合成生物学 ›› 2021, Vol. 2 ›› Issue (6): 876-885.DOI: 10.12211/2096-8280.2020-053
微生物共培养生产化学品的研究进展
李向来, 申晓林, 王佳, 袁其朋, 孙新晓
- 北京化工大学,化工资源有效利用国家重点实验室,北京 100029
-
收稿日期:2020-04-18修回日期:2021-01-15出版日期:2021-12-31发布日期:2022-01-21 -
通讯作者:孙新晓 -
作者简介:李向来 (1993—),男,博士研究生。研究方向为代谢工程及合成生物学。E-mail:1003277591@qq.com孙新晓 (1985—),男,博士,副教授。研究方向为代谢工程及合成生物学。E-mail:sunxx@mail.buct.edu.cn -
基金资助:国家重点研发计划(2018YFA0901800);北京化工大学双一流项目(XK1802-8)
Recent advances in biosynthesis of chemicals by microbial co-culture
LI Xianglai, SHEN Xiaolin, WANG Jia, YUAN Qipeng, SUN Xinxiao
- State Key Laboratory of Chemical Resources Engineering,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2020-04-18Revised:2021-01-15Online:2021-12-31Published:2022-01-21 -
Contact:SUN Xinxiao
摘要:
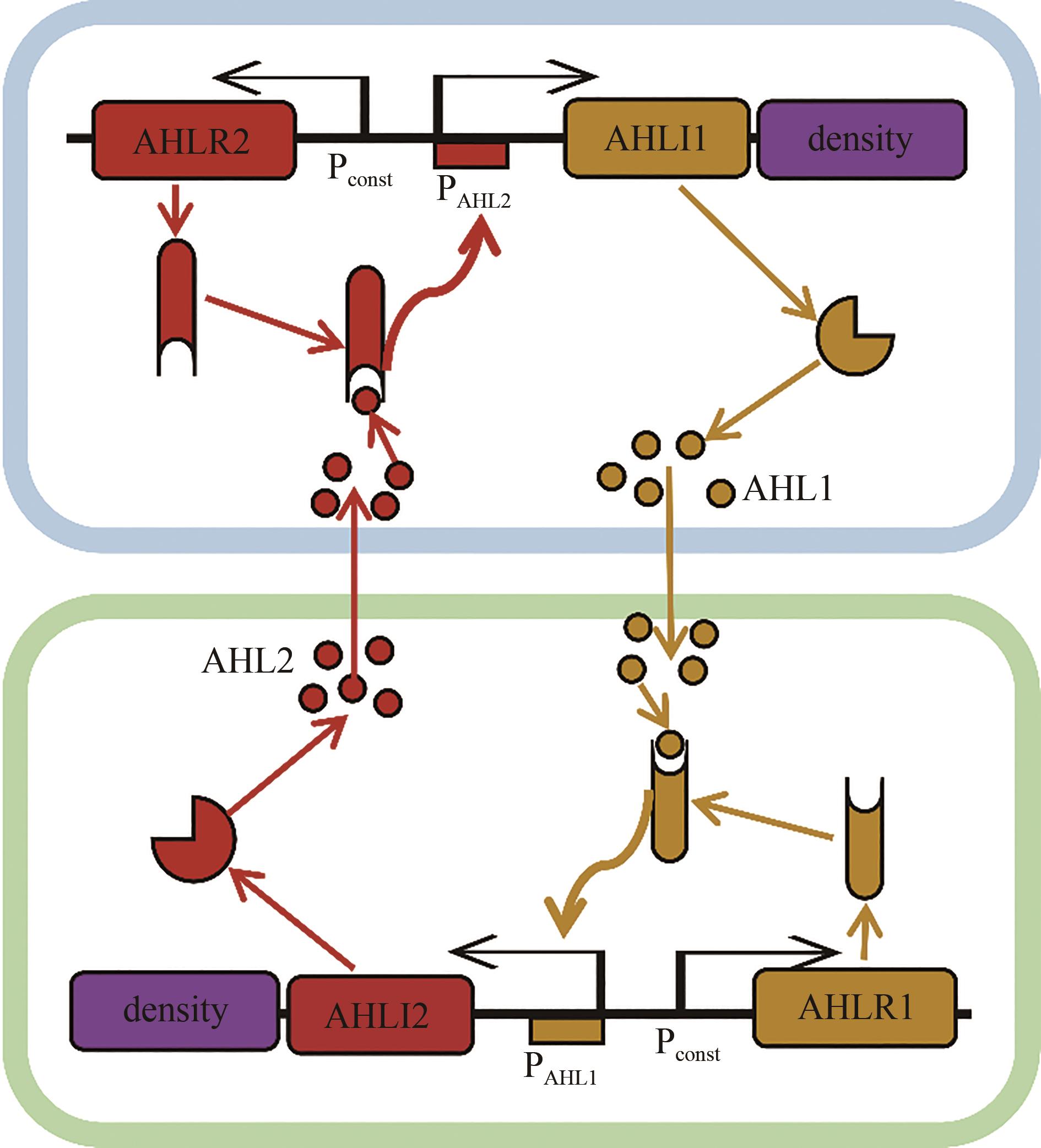
生物合成已成为化学品绿色制造的重要方式。传统上,微生物合成化学品以单菌株培养为主。然而,单培养经常存在引入复杂途径造成沉重代谢负担、细胞微环境无法满足所有酶的功能性表达以及不同途径模块之间相互干扰等问题。借鉴自然界中普遍存在的共生现象,研究者开发了共培养技术,通过在同一体系中培养两种或多种细胞,以充分模拟自然共生环境,实现不同物种之间能量、物质及信号的交流,达到劳动分工以及代谢分区的目的。该技术在减轻宿主代谢负担、提供适宜的酶催化环境以及底物共利用方面表现出突出优势。不过作为一种新兴技术,微生物共培养技术在菌群稳定性、物种兼容性以及菌群比例调控等方面还存在一些挑战。本文列举了近年来微生物共培养划分长途径减轻代谢负担以及利用复杂、混合、非常规底物生产化学品和扩大化学品多样性的成功案例,总结了通过群体感应调控菌群比例以及通过计算机模拟工具预测菌群动态变化的研究进展,并对设计复杂稳定可控的共培养体系在高效生产化学品方面的应用前景和挑战进行了讨论。共培养技术有望成为合成复杂化学品的重要策略,并推动合成生物学的发展。
中图分类号:
引用本文
李向来, 申晓林, 王佳, 袁其朋, 孙新晓. 微生物共培养生产化学品的研究进展[J]. 合成生物学, 2021, 2(6): 876-885.
LI Xianglai, SHEN Xiaolin, WANG Jia, YUAN Qipeng, SUN Xinxiao. Recent advances in biosynthesis of chemicals by microbial co-culture[J]. Synthetic Biology Journal, 2021, 2(6): 876-885.
| 共培养体系 | 产物 | 产量/(mg/L) | 较单培养提高 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌/酿酒酵母 | 氧化紫杉烷 | 33 | 单培养无产物 | [ |
| 大肠杆菌 | 黄烷醇 | 40.7 | 970倍 | [ |
| 大肠杆菌 | 迷迭香酸 | 172 | 38倍 | [ |
| 大肠杆菌 | 红景天苷 | 6030(罐) | 20倍 | [ |
| 大肠杆菌 | 咖啡醇 | 401 | 12倍 | [ |
| 大肠杆菌 | 黏糠酸 | 682 | 19倍 | [ |
| 大肠杆菌 | 苯酚 | 210 | 3.9倍 | [ |
| 大肠杆菌 | 柚皮素 | 24 | 35% | [ |
表1 共培养生产化学品的代表性研究进展汇总
Tab. 1 Summary of representative research progresses on microbial co-culture to produce chemicals
| 共培养体系 | 产物 | 产量/(mg/L) | 较单培养提高 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌/酿酒酵母 | 氧化紫杉烷 | 33 | 单培养无产物 | [ |
| 大肠杆菌 | 黄烷醇 | 40.7 | 970倍 | [ |
| 大肠杆菌 | 迷迭香酸 | 172 | 38倍 | [ |
| 大肠杆菌 | 红景天苷 | 6030(罐) | 20倍 | [ |
| 大肠杆菌 | 咖啡醇 | 401 | 12倍 | [ |
| 大肠杆菌 | 黏糠酸 | 682 | 19倍 | [ |
| 大肠杆菌 | 苯酚 | 210 | 3.9倍 | [ |
| 大肠杆菌 | 柚皮素 | 24 | 35% | [ |
| 1 | SUN X X, SHEN X L, JAIN R, et al. Synthesis of chemicals by metabolic engineering of microbes[J]. Chemical Society Reviews, 2015, 44(11): 3760-3785. |
| 2 | DE LIMA BROSSI M J, JIMÉNEZ D J, CORTES-TOLALPA L, et al. Soil-derived microbial consortia enriched with different plant biomass reveal distinct players acting in lignocellulose degradation[J]. Microbial Ecology, 2016, 71(3): 616-627. |
| 3 | BILIOURIS K, BABSON D, SCHMIDT-DANNERT C, et al. Stochastic simulations of a synthetic bacteria-yeast ecosystem[J]. BMC Systems Biology, 2012, 6: 58. |
| 4 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 5 | HANLY T J, HENSON M A. Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures[J]. Biotechnology and Bioengineering, 2011, 108(2): 376-385. |
| 6 | WANG Z Y, CAO G L, ZHENG J, et al. Developing a mesophilic co-culture for direct conversion of cellulose to butanol in consolidated bioprocess[J]. Biotechnology for Biofuels, 2015, 8(1):84. |
| 7 | SCHOLZ S A, GRAVES I, MINTY J J, et al. Production of cellulosic organic acids via synthetic fungal consortia[J]. Biotechnology and Bioengineering, 2018, 115(4): 1096-1100. |
| 8 | JIANG Y J, ZHANG T, LU J S, et al. Microbial co-culturing systems: butanol production from organic wastes through consolidated bioprocessing[J]. Applied Microbiology and Biotechnology, 2018, 102(13): 5419-5425. |
| 9 | JIANG Y J, GUO D, LU J S, et al. Consolidated bioprocessing of butanol production from xylan by a thermophilic and butanologenic Thermoanaerobacterium sp. M5[J]. Biotechnology for Biofuels, 2018, 11: 89. |
| 10 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor[J]. Microbial Cell Factories, 2019, 18(1): 1-11. |
| 11 | ZHANG H R, WANG X N. Modular co-culture engineering, a new approach for metabolic engineering[J]. Metabolic Engineering, 2016, 37: 114-121. |
| 12 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids[J]. Metabolic Engineering, 2016, 35: 55-63. |
| 13 | LI Z H, WANG X N, ZHANG H R. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering[J]. Metabolic Engineering, 2019, 54: 1-11. |
| 14 | LIU X, LI X B, JIANG J, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metabolic Engineering, 2018, 47: 243-253. |
| 15 | CHEN Z Y, SUN X X, LI Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols[J]. Metabolic Engineering, 2017, 39: 102-109. |
| 16 | ZHANG H R, PEREIRA B, LI Z J, et al. Engineering Escherichia coli coculture systems for the production of biochemical products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(27): 8266-8271. |
| 17 | GUO X Y, LI Z H, WANG X N, et al. De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering[J]. Biotechnology and Bioengineering, 2019, 116(12): 3349-3359. |
| 18 | GANESAN V, LI Z H, WANG X N, et al. Heterologous biosynthesis of natural product naringenin by co-culture engineering[J]. Synthetic and Systems Biotechnology, 2017, 2(3): 236-242. |
| 19 | THUAN N H, TRUNG N T, CUONG N X, et al. Escherichia coli modular coculture system for resveratrol glucosides production[J]. World Journal of Microbiology and Biotechnology, 2018, 34(6): 1-13. |
| 20 | THUAN N H, CHAUDHARY A K, CUONG D V, et al. Engineering co-culture system for production of apigetrin in Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(3): 175-185. |
| 21 | CAMACHO-ZARAGOZA J M, HERNÁNDEZ-CHÁVEZ G, MORENO-AVITIA F, et al. Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol[J]. Microbial Cell Factories, 2016, 15(1): 1-11. |
| 22 | AKDEMIR H, SILVA A, ZHA J, et al. Production of pyranoanthocyanins using Escherichia coli co-cultures[J]. Metabolic Engineering, 2019, 55: 290-298. |
| 23 | SUN J, RAZA M, SUN X X, et al. Biosynthesis of adipic acid via microaerobic hydrogenation of cis,cis-muconic acid by oxygen-sensitive enoate reductase[J]. Journal of Biotechnology, 2018, 280: 49-54. |
| 24 | FAN L H, ZHANG Z J, YU X Y, et al. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(33): 13260-13265. |
| 25 | HE Q, HEMME C L, JIANG H L, et al. Mechanisms of enhanced cellulosic bioethanol fermentation by co-cultivation of Clostridium and Thermoanaerobacter spp.[J]. Bioresource Technology, 2011, 102(20):9586-9592. |
| 26 | NAKAYAMA S, KIYOSHI K, KADOKURA T, et al. Butanol production from crystalline cellulose by cocultured Clostridium thermocellum and Clostridium saccharoperbutylacetonicum N1-4[J]. Applied and Environmental Microbiology, 2011, 77(18): 6470-6475. |
| 27 | MINTY J J, SINGER M E, SCHOLZ S A, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(36): 14592-14597. |
| 28 | WANG M, YU C Z, ZHAO H M. Directed evolution of xylose specific transporters to facilitate glucose-xylose co-utilization[J]. Biotechnology and Bioengineering, 2016, 113(3): 484-491. |
| 29 | SEDLAK M, HO N W. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast[J]. Yeast, 2004, 21(8):671-684. |
| 30 | LI H B, SCHMITZ O, ALPER H S. Enabling glucose/xylose co-transport in yeast through the directed evolution of a sugar transporter[J]. Applied Microbiology and Biotechnology, 2016, 100(23): 10215-10223. |
| 31 | VERHOEVEN M D, DE VALK S C, DARAN J M G, et al. Fermentation of glucose-xylose-arabinose mixtures by a synthetic consortium of single-sugar-fermenting Saccharomyces cerevisiae strains[J]. FEMS Yeast Research, 2018, 18(8): foy075. |
| 32 | HILL E A, CHRISLER W B, BELIAEV A S, et al. A flexible microbial co-culture platform for simultaneous utilization of methane and carbon dioxide from gas feedstocks[J]. Bioresource Technology, 2017, 228: 250-256. |
| 33 | LEE C R, KIM C, SONG Y E, et al. Co-culture-based biological carbon monoxide conversion by Citrobacter amalonaticus Y19 and Sporomusa ovata via a reducing-equivalent transfer mediator[J]. Bioresource Technology, 2018, 259: 128-135. |
| 34 | WEISS T L, YOUNG E J, DUCAT D C. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production[J]. Metabolic Engineering, 2017, 44: 236-245. |
| 35 | DUCAT D C, AVELAR-RIVAS J A, WAY J C, et al. Rerouting carbon flux to enhance photosynthetic productivity[J]. Applied and Environmental Microbiology, 2012, 78(8): 2660-2668. |
| 36 | SMITH M J, FRANCIS M B. A designed A. vinelandii-S. elongatus coculture for chemical photoproduction from air, water, phosphate, and trace metals[J]. ACS Synthetic Biology, 2016, 5(9): 955-961. |
| 37 | HAYS S G, YAN L L W, SILVER P A, et al. Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction[J]. Journal of Biological Engineering, 2017, 11: 4. |
| 38 | LI T, LI C-T, BUTLER K, et al. Mimicking lichens: incorporation of yeast strains together with sucrose-secreting cyanobacteria improves survival, growth, ROS removal, and lipid production in a stable mutualistic co-culture production platform[J]. Biotechnology for Biofuels, 2017, 10(1):55. |
| 39 | LI T T, JIANG L Q, HU Y F, et al. Creating a synthetic lichen: mutualistic co-culture of fungi and extracellular polysaccharide-secreting cyanobacterium Nostoc PCC 7413[J]. Algal Research, 2020, 45:101755. |
| 40 | GOODWIN C R, COVINGTON B C, DEREWACZ D K, et al. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps[J]. Chemistry & Biology, 2015, 22(5): 661-670. |
| 41 | PONOMAROVA O, PATIL K R. Metabolic interactions in microbial communities: untangling the Gordian knot[J]. Current Opinion in Microbiology, 2015, 27: 37-44. |
| 42 | BERTRAND S, BOHNI N, SCHNEE S, et al. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery[J]. Biotechnology Advances, 2014, 32(6): 1180-1204. |
| 43 | HARWANI D, BEGANI J, LAKHANI J. Co-cultivation strategies to induce de novo synthesis of novel chemical scaffolds from cryptic secondary metabolite gene clusters [M]// GEHLOT P, SINGH J. Fungi and their role in sustainable development: current perspectives. Springer, 2018: 617-631. |
| 44 | TAN Z Q, LEOW H Y, LEE D C W, et al. Co-culture systems for the production of secondary metabolites: current and future prospects[J]. The Open Biotechnology Journal, 2019, 13(1): 18-26. |
| 45 | MOUSSA M, EBRAHIM W, BONUS M, et al. Co-culture of the fungus Fusarium tricinctum with Streptomyces lividans induces production of cryptic naphthoquinone dimers[J]. RSC Advances, 2019, 9(3): 1491-1500. |
| 46 | MA Y J, ZHENG L P, WANG J W. Inducing perylenequinone production from a bambusicolous fungus Shiraia sp. S9 through co-culture with a fruiting body-associated bacterium Pseudomonas fulva SB1 [J]. Microbial Cell Factories, 2019, 18(1): 121. |
| 47 | HOSHINO S, WONG C P, OZEKI M, et al. Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium[J]. The Journal of Antibiotics, 2018, 71(7):653-657. |
| 48 | KHALIL Z G, CRUZ-MORALES P, LICONA-CASSANI C, et al. Inter-Kingdom beach warfare: microbial chemical communication activates natural chemical defences[J]. The ISME Journal, 2019, 13(1): 147-158. |
| 49 | ARORA D, CHASHOO G, SINGAMANENI V, et al. Bacillus amyloliquefaciens induces production of a novel blennolide K in coculture of Setophoma terrestris [J]. Journal of Applied Microbiology, 2018, 124(3):730-739. |
| 50 | ZHOU Q Y, YANG X Q, ZHANG Z X, et al. New azaphilones and tremulane sesquiterpene from endophytic Nigrospora oryzae cocultured with Irpex lacteus [J]. Fitoterapia, 2018, 130: 26-30. |
| 51 | NG W L, BASSLER B L. Bacterial quorum-sensing network architectures[J]. Annual Review of Genetics, 2009, 43: 197-222. |
| 52 | DAVIS R M, MULLER R Y, HAYNES K A. Can the natural diversity of quorum-sensing advance synthetic biology?[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 30. |
| 53 | KYLILIS N, TUZA Z A, STAN G B, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia[J]. Nature Communications, 2018, 9(1): 2677. |
| 54 | SCOTT S R, HASTY J. Quorum sensing communication modules for microbial consortia[J]. ACS Synthetic Biology, 2016, 5(9): 969-977. |
| 55 | CHEN Y, KIM J K, HIRNING A J, et al. Emergent genetic oscillations in a synthetic microbial consortium[J]. Science, 2015, 349(6251):986-989. |
| 56 | SCOTT S R, DIN M O, BITTIHN P, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis[J]. Nature Microbiology, 2017, 2: 17083. |
| 57 | MCCARDELL R D, HUANG S, GREEN L N, et al. Control of bacterial population density with population feedback and molecular sequestration[EB/OL]. bioRxiv, 2017, DOI:10.1101/225045 . |
| 58 | THOMPSON J A, OLIVEIRA R A, DJUKOVIC A, et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota[J]. Cell Reports, 2015, 10(11):1861-1871. |
| 59 | STEPHENS K, POZO M, TSAO C Y, et al. Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition[J]. Nature Communications, 2019, 10(1): 4129. |
| 60 | AUTEBERT J, COUDERT B, F-C BIDARD, et al. Microfluidic: an innovative tool for efficient cell sorting[J]. Methods, 2012, 57(3):297-307. |
| 61 | KOCH S, BENNDORF D, FRONK K, et al. Predicting compositions of microbial communities from stoichiometric models with applications for the biogas process[J]. Biotechnology for Biofuels, 2016, 9(1):17. |
| 62 | MILNE C B, KIM P J, EDDY J A, et al. Accomplishments in genome-scale in silico modeling for industrial and medical biotechnology[J]. Biotechnology Journal, 2009, 4(12): 1653-1670. |
| 63 | MILLER I J, VANEE N, FONG S S, et al. Lack of overt genome reduction in the bryostatin-producing bryozoan symbiont ''candidatus endobugula sertula''[J]. Applied and Environmental Microbiology, 2016, 82(22): 6573-6583. |
| 64 | STOLYAR S, DIEN S VAN, HILLESLAND K L, et al. Metabolic modeling of a mutualistic microbial community[J]. Molecular System Biology, 2007, 3(1): 92. |
| 65 | SALIMI F, ZHUANG K, MAHADEVAN R. Genome‐scale metabolic modeling of a clostridial co‐culture for consolidated bioprocessing[J]. Biotechnology Journal, 2010, 5(7):726-738. |
| 66 | KONG W, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14(8): 821-829. |
| 67 | MCCULLY A L, LASARRE B, MCKINLAY J B. Growth-independent cross-feeding modifies boundaries for coexistence in a bacterial mutualism[J]. Environmental Microbiology, 2017, 19(9): 3538-3550. |
| 68 | TSOI R, WU F L, ZHANG C, et al. Metabolic division of labor in microbial systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2526-2531. |
| 69 | GUERRA-BUBB J, CROTEAU R, WILLIAMS R M. The early stages of taxol biosynthesis: an interim report on the synthesis and identification of early pathway metabolites[J]. Natural Product Reports, 2012, 29(6): 683-696. |
| 70 | NITSCHKE M, PASTORE G M. Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater[J]. Bioresource Technology, 2006, 97(2): 336-341. |
| 71 | TAO K Y, LIU X Y, CHEN X P, et al. Biodegradation of crude oil by a defined co-culture of indigenous bacterial consortium and exogenous Bacillus subtilis [J]. Bioresource Technology, 2017, 224:327-332. |
| [1] | 仲泉周, 单依怡, 裴清云, 金艳芸, 王艺涵, 孟璐远, 王歆韵, 张雨鑫, 刘坤媛, 王慧中, 冯尚国. 生物合成法生产α-熊果苷的研究进展[J]. 合成生物学, 2025, 6(1): 118-135. |
| [2] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [3] | 刘益宁, 蒲伟, 杨金星, 王钰. ω-氨基酸与内酰胺的生物合成研究进展[J]. 合成生物学, 2024, 5(6): 1350-1366. |
| [4] | 李庚, 申晓林, 孙新晓, 王佳, 袁其朋. 过氧化物酶的重组表达和应用研究进展[J]. 合成生物学, 2024, 5(6): 1498-1517. |
| [5] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [6] | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| [7] | 刘子健, 穆柏杨, 段志强, 王璇, 陆晓杰. 与核酸兼容的化学反应开发进展[J]. 合成生物学, 2024, 5(5): 1102-1124. |
| [8] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| [9] | 谢向前, 郭雯, 王欢, 李进. 含氨基乙烯半胱氨酸核糖体肽的生物合成与化学合成[J]. 合成生物学, 2024, 5(5): 981-996. |
| [10] | 汤志军, 胡友财, 刘文. 酶促4+2和2+2环加成反应:区域与立体选择性的理解与应用[J]. 合成生物学, 2024, 5(3): 401-407. |
| [11] | 张俊, 金诗雪, 云倩, 瞿旭东. 聚酮化合物非天然延伸单元的生物合成与结构改造应用[J]. 合成生物学, 2024, 5(3): 561-570. |
| [12] | 陈锡玮, 张华然, 邹懿. 真菌源非核糖体肽类药物生物合成及代谢工程[J]. 合成生物学, 2024, 5(3): 571-592. |
| [13] | 冯金, 潘海学, 唐功利. 近十年天然产物药物的生物合成研究进展[J]. 合成生物学, 2024, 5(3): 408-446. |
| [14] | 奚萌宇, 胡逸灵, 顾玉诚, 戈惠明. 基因组挖掘指导天然药物分子的发现[J]. 合成生物学, 2024, 5(3): 447-473. |
| [15] | 施鑫杰, 杜艺岭. 双嵌入家族抗肿瘤非核糖体肽的生物合成研究进展[J]. 合成生物学, 2024, 5(3): 593-611. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||