 | GARCIA F C, BESTION E, WARFIELD R, et al. Changes in temperature alter the relationship between biodiversity and ecosystem functioning[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(43):10989-10999. |
| 46 | WANG X F, WEI Z, YANG K M, et al. Phage combination therapies for bacterial wilt disease in tomato[J]. Nature Biotechnology, 2019, 37(12):1513-1520. |
| 47 | HSU B B, GIBSON T E, YELISEYEV V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model[J]. Cell Host & Microbe, 2019, 25(6):803-814. |
| 48 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12):725-741. |
| 49 | STEIN R R, TANOUE T, SZABADY R L, et al. Computer-guided design of optimal microbial consortia for immune system modulation [J]. eLife, 2018, 7:e30916. |
| 50 | ZHAO C H, SINUMVAYO J P, ZHANG Y P, et al. Design and development of a ‘Y-shaped’ microbial consortium capable of simultaneously utilizing biomass sugars for efficient production of butanol [J]. Metabolic Engineering, 2019, 55:111-119. |
| 51 | PAPENFORT K, BASSLER B L. Quorum sensing signal-response systems in Gram-negative bacteria [J]. Nature Reviews Microbiology, 2016, 14(9):576-588. |
| 52 | MARTELLACCI L, QUARANTA G, PATINI R, et al. A Literature review of metagenomics and culturomics of the peri-implant microbiome: current evidence and future perspectives[J]. Materials, 2019, 12(18):e3010. |
| 53 | LAGIER J C, DUBOURG G, MILLION M, et al. Culturing the human microbiota and culturomics[J]. Nature Reviews Microbiology, 2018, 16(9):540-550. |
| 54 | SARHAN M S, HAMZA M A, YOUSSEF H H, et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media - a review[J]. Journal of Advanced Research, 2019, 19:15-27. |
| 55 | LAGIER J C, ARMOUGOM F, MILLION M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study[J]. Clinical Microbiology and Infection, 2012, 18(12):1185-1193. |
| 56 | BAI Y, MULLER D B, SRINIVAS G, et al. Functional overlap of the Arabidopsis leaf and root microbiota [J]. Nature, 2015, 528(7582):364-369. |
| 57 | OBERHARDT M A, ZARECKI R, GRONOW S, et al. Harnessing the landscape of microbial culture media to predict new organism-media pairings[J]. Nature Communications, 2015, 6:8493. |
| 58 | WARD T L, LARSON J, MEULEMANS J, et al. BugBase predicts organism-level microbiome phenotypes [EB/OL]. [2017-05-02]. . |
| 59 | KAMINSKI T S, SCHELER O, GARSTECKI P. Droplet microfluidics for microbiology: techniques, applications and challenges[J]. Lab on a Chip, 2016, 16(12):2168-2187. |
| 60 | BEIN A, SHIN W, JALILI F S, et al. Microfluidic organ-on-a-chip models of human Intestine[J]. Cellular and Molecular Gastroenterology and Hepatology, 2018, 5(4):659-668. |
| 61 | ALEKLETT K, KIERS E T, OHLSSON P, et al. Build your own soil: exploring microfluidics to create microbial habitat structures[J]. Isme Journal, 2018, 12(2):312-319. |
| 62 | KEHE J, KULESA A, ORTIZ A, et al. Massively parallel screening of synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(26):12804-12809. |
| 63 | KEHE J, ORTIZ A, KULESA A, et al. Positive interactions are common among culturable bacteria [EB/OL]. [2020-06-25]. . |
| 64 | ALBERTSEN M, HUGENHOLTZ P, SKARSHEWSKI A, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes[J]. Nature Biotechnology, 2013, 31(6):533-538. |
| 65 | HAWLEY A K, BREWER H M, NORBECK A D, et al. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31):11395-11400. |
| 66 | SWENSON T L, KARAOZ U, SWENSON J M, et al. Linking soil biology and chemistry in biological soil crust using isolate exometabolomics[J]. Nature Communications, 2018, 9(1):19. |
| 67 | PERNTHALER J, GLOCKNER F O, SCHONHUBER W, et al. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes[J]. Methods in Microbiology, 2001, 30:207-226. |
| 68 | GHOSH A, NILMEIER J, WEAVER D, et al. A peptide-based method for C-13 metabolic flux analysis in microbial communities[J]. PLoS Computational Biology, 2014, 10(9):e1003827. |
| 69 | GEBRESELASSIE N A, ANTONIEWICZ M R. C-13-metabolic flux analysis of co-cultures: a novel approach[J]. Metabolic Engineering, 2015, 31:132-139. |
| 70 | GILMORE I S, HEILES S, PIETERSE C L. Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging[J]. Annual Review of Analytical Chemistry, 2019, 12(1):201-224. |
| 71 | DUNHAM S J B, ELLIS J F, LI B, et al. Mass spectrometry imaging of complex microbial communities[J]. Accounts of Chemical Research, 2017, 50(1):96-104. |
| 72 | NUNEZ J, RENSLOW R, CLIFF J B, et al. NanoSIMS for biological applications: current practices and analyses[J]. Biointerphases, 2018, 13(3):03B301. |
| 73 | VRANCKEN G, GREGORY A C, HUYS G R B, et al. Synthetic ecology of the human gut microbiota[J]. Nature Reviews Microbiology, 2019, 17(12)754-763. |
| 74 | JO J, OH J, PARK C. Microbial community analysis using high-throughput sequencing technology: a beginner's guide for microbiologists[J]. Journal of Microbiology, 2020, 58(3):176-192. |
| 75 | ZHANG X, DEEKE S A, NING Z B, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease[J]. Nature Communications, 2018, 9(1):2873. |
| 76 | HEYER R, SCHALLERT K, SIEWERT C, et al. Metaproteome analysis reveals that syntrophy, competition, and phage-host interaction shape microbial communities in biogas plants[J]. Microbiome, 2019, 7(1):69. |
| 77 | FRANZOSA E A, MCIVER L J, RAHNAVARD Gholamali, et al. Species-level functional profiling of metagenomes and metatranscriptomes[J]. Nature Methods, 2018, 15(11):962-968. |
| 78 | KNIEF C, DELMOTTE N, CHAFFRON S, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice[J]. ISME Journal, 2012, 6(7):1378-1390. |
| 79 | BROBERG M, DOONAN J, MUNDT F, et al. Integrated multi-omic analysis of host-microbiota interactions in acute oak decline[J]. Microbiome, 2018, 6(1):21. |
| 80 | PERNTHALER A, DEKAS A E, BROWN C T, et al. Diverse syntrophic partnerships from-deep-sea methane vents revealed by direct cell capture and metagenomics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(19):7052-7057. |
| 81 | SEKIGUCHI Y, KAMAGATA Y, NAKAMURA K, et al. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules[J]. Applied and Environmental Microbiology, 1999, 65(3):1280-1288. |
| 82 | NAVA G M, FRIEDRICHSEN H J, STAPPENBECK T S. Spatial organization of intestinal microbiota in the mouse ascending colon[J]. ISME Journal, 2011, 5(4):627-638. |
| 83 | SWIDSINSKI A, WEBER J, LOENING-BAUCKE V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease[J]. Journal of Clinical Microbiology, 2005, 43(7):3380-3389. |
| 84 | WHITAKER W R, SHEPHERD E S, SONNENBURG J L. Tunable expression tools enable single-cell strain distinction in the gut microbiome[J]. Cell, 2017, 169(3):538-546. |
| 85 | GEVA-ZATORSKY N, ALVAREZ D, HUDAK J E, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria[J]. Nature Medicine, 2015, 21(9):1091-1100. |
| 86 | PRAVESCHOTINUNT P, DORVAL C N M, DEN H I, et al. Tracking of engineered bacteria in vivo using nonstandard amino acid incorporation[J]. ACS Synthetic Biology, 2018, 7(6):1640-1650. |
| 87 | EARLE K A, BILLINGS G, SIGAL M, et al. Quantitative imaging of gut microbiota spatial organization[J]. Cell Host Microbe, 2015, 18(4):478-488. |
| 88 | MARK WELCH J. L., HASEGAWA Y., MCNULTY N. P., et al. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(43):9105-9114. |
| 89 | WELCH J L M, ROSSETTI Blair J, RIEKEN C W, et al. Biogeography of a human oral microbiome at the micron scale[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(6):791-800. |
| 90 | WILBERT S A, WELCH J L M, BORISY G G. Spatial ecology of the human tongue dorsum microbiome[J]. Cell Host & Microbe, 2020, 30(12):4003-4015. |
| 91 | PINTO F, MEDINA D A, PEREZ-CORREA J R, et al. Modeling metabolic interactions in a consortium of the infant gut microbiome[J]. Frontiers in Microbiology, 2017, 8:e2507. |
| 92 | THIELE I, PALSSON B O. A protocol for generating a high-quality genome-scale metabolic reconstruction[J]. Nature Protocols, 2010, 5(1):93-121. |
| 93 | GONZE D, LAHTI L, RAES J, et al. Multi-stability and the origin of microbial community types[J]. ISME Journal, 2017, 11(10):2159-2166. |
| 94 | GOYAL A, DUBINKINA V, MASLOV S. Multiple stable states in microbial communities explained by the stable marriage problem[J]. ISME Journal, 2018, 12(12):2823-2834. |
| 95 | GIBSON T E, BASHAN A, CAO H T, et al. On the origins and control of community types in the human microbiome[J]. PLoS Computational Biology, 2016, 12(2):e1004688. |
| 96 | CARLSTRÖM C I, FIELD C M, BORTFELD-MILLER M, et al. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere[J]. Nature Ecology & Evolution, 2019, 3(10):1445-1454. |
| 97 | GONZE D, COYTE Katharine Z, LAHTI L, et al. Microbial communities as dynamical systems[J]. Current Opinion in Microbiology, 2018, 44:41-49. |
| 98 | CAO H T, GIBSON T E, BASHAN A, et al. Inferring human microbial dynamics from temporal metagenomics data: pitfalls and lessons[J]. Bioessays, 2017, 39(2):e1600188. |
| 99 | XIAO Y D, TULIO A M, FRIEDMAN J, et al. Mapping the ecological networks of microbial communities[J]. Nature Communications, 2017, 8(1):2042. |
| 100 | FRIEDMAN J, HIGGINS L M, GORE J. Community structure follows simple assembly rules in microbial microcosms [J]. Nature Ecology & Evolution, 2017, 1(5):0109. |
| 101 | LI C, CHNG K R, KWAH J S, et al. An expectation-maximization algorithm enables accurate ecological modeling using longitudinal microbiome sequencing data [J]. Microbiome, 2019, 7(1):118. |
| 102 | BUCCI V, TZEN B, LI N, et al. MDSINE: Microbial dynamical systems inference engine for microbiome time-series analyses[J]. Genome Biology, 2016, 17(1):e121. |
| 103 | GOTTSTEIN W, OLIVIER B G, BRUGGEMAN F J, et al. Constraint-based stoichiometric modelling from single organisms to microbial communities[J]. Journal of the Royal Society Interface, 2016, 13(124):e20160627. |
| 104 | KETTLE H, LOUIS P, HOLTROP G, et al. Modelling the emergent dynamics and major metabolites of the human colonic microbiota[J]. Environmental Microbiology, 2015, 17(5):1615-1630. |
| 105 | PACHECO Alan R, MOEL Mauricio, SEGRE Daniel. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems[J]. Nature Communications, 2019, 10(1):103. |
| 106 | GOLDFORD J E, LU N X, BAJIC D, et al. Emergent simplicity in microbial community assembly[J]. Science, 2018, 361(6401):469-474. |
| 107 | HUTTENHOWER C. Structure, function and diversity of the healthy human microbiome[J]. Nature, 2012, 486:207-214. |
| 108 | LLOYD-PRICE J, ABU-ALI, G, HUTTENHOWER C. The healthy human microbiome[J]. Genome Medicine, 2016, 8:51-62. |
| 109 | GENSOLLEN T, IYER S S, KASPER D L, et al. How colonization by microbiota in early life shapes the immune system[J]. Science, 2016, 352:539-544. |
| 110 | HONDA K, LITTMAN D R. The microbiota in adaptive immune homeostasis and disease[J]. Nature, 2016, 535:75-84. |
| 111 | CLEMENTE J C, URSELL L K, PARFREY L W, et al. The impact of the gut microbiota on human health: an integrative view[J]. Cell, 2012, 148(6):1258-1270. |
| 112 | BÄCKHED F, DING H, WANG T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101:15718-15723. |
| 113 | LEY R E, BÄCKHED F, TURNBAUGH P, et al. Obesity alters gut microbial ecology[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102:11070-11075. |
| 114 | RIDAURA V K. Gut microbiota from twins discordant for obesity modulate metabolism in mice[J]. Science, 2013, 341(6150):e1241214. |
| 115 | TURNBAUGH P J. A core gut microbiome in obese and lean twins[J]. Nature, 2009, 457:480-484. |
| 116 | KARLSSON F H. Gut metagenome in European women with normal impaired and diabetic glucose control[J]. Nature, 2013, 498:99-103. |
| 117 | LARSEN N. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults[J]. PLoS One, 2010, 5:9085. |
| 118 | QIN J J, LI R Q, RAES J, et al. A human gut microbial gene catalogue established by metagenomic sequencing[J]. Nature, 2010, 464:59-65. |
| 119 | TILG H, MOSCHEN A R. Microbiota and diabetes: an evolving relationship[J]. Gut Microbes, 2014, 63:1513-1521. |
| 120 | CANI P D. Human gut microbiome: hopes, threats and promises[J]. Gut Microbes, 2018, 67:1716-1725. |
| 121 | SCHMIDT T S B, RAES J, BORK P. The human gut microbiome: from association to modulation[J]. Cell, 2018, 172:1198-1215. |
| 122 | GIANOTTI R J, MOSS A C. Fecal microbiota transplantation: from Clostridium difficile to inflammatory bowel disease[J]. Gastroenterology & Hepatology, 2017, 13(4):209-213. |
| 123 | ANDERSON J L, EDNEY R J, WHELAN K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease[J]. Alimentary Pharmacology & Therapeutics, 2012, 36(6):503-516. |
| 124 | ZHANG F M, WANG H G, WANG M, et al. Fecal microbiota transplantation for severe enterocolonic fistulizing Crohn's disease[J]. World Journal of Gastroenterology, 2013, 19(41):7213-7216. |
| 125 | WILLEM M V. Fame and future of faecal transplantations - developing next-generation therapies with synthetic microbiomes[J]. Microbial Biotechnology, 2013, 6(4):316-325. |
| 126 | ATARASHI K, TANOUE T, SHIMA T, et al. Induction of colonic regulatory T cells by indigenous clostridium species[J]. Science, 2011, 331(6015):337-341. |
| 127 | CHIBA T, SENO H. Indigenous clostridium species regulate systemic immune responses by induction of colonic regulatory T cells[J]. Gastroenterology, 2011, 141(3):1114-1116. |
| 128 | TANOUE T, MORITA S, PLICHTA D R, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity[J]. Nature, 2019, 565(7741):600-605. |
| 129 | BERENDSEN R L, PIETERSE C M J, BAKKER P. The rhizosphere microbiome and plant health[J]. Trends in Plant Science, 2012, 17(8):478-486. |
| 130 | ZHANG J Y, LIU Y X, ZHANG N, et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice[J]. Nature Biotechnology, 2019, 37(6):676-684. |
| 131 | FINKEL O M, SALAS-GONZALEZ I, CASTRILLO G, et al. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response[J]. PLoS Biology, 2019, 17(11):e3000534. |
| 132 | LEBEIS S L, PAREDES S H, LUNDBERG D S, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa[J]. Science, 2015, 349(6250):860-864. |
| 133 | CASTRILLO G, TEIXEIRA P J P L, PAREDES S H, et al. Root microbiota drive direct integration of phosphate stress and immunity[J]. Nature, 2017, 543(7646):513-518. |
| 134 | WEI Z, GU Y, FRIMAN V P, et al. Initial soil microbiome composition and functioning predetermine future plant health[J]. Science Advances, 2019, 5(9):eaaw0759. |
| 135 | DURÁN P, THIERGART T, GARRIDO-OTER R, et al. Microbial interkingdom interactions in roots promote Arabidopsis survival[J]. Cell, 2018, 175(4):973-983. |
| 136 | LI M, WEI Z, WANG J N, et al. Facilitation promotes invasions in plant-associated microbial communities[J]. Ecology Letters, 2019, 22(1):149-158. |
| 137 | LIU Y X, QIN Y, BAI Y. Reductionist synthetic community approaches in root microbiome research[J]. Current Opinion in Microbiology, 2019, 49:97-102. |
| 138 | SYED AB RAHMAN S F, SINGH E, PIETERSE C M J, et al. Emerging microbial biocontrol strategies for plant pathogens[J]. Plant Science : an International Journal of Experimental Plant Biology, 2018, 267:102-111. |
| 139 | AZIZOGLU U. Bacillus thuringiensis as a biofertilizer and biostimulator: a mini-review of the little-known plant growth-promoting properties of Bt[J]. Current Microbiology, 2019, 76(11):1379-1385. |
| 140 | 刘炜伟, 吴冰, 向梅春, 等. 从微生物组到合成功能菌群[J]. 微生物学通报, 2017, 44(4):881-889. |
| LIU W W, WU B, XIANG M C, et al. From microbiome to synthetic microbial community[J]. Microbiology China, 2017, 44(4):881-889. | |
| 141 | 王丽娟. 产乙偶姻功能微生物群落的分离及其在食醋酿造中的应用 [D]. 无锡:江南大学, 2018. |
| WANG L J. Isolation of acetoin-producing microbial community in vinegar Pei and its application in vinegar brewing process[D]. Wuxi: Jiangnan University, 2018. | |
| 142 | WU Q, LING J, XU Y. Starter culture selection for making Chinese sesame-flavored liquor based on microbial metabolic activity in mixed-culture fermentation[J]. Applied and Environmental Microbiology, 2014, 80(14):4450-4459. |
| 143 | CHEN Y L. Development and application of co-culture for ethanol production by co-fermentation of glucose and xylose: a systematic review[J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(5):581-597. |
| 144 | ABDALLA M A, SULIEMAN S, MCGAW L J. Microbial communication: a significant approach for new leads[J]. South African Journal of Botany, 2017, 113:461-470. |
| 145 | SCHROECKH V, SCHERLACH K, NUTZMANN H W, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(34):14558-14563. |
| 146 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4):377-383. |
| 147 | RICHARDSON M. Pesticides - friend or foe?[J]. Water Science and Technology, 1998, 37(8):19-25. |
| 148 | DEJONGHE W, BERTELOOT E, GORIS J, et al. Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain[J]. Applied and Environmental Microbiology, 2003, 69(3):1532-1541. |
| 149 | HEIJDEN M G A VAN DER, KLIRONOMOS J N, URSIC M, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity[J]. Nature, 1998, 396(6706):69-72. |
| 150 | BRENNER K, YOU L C, ARNOLD F H. Engineering microbial consortia: a new frontier in synthetic biology[J]. Trends in Biotechnology, 2008, 26(9):483-489. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||


