合成生物学 ›› 2023, Vol. 4 ›› Issue (4): 840-851.DOI: 10.12211/2096-8280.2022-048
• 研究论文 • 上一篇
限制性内切酶的无细胞快速制备研究
刘晚秋, 季向阳, 许慧玲, 卢屹聪, 李健
- 上海科技大学物质科学与技术学院,上海 201210
-
收稿日期:2022-09-06修回日期:2022-12-01出版日期:2023-08-31发布日期:2023-09-14 -
通讯作者:李健 -
作者简介:刘晚秋 (1986—),女,博士,副研究员。研究方向为无细胞合成生物学。E-mail:liuwq@shanghaitech.edu.cn李健 (1982—),男,博士,研究员,博士生导师。研究方向为无细胞合成生物学。E-mail:lijian@shanghaitech.edu.cn -
基金资助:国家自然科学基金(32171427)
Cell-free protein synthesis system enables rapid and efficient biosynthesis of restriction endonucleases
LIU Wanqiu, JI Xiangyang, XU Huiling, LU Yicong, LI Jian
- School of Physical Science and Technology,ShanghaiTech University,Shanghai 201210,China
-
Received:2022-09-06Revised:2022-12-01Online:2023-08-31Published:2023-09-14 -
Contact:LI Jian
摘要:
限制性内切酶在分子生物学研究中是一类重要的工具酶,目前主要由异源生物合成的方式进行表达与生产,由于它们对特定的DNA序列(即酶切位点)具有切割活性,在异源表达时会对宿主产生较高的细胞毒性。而无细胞生物合成体系具有操作快捷、灵活高效、无细胞毒性等优势,因此,本研究利用无细胞蛋白合成(cell-free protein synthesis, CFPS)技术进行限制性内切酶的表达制备。本课题组选择3种限制性内切酶EcoRⅠ、BamHⅠ和BsaⅠ作为研究对象,构建线性DNA为表达模板,无需甲基化酶对宿主的保护,在6 h内即可完成蛋白表达。经亲和色谱与凝胶色谱两步纯化,得到了纯度高(95%左右)、酶活相当(EcoRⅠ 3.7 × 105~3.7 × 106 U/mg,BamHⅠ 8.3 × 102~4.1 × 103 U/mg,BsaⅠ 4.4 × 105 ~ 4.4 × 106 U/mg)的目标蛋白。同时,建立了限制性内切酶的实时酶活检测方法,将有助于限制性内切酶的催化和快速筛选研究。本研究所开发的限制性内切酶无细胞表达制备体系,从基因模板构建到纯化蛋白所需时间短(1~2 d)、蛋白产量高(32.5~130 mg/L无细胞反应)、制备效率高(1.3 × 105 ~ 5.7 × 108 U/L无细胞反应),具有较好的普适性,为限制性内切酶的研发与制备生产提供了新的思路。
中图分类号:
引用本文
刘晚秋, 季向阳, 许慧玲, 卢屹聪, 李健. 限制性内切酶的无细胞快速制备研究[J]. 合成生物学, 2023, 4(4): 840-851.
LIU Wanqiu, JI Xiangyang, XU Huiling, LU Yicong, LI Jian. Cell-free protein synthesis system enables rapid and efficient biosynthesis of restriction endonucleases[J]. Synthetic Biology Journal, 2023, 4(4): 840-851.
| 引物名称 | 序列 | 用途 |
|---|---|---|
| linearPro_F | CCTACAGCGTGAGCATTG | 扩增启动子片段 |
| linearPro_R | CATATGGTGATGATGATG | |
| linearTer_F | GTCGACCGGCTGCTAACA | 扩增终止子片段 |
| linearTer_R | CGGATTCAGTCGTCACTCA | |
| SUMOPro_F | GGATCTCGACGCTCTCCCT | 扩增启动子+SUMO标签片段 |
| SUMOPro_R | AGGTCCCTGAAACAGGACCTCTAAACCACCAATCTGTTCTCTG | |
| SUMO_mut_F | TACGACGGTATTCGTATTCAAGCTGATCAGAC | 去除pSUMO质粒中的EcoRⅠ酶切位点 |
| SUMO_mut_R | CAGCTTGAATACGAATACCGTCGTACAAGAATC | |
| EcoRⅠ_SUMO_F | TTAGAGGTCCTGTTTCAGGGACCTAGCAACAAAAAACAGAGC | 扩增EcoRⅠ基因片段 |
| EcoRⅠ_SUMO_R | CATCATCATCACCATATGAGCAACAAAAAACAGAGC | |
| sfGFP_SUMO_F | ATTGGTGGTACCGAGCTCATGAGCAAAGGTGAAGAA | 构建质粒pSUMO-sfGFP |
| sfGFP_SUMO_R | GAGTGCGGCCGCAAGCTTTTATTTTTCGAACTGCGG | |
| BamHⅠ_F | CATCATCATCACCATATGAAAGTGGAAAAAGA | 扩增BamHⅠ基因片段 |
| BamHⅠ_R | TGTTAGCAGCCGGTCGACTTATTTGTTTTCCACTTTATC | |
| BsaⅠ_F | TTAGAGGTCCTGTTTCAGGGACCTATGGCAAAAAAGCGGAA | 扩增BsaⅠ基因片段 |
| BsaⅠ_R | TGTTAGCAGCCGGTCGACTTAATCCAGATCCGCAAA |
表1 本研究中用到的引物序列
Table 1 Oligonucleotide primers used in this study
| 引物名称 | 序列 | 用途 |
|---|---|---|
| linearPro_F | CCTACAGCGTGAGCATTG | 扩增启动子片段 |
| linearPro_R | CATATGGTGATGATGATG | |
| linearTer_F | GTCGACCGGCTGCTAACA | 扩增终止子片段 |
| linearTer_R | CGGATTCAGTCGTCACTCA | |
| SUMOPro_F | GGATCTCGACGCTCTCCCT | 扩增启动子+SUMO标签片段 |
| SUMOPro_R | AGGTCCCTGAAACAGGACCTCTAAACCACCAATCTGTTCTCTG | |
| SUMO_mut_F | TACGACGGTATTCGTATTCAAGCTGATCAGAC | 去除pSUMO质粒中的EcoRⅠ酶切位点 |
| SUMO_mut_R | CAGCTTGAATACGAATACCGTCGTACAAGAATC | |
| EcoRⅠ_SUMO_F | TTAGAGGTCCTGTTTCAGGGACCTAGCAACAAAAAACAGAGC | 扩增EcoRⅠ基因片段 |
| EcoRⅠ_SUMO_R | CATCATCATCACCATATGAGCAACAAAAAACAGAGC | |
| sfGFP_SUMO_F | ATTGGTGGTACCGAGCTCATGAGCAAAGGTGAAGAA | 构建质粒pSUMO-sfGFP |
| sfGFP_SUMO_R | GAGTGCGGCCGCAAGCTTTTATTTTTCGAACTGCGG | |
| BamHⅠ_F | CATCATCATCACCATATGAAAGTGGAAAAAGA | 扩增BamHⅠ基因片段 |
| BamHⅠ_R | TGTTAGCAGCCGGTCGACTTATTTGTTTTCCACTTTATC | |
| BsaⅠ_F | TTAGAGGTCCTGTTTCAGGGACCTATGGCAAAAAAGCGGAA | 扩增BsaⅠ基因片段 |
| BsaⅠ_R | TGTTAGCAGCCGGTCGACTTAATCCAGATCCGCAAA |

图2 限制性内切酶EcoRⅠ的无细胞制备(Western blot和SDS-PAGE检测无细胞表达、纯化的限制性内切酶EcoRⅠ)CE—cell extract,以BL21 Star (DE3)制备的细胞提取物;CE+ —含有分子伴侣(pG-KJE8)的BL21 Star(DE3)细胞提取物;T—全菌蛋白;S—可溶蛋白;M—蛋白标准样品;FT—纯化上样穿出液;W—洗杂流出液;Eluate—洗脱液
Fig. 2 Cell-free production of restriction endonuclease EcoRⅠ(Western blot and SDS-PAGE analyses of cell-free expressed and purified restriction endonuclease EcoRⅠ) CE—cell extract of BL21 Star (DE3); CE+ —cell extract of BL21 Star (DE3) with chaperone (pG-KJE8); T—total protein; S—soluble protein; M—marker; FT—flow throughout sample; W—washed sample; Eluate—eluted sample
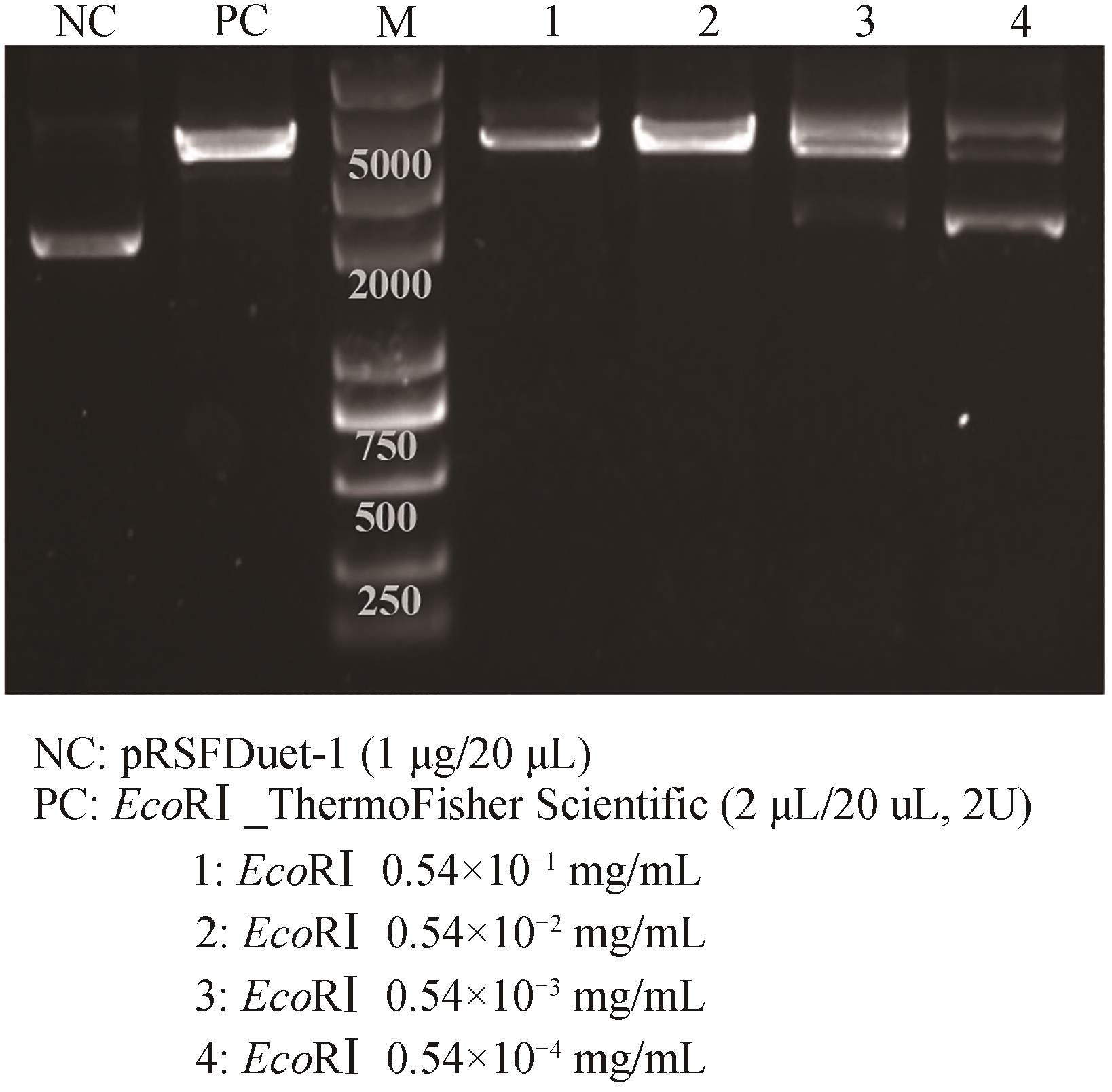
图4 无细胞制备限制性内切酶EcoRⅠ的酶活测定NC—阴性对照;PC—阳性对照
Fig. 4 Catalytic activity determination of cell-free produced restriction endonuclease EcoRⅠNC—negative control; PC—positive control
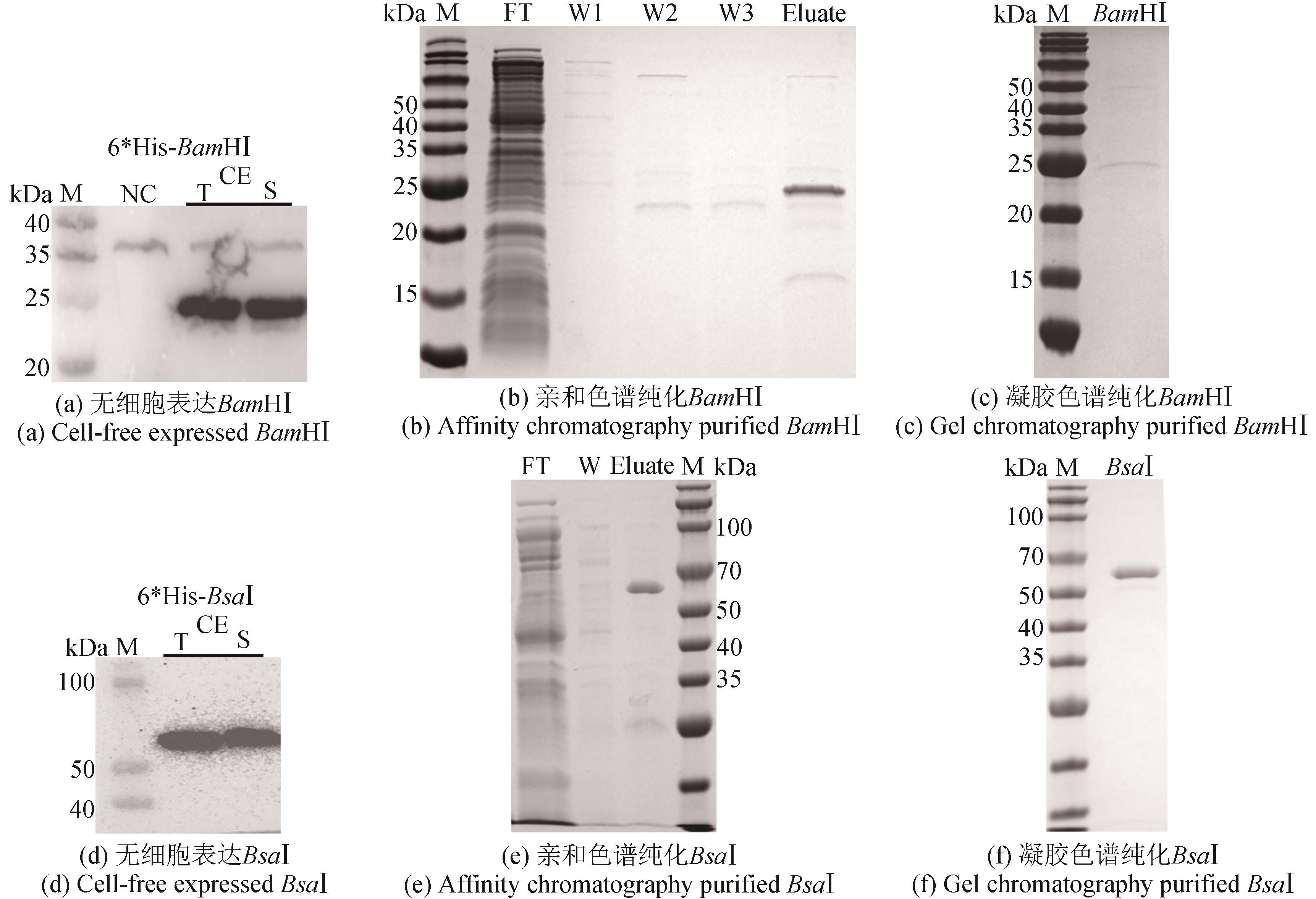
图5 限制性内切酶BamHⅠ和BsaⅠ的无细胞制备(Western blot和SDS-PAGE检测无细胞表达纯化的限制性内切酶BamHⅠ和BsaⅠ)
Fig. 5 Cell-free production of restriction endonucleases BamHⅠ and BsaⅠ(Western Blot and SDS-PAGE analyses of cell-free expressed and purified restriction endonucleases BamHⅠ and BsaⅠ)
| 1 | LURIA S E, HUMAN M L. A nonhereditary, host-induced variation of bacterial viruses[J]. Journal of Bacteriology, 1952, 64(4): 557-569. |
| 2 | BERTANI G, WEIGLE J J. Host controlled variation in bacterial viruses[J]. Journal of Bacteriology, 1953, 65(2): 113-121. |
| 3 | ARBER W. Host-controlled modification of bacteriophage[J]. Annual Review of Microbiology, 1965, 19: 365-378. |
| 4 | DANNA K, NATHANS D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae [J]. Proceedings of the National Academy of Sciences of the United States of America, 1971, 68(12): 2913-2917. |
| 5 | KELLY T J JR, SMITH H O. A restriction enzyme from Hemophilus influenzae.Ⅱ[J]. Journal of Molecular Biology, 1970, 51(2): 393-409. |
| 6 | WILLIAMS R J. Restriction endonuclease[J]. Molecular Biotechnology, 2003, 23(3): 225-243. |
| 7 | PINGOUD A M. Restriction Endonucleases[M]. Berlin, Heidelberg: Springer Berlin Heidelberg, 2004. |
| 8 | PINGOUD A, WILSON G G, WENDE W. TypeⅡrestriction endonucleases—a historical perspective and more[J]. Nucleic Acids Research, 2014, 42(12): 7489-7527. |
| 9 | ROBERTS R J. How restriction enzymes became the workhorses of molecular biology[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(17): 5905-5908. |
| 10 | KNIZEWSKI L, KINCH L N, GRISHIN N V, et al. Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches[J]. BMC Structural Biology, 2007, 7: 40. |
| 11 | GUPTA R, CAPALASH N, SHARMA P. Restriction endonucleases: natural and directed evolution[J]. Applied Microbiology and Biotechnology, 2012, 94(3): 583-599. |
| 12 | DI FELICE F, MICHELI G, CAMILLONI G. Restriction enzymes and their use in molecular biology: an overview[J]. Journal of Biosciences, 2019, 44(2): 38. |
| 13 | CHENG S C, KIM R, KING K, et al. Isolation of gram quantities of EcoRⅠ restriction and modification enzymes from an overproducing strain[J]. Journal of Biological Chemistry, 1984, 259(18): 11571-11575. |
| 14 | HOCHULI E, BANNWARTH W, DÖBELI H, et al. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent[J]. Nature Biotechnology, 1988, 6(11): 1321-1325. |
| 15 | WATANABE N, TAKASAKI Y, SATO C, et al. Structures of restriction endonuclease HindⅢ in complex with its cognate DNA and divalent cations[J]. Acta Crystallographica Section D, 2009, 65(12): 1326-1333. |
| 16 | 朱化星, 邹媛华, 汤玉洁, 等. 一种制备限制性内切酶类产品的方法: CN113652412A[P]. 2021-11-16. |
| ZHU H X, ZOU Y H, TANG Y J, et al. Method for preparing restriction enzyme product: CN113652412A[P]. 2021-11-16. | |
| 17 | 于博文, 李建辉, 单永超. 一种SalⅠ限制性内切酶的制备方法: CN112813087A[P]. 2021-05-18. |
| YU B W, LI J H, SHAN Y C. Preparation method of SalⅠ restriction endonuclease: CN112813087A[P]. 2021-05-18. | |
| 18 | ORLOWSKI J, BUJNICKI J M. Structural and evolutionary classification of type Ⅱ restriction enzymes based on theoretical and experimental analyses[J]. Nucleic Acids Research, 2008, 36(11): 3552-3569. |
| 19 | LIU W Q, ZHANG L, CHEN M, et al. Cell-free protein synthesis: recent advances in bacterial extract sources and expanded applications[J]. Biochemical Engineering Journal, 2019, 141: 182-189. |
| 20 | LIU W Q, WU C Z, JEWETT M C, et al. Cell-free protein synthesis enables one-pot cascade biotransformation in an aqueous-organic biphasic system[J]. Biotechnology and Bioengineering, 2020, 117(12): 4001-4008. |
| 21 | XU H L, YANG C, TIAN X T, et al. Regulatory part engineering for high-yield protein synthesis in an all-Streptomyces-based cell-free expression system[J]. ACS Synthetic Biology, 2022, 11(2): 570-578. |
| 22 | JI X Y, LIU W Q, LI J. Recent advances in applying cell-free systems for high-value and complex natural product biosynthesis[J]. Current Opinion in Microbiology, 2022, 67: 102142. |
| 23 | LI J, LAWTON T J, KOSTECKI J S, et al. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase[J]. Biotechnology Journal, 2016, 11(2): 212-218. |
| 24 | MARTIN R W, DES SOYE B J, KWON Y C, et al. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids[J]. Nature Communications, 2018, 9: 1203. |
| 25 | ZHANG L Y, GUO W, LU Y. Advances in cell-free biosensors: principle, mechanism, and applications[J]. Biotechnology Journal, 2020, 15(9): 2000187. |
| 26 | SILVERMAN A D, KARIM A S, JEWETT M C. Cell-free gene expression: an expanded repertoire of applications[J]. Nature Reviews Genetics, 2020, 21(3): 151-170. |
| 27 | SI Y Y, KRETSCH A M, DAIGH L M, et al. Cell-free biosynthesis to evaluate lasso peptide formation and enzyme-substrate tolerance[J]. Journal of the American Chemical Society, 2021, 143(15): 5917-5927. |
| 28 | RASOR B J, VÖGELI B, LANDWEHR G M, et al. Toward sustainable, cell-free biomanufacturing[J]. Current Opinion in Biotechnology, 2021, 69: 136-144. |
| 29 | ZAWADA J F, BURGENSON D, YIN G, et al. Cell-free technologies for biopharmaceutical research and production[J]. Current Opinion in Biotechnology, 2022, 76: 102719. |
| 30 | BERNARDI A, BERNARDI G. Cloning of all EcoRⅠ fragments from phage λ in E. coli [J]. Nature, 1976, 264(5581): 89-90. |
| 31 | BROOKS J E, BENNER J S, HEITER D F, et al. Cloning the BamHⅠ restriction modification system[J]. Nucleic Acids Research, 1989, 17(3): 979-997. |
| 32 | ZHU Z Y, SAMUELSON J C, ZHOU J, et al. Engineering strand-specific DNA nicking enzymes from the type ⅡS restriction endonucleases BsaⅠ, BsmBⅠ, and BsmAⅠ[J]. Journal of Molecular Biology, 2004, 337(3): 573-583. |
| 33 | CASINI A, STORCH M, BALDWIN G S, et al. Bricks and blueprints: methods and standards for DNA assembly[J]. Nature Reviews Molecular Cell Biology, 2015, 16(9): 568-576. |
| 34 | KHORASANIZADEH S, PETERS I D, RODER H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues[J]. Nature Structural Biology, 1996, 3(2): 193-205. |
| 35 | CREIGHTON T E. How important is the molten globule for correct protein folding?[J]. Trends in Biochemical Sciences, 1997, 22(1): 6-10. |
| 36 | ENGLANDER S W. Protein folding intermediates and pathways studied by hydrogen exchange[J]. Annual Review of Biophysics and Biomolecular Structure, 2000, 29: 213-238. |
| 37 | MARBLESTONE J G, EDAVETTAL S C, LIM Y, et al. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO[J]. Protein Science, 2006, 15(1): 182-189. |
| 38 | 高嵩, 张坤晓, 许恒皓, 等. 一种高效重组表达限制性内切酶的方法: CN107058260A[P]. 2017-08-18. |
| GAO S, ZHANG K X, XU H H, et al. Method for efficient recombinant expression of restriction endonuclease: CN107058260A[P]. 2017-08-18. | |
| 39 | 于博文, 李建辉, 单永超. 重组NcoⅠ限制性内切酶的制备方法: CN112662647A[P]. 2021-04-16. |
| YU B W, LI J H, SHAN Y C. Preparation method of recombinant NcoⅠ restriction endonuclease: CN112662647A[P]. 2021-04-16. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||



