合成生物学 ›› 2023, Vol. 4 ›› Issue (5): 1000-1019.DOI: 10.12211/2096-8280.2023-031
基于高通量液相色谱质谱技术的菌株筛选与关键分子定量分析研究进展
吴玉洁1,2,3, 刘欣欣1, 刘健慧1, 杨开广1, 随志刚1, 张丽华1, 张玉奎1
- 1.中国科学院大连化学物理研究所,中国科学院分离分析化学重点实验室,辽宁 大连 116023
2.大连理工大学化学学院,辽宁 大连 116024
3.中国科学院大学,北京 100049
-
收稿日期:2023-04-17修回日期:2023-06-26出版日期:2023-10-31发布日期:2023-11-15 -
通讯作者:杨开广,随志刚 -
作者简介:吴玉洁 (1998—),女,博士研究生。研究方向为面向合成生物学的蛋白质组学分析方法。E-mail:yujiewu@dicp.ac.cn杨开广 (1981—),男,研究员,博士生导师。研究方向为蛋白质组学新技术新方法。E-mail:yangkaiguang@dicp.ac.cn随志刚 (1979—),男,副研究员,硕士生导师。研究方向为基于质谱的蛋白质及代谢物高通量分析方法开发。E-mail:zhigangsui@dicp.ac.cn -
基金资助:国家自然科学基金(22104138);中国科学院“青年创新促进会”优秀会员基金(Y2021058)
Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry
WU Yujie1,2,3, LIU Xinxin1, LIU Jianhui1, Yang Kaiguang1, SUI Zhigang1, ZHANG Lihua1, ZHANG Yukui1
- 1.Key Laboratory of Separation Science for Analytical Chemistry,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian 116023,Liaoning,China
2.School of Chemistry,Dalian University of Technology,Dalian 116024,Liaoning,China
3.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2023-04-17Revised:2023-06-26Online:2023-10-31Published:2023-11-15 -
Contact:Yang Kaiguang, SUI Zhigang
摘要:
合成生物学是21世纪新兴的交叉学科,已经为医药、化工、能源、食品和农业等领域带来了显著的改变。微生物细胞工厂(microbial cell factory,MCF)的构建和应用是合成生物学的重要研究内容,正迅速向实用化、产业化的方向发展。得益于基因编辑技术的进步,MCF菌株文库构建的能力大幅提升,亦产生了数量庞大的待测样本,亟需发展高通量自动化的分析检测方法与之匹配。此外,针对关键代谢酶或代谢物的高通量检测技术,也对MCF改造乃至工业化生产有重要的指导意义。液相色谱质谱技术是生物分子检测分析的重要手段,在蛋白质、代谢物定量分析方面占据重要地位。因此本文总结回顾了近年来高通量液相色谱质谱技术在微生物菌株筛选及关键分子定量分析方面的研究进展,从样品制备、色谱分离、质谱分析及数据处理等层面展开阐述,并对这一领域与自动化平台结合的前景及如何深度融合的挑战进行简要概括,为推动基于合成生物学生物智造领域的快速发展提供指导。
中图分类号:
引用本文
吴玉洁, 刘欣欣, 刘健慧, 杨开广, 随志刚, 张丽华, 张玉奎. 基于高通量液相色谱质谱技术的菌株筛选与关键分子定量分析研究进展[J]. 合成生物学, 2023, 4(5): 1000-1019.
WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry[J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019.
| 质谱 技术 | 优缺点 | 菌株培养 方式 | 筛选通量 | 适用对象 | 应用示例 | 参考 文献 |
|---|---|---|---|---|---|---|
| MALDI | 优点:高耐盐性,易于操作,样品用量少,分析速度快,通量高 缺点:需要添加基质,可能引入基质峰干扰 | 琼脂平板 | 约2 s/样品 | 可用于分析脂肪酸等小分子及蛋白质等生物大分子 | 通过检测酰基链磷脂酰胆碱,以筛选高产中链脂肪酸的酿酒酵母菌株 | [ |
| 琼脂平板 | <2.5 s/样品 | 机器视觉算法辅助识别随机分布菌落,应用于大肠杆菌天然产物文库筛选 | [ | |||
| 琼脂平板/ 微孔板 | <5 s/样品 | 开发了菌落位置转换MALDI坐标算法,用于酿酒酵母、大肠杆菌及荧光假单胞菌文库筛选 | [ | |||
| 微孔板 | 约5 s/样品 | 筛选大肠杆菌环二肽合酶突变体文库及羊毛硫肽化合物文库,基本实现整个流程自动化 | [ | |||
| 液滴 | 约5 s/样品 | 酵母甜蛋白及植酸酶产物文库筛选 | [ | |||
| SAMDI | 优点:特异性强,可用于检测复杂体系中的目标分子 缺点:无法区分分子量相同的产物和底物 | 琼脂平板 | <1 s/样品 | 可用于分析小分子 | 利用自组转单层技术在靶板上修饰马来酰亚胺基团,筛选大肠杆菌细胞色素P411突变体文库 | [ |
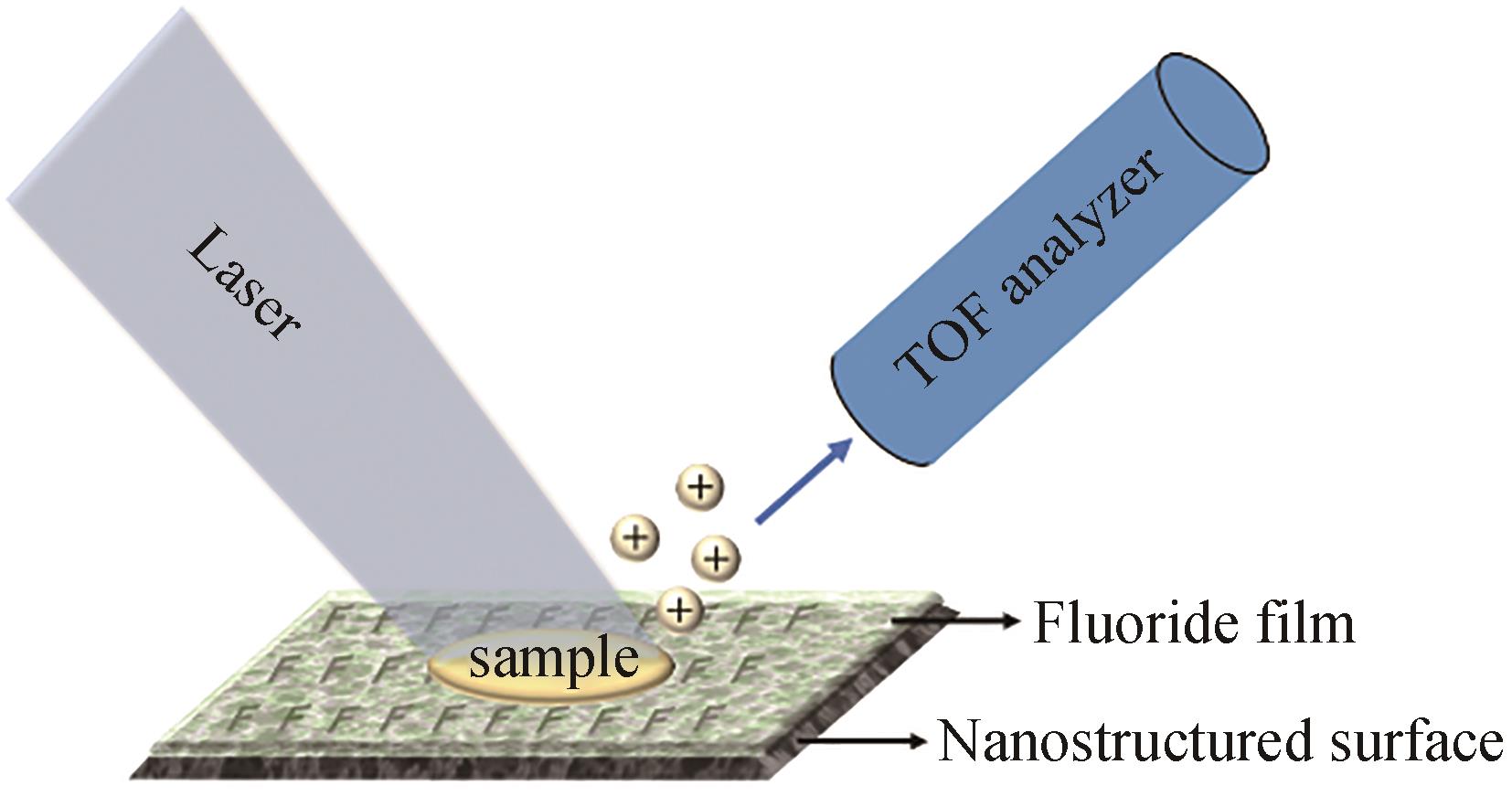
| NIMS | 优点:灵敏度高,无需添加基质 缺点:需要构建纳米结构表面 | 琼脂平板 | <1 s/样品 | 可用于分析小分子、脂质和肽 | 结合点击化学,筛选大肠杆菌细胞色素P450突变体文库 | [ |
| ESI | 优点:采用软电离方式,基本不产生碎片峰,分辨率高 缺点:耐盐性较差,取样效率低 | 微孔板 | 约84 s/样品 | 可用于分析极性强、热稳定性差的小分子和生物大分子 | 液相色谱质谱联用快速筛选三萜桦木酸产量提高的酿酒酵母菌株 | [ |
| 液滴 | 约1 s/样品 | 在线筛选谷氨酸棒杆菌文库 | [ | |||
| 微孔板 | 约60 s/样品 | 融合LESA技术,用于酵母菌株衣康酸、三乙酸内酯和棕榈酸产物定量筛选分析 | [ | |||
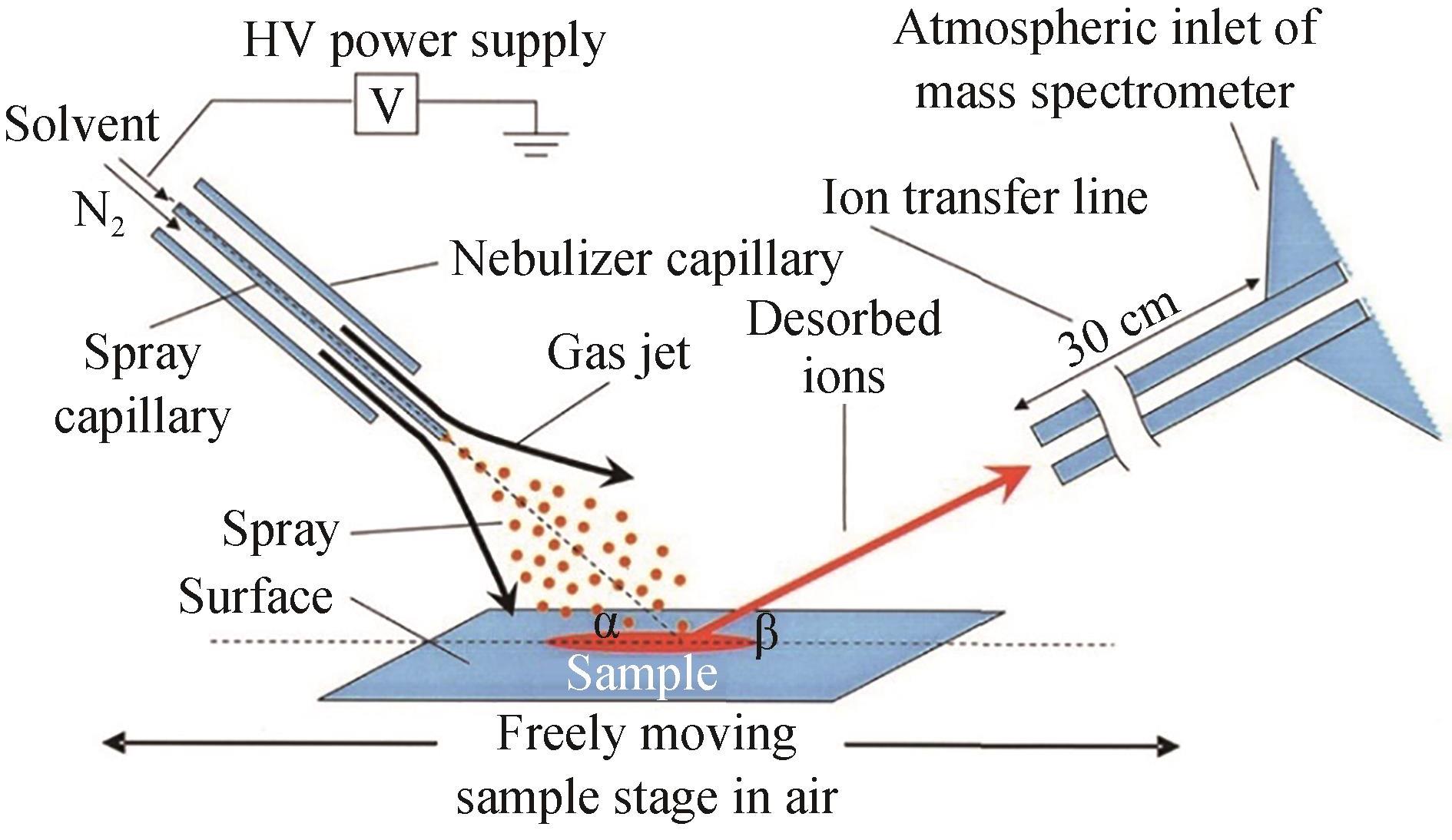
| DESI | 优点:高耐盐性,可实现原位实时分析,通量高 缺点:电喷雾溶剂组成可能影响样品溶解与电离 | 琼脂平板 | <1 s/样品 | 可用于分析非极性小分子及极性大分子 | 与成像技术结合,快速筛选大肠杆菌突变体文库 | [ |
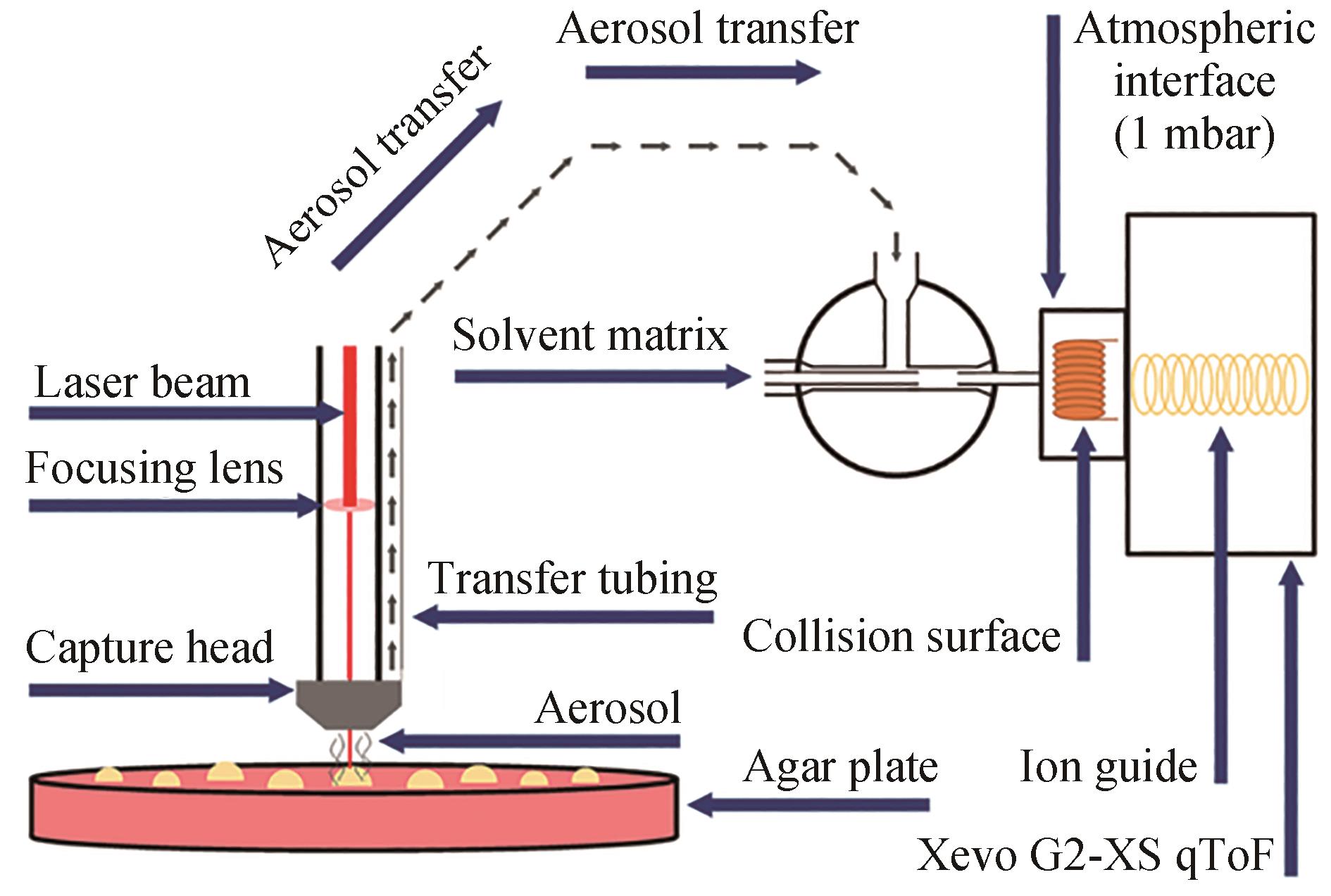
| LA-REI | 优点:易于操作,无需样品制备,快速分析 缺点:无法区分异构体 | 琼脂平板 | 约10 s/样品 | 可用于分析小分子代谢物 | 激光辅助实现酵母白桦酸产物文库筛选 | [ |
表1 基于高通量质谱技术的菌株筛选研究汇总
Table 1 Summary of strain screening studies based on high-throughput mass spectrometry
| 质谱 技术 | 优缺点 | 菌株培养 方式 | 筛选通量 | 适用对象 | 应用示例 | 参考 文献 |
|---|---|---|---|---|---|---|
| MALDI | 优点:高耐盐性,易于操作,样品用量少,分析速度快,通量高 缺点:需要添加基质,可能引入基质峰干扰 | 琼脂平板 | 约2 s/样品 | 可用于分析脂肪酸等小分子及蛋白质等生物大分子 | 通过检测酰基链磷脂酰胆碱,以筛选高产中链脂肪酸的酿酒酵母菌株 | [ |
| 琼脂平板 | <2.5 s/样品 | 机器视觉算法辅助识别随机分布菌落,应用于大肠杆菌天然产物文库筛选 | [ | |||
| 琼脂平板/ 微孔板 | <5 s/样品 | 开发了菌落位置转换MALDI坐标算法,用于酿酒酵母、大肠杆菌及荧光假单胞菌文库筛选 | [ | |||
| 微孔板 | 约5 s/样品 | 筛选大肠杆菌环二肽合酶突变体文库及羊毛硫肽化合物文库,基本实现整个流程自动化 | [ | |||
| 液滴 | 约5 s/样品 | 酵母甜蛋白及植酸酶产物文库筛选 | [ | |||
| SAMDI | 优点:特异性强,可用于检测复杂体系中的目标分子 缺点:无法区分分子量相同的产物和底物 | 琼脂平板 | <1 s/样品 | 可用于分析小分子 | 利用自组转单层技术在靶板上修饰马来酰亚胺基团,筛选大肠杆菌细胞色素P411突变体文库 | [ |
| NIMS | 优点:灵敏度高,无需添加基质 缺点:需要构建纳米结构表面 | 琼脂平板 | <1 s/样品 | 可用于分析小分子、脂质和肽 | 结合点击化学,筛选大肠杆菌细胞色素P450突变体文库 | [ |
| ESI | 优点:采用软电离方式,基本不产生碎片峰,分辨率高 缺点:耐盐性较差,取样效率低 | 微孔板 | 约84 s/样品 | 可用于分析极性强、热稳定性差的小分子和生物大分子 | 液相色谱质谱联用快速筛选三萜桦木酸产量提高的酿酒酵母菌株 | [ |
| 液滴 | 约1 s/样品 | 在线筛选谷氨酸棒杆菌文库 | [ | |||
| 微孔板 | 约60 s/样品 | 融合LESA技术,用于酵母菌株衣康酸、三乙酸内酯和棕榈酸产物定量筛选分析 | [ | |||
| DESI | 优点:高耐盐性,可实现原位实时分析,通量高 缺点:电喷雾溶剂组成可能影响样品溶解与电离 | 琼脂平板 | <1 s/样品 | 可用于分析非极性小分子及极性大分子 | 与成像技术结合,快速筛选大肠杆菌突变体文库 | [ |
| LA-REI | 优点:易于操作,无需样品制备,快速分析 缺点:无法区分异构体 | 琼脂平板 | 约10 s/样品 | 可用于分析小分子代谢物 | 激光辅助实现酵母白桦酸产物文库筛选 | [ |
| 质谱采集类型 | 菌株样本 | 分析通量(每天检测样本数估值) | 参考文献 |
|---|---|---|---|
| 靶向采集 | 大肠杆菌E.coil | 25 min分离梯度定量大肠杆菌中600多条肽(约50样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 19 min有效分离梯度定量检测796个酵母菌株蛋白质组(约70样品/天) | [ |
| 非靶向采集 | 大肠杆菌E.coil | 4 min分离梯度定量226个脂类分子(约320样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 15 min分离梯度结合双分析柱系统检测蛋白(约90样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 23 min分离梯度定量检测100个酵母蛋白质组(约45样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 5 min分离梯度定量1900种蛋白(约280样品/天) | [ |
| 靶向采集 | 大肠杆菌E.coil | 10 min分离梯度定量400多种蛋白质(约130样品/天) | [ |
| 靶向采集 | 恶臭假单胞菌P.putida KT2440 | 32 min分离梯度定量132种蛋白质(约40样品/天) | [ |
| 靶向采集 | 大肠杆菌E.coil | 5 min分离梯度检测102种代谢物(约280样品/天) | [ |
表2 基于液相色谱质谱技术的菌株关键分子定量研究汇总
Table 2 Summary of quantitative studies of key molecules of strains based on liquid chromatography mass spectrometry
| 质谱采集类型 | 菌株样本 | 分析通量(每天检测样本数估值) | 参考文献 |
|---|---|---|---|
| 靶向采集 | 大肠杆菌E.coil | 25 min分离梯度定量大肠杆菌中600多条肽(约50样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 19 min有效分离梯度定量检测796个酵母菌株蛋白质组(约70样品/天) | [ |
| 非靶向采集 | 大肠杆菌E.coil | 4 min分离梯度定量226个脂类分子(约320样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 15 min分离梯度结合双分析柱系统检测蛋白(约90样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 23 min分离梯度定量检测100个酵母蛋白质组(约45样品/天) | [ |
| 非靶向采集 | 酿酒酵母S.cerevistae | 5 min分离梯度定量1900种蛋白(约280样品/天) | [ |
| 靶向采集 | 大肠杆菌E.coil | 10 min分离梯度定量400多种蛋白质(约130样品/天) | [ |
| 靶向采集 | 恶臭假单胞菌P.putida KT2440 | 32 min分离梯度定量132种蛋白质(约40样品/天) | [ |
| 靶向采集 | 大肠杆菌E.coil | 5 min分离梯度检测102种代谢物(约280样品/天) | [ |
| 软件工具 | 分析对象 | 网址 | 参考文献 |
|---|---|---|---|
| AB3D | 蛋白质组 | http://www.first-ms3d.jp/english/(Mass++插件) | [ |
| DynaMet | 代谢组 | https://pypi.python.org/pypi/dynamet/ | [ |
| DIA-NN | 蛋白质组 | https://github.com/vdemichev/diann | [ |
| Tric | 蛋白质组 | http://proteomics.ethz.ch/tric/ | [ |
| MRMAnalyzer | 代谢组 | http://link.springer.com/10.1007/s11306-015-0809-4(R包) | [ |
| MESSI | 代谢组 | http://sbb.hku.hk/MESSI/ | [ |
| PTools | 代谢组 | https://brg.ai.sri.com/ptools/ | [ |
表3 关键分子定量分析中数据处理软件及生物信息学工具汇总
Table 3 Summary of data processing software and bioinformatics tools in the quantitative analysis of key molecules
| 软件工具 | 分析对象 | 网址 | 参考文献 |
|---|---|---|---|
| AB3D | 蛋白质组 | http://www.first-ms3d.jp/english/(Mass++插件) | [ |
| DynaMet | 代谢组 | https://pypi.python.org/pypi/dynamet/ | [ |
| DIA-NN | 蛋白质组 | https://github.com/vdemichev/diann | [ |
| Tric | 蛋白质组 | http://proteomics.ethz.ch/tric/ | [ |
| MRMAnalyzer | 代谢组 | http://link.springer.com/10.1007/s11306-015-0809-4(R包) | [ |
| MESSI | 代谢组 | http://sbb.hku.hk/MESSI/ | [ |
| PTools | 代谢组 | https://brg.ai.sri.com/ptools/ | [ |
| 1 | DRUBIN D A, WAY J C, SILVER P A. Designing biological systems[J]. Genes & Development, 2007, 21(3): 242-254. |
| 2 | CAMERON D E, BASHOR C J, COLLINS J J. A brief history of synthetic biology[J]. Nature Reviews Microbiology, 2014, 12(5): 381-390. |
| 3 | 唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台[J]. 科学通报, 2021, 66(3): 300-309. |
| TANG T, FU L H, GUO E P, et al. Automation in synthetic biology using biological foundries[J]. Chinese Science Bulletin, 2021, 66(3): 300-309. | |
| 4 | CLARKE L, KITNEY R. Developing synthetic biology for industrial biotechnology applications[J]. Biochemical Society Transactions, 2020, 48(1): 113-122. |
| 5 | VOIGT C A. Synthetic biology 2020-2030: six commercially-available products that are changing our world[J]. Nature Communications, 2020, 11: 6379. |
| 6 | 熊燕, 陈大明, 杨琛, 等. 合成生物学发展现状与前景[J]. 生命科学, 2011, 23(9): 826-837. |
| XIONG Y, CHEN D M, YANG C, et al. Progress and perspective of synthetic biology[J]. Chinese Bulletin of Life Sciences, 2011, 23(9): 826-837. | |
| 7 | 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673. |
| YUAN Y M, XING X H, ZHANG C. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale[J]. Synthetic Biology Journal, 2020, 1(6): 656-673. | |
| 8 | 杨建花, 苏晓岚, 朱蕾蕾. 高通量筛选系统在定向改造中的新进展[J]. 生物工程学报, 2021, 37(7): 2197-2210. |
| YANG J H, SU X L, ZHU L L. Advances of high-throughput screening system in reengineering of biological entities[J]. Chinese Journal of Biotechnology, 2021, 37(7): 2197-2210. | |
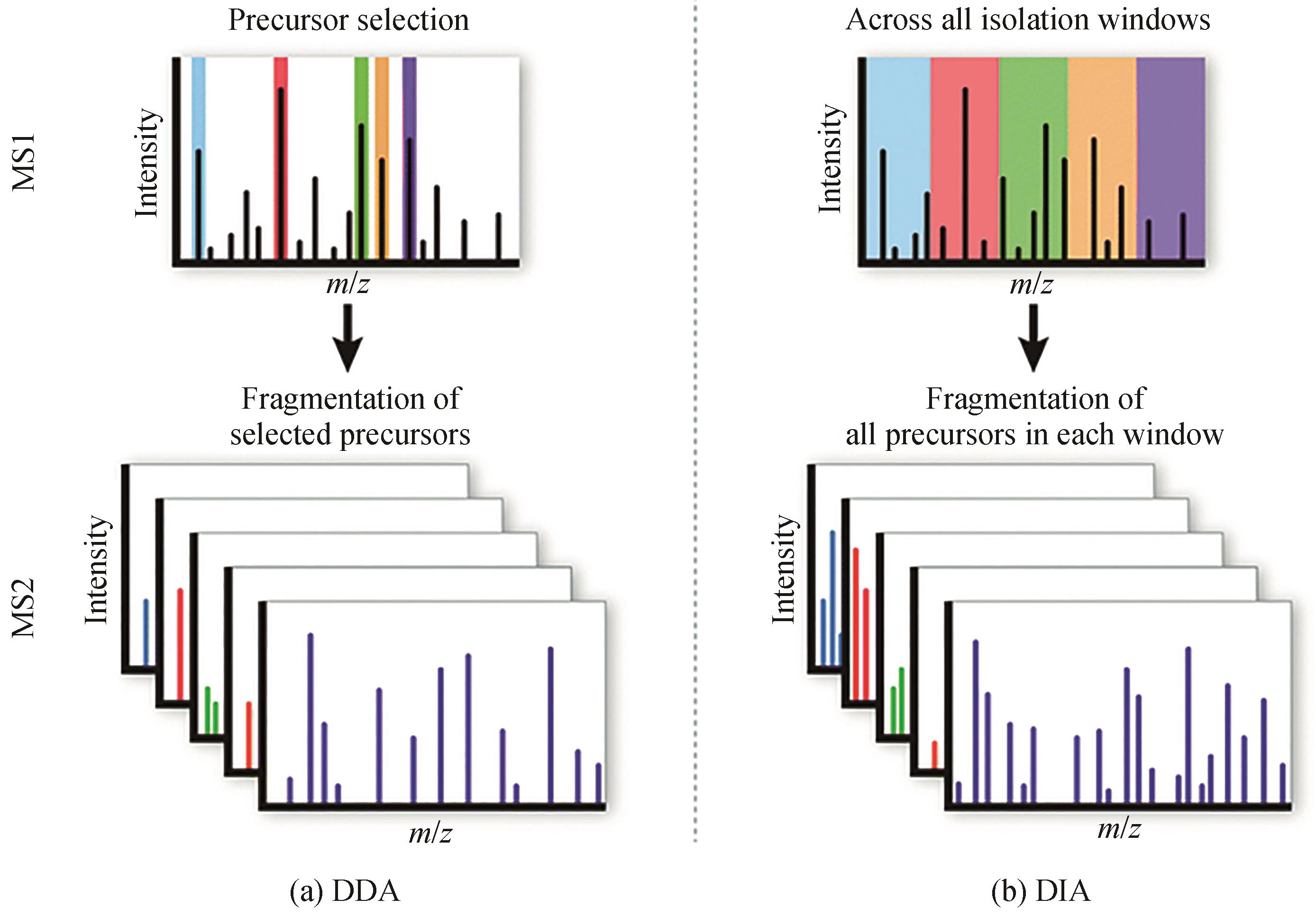
| 9 | 杨祖明, 王颖, 姚明东, 等. 高通量筛选技术在菌种进化中的研究进展[J]. 化工进展, 2019, 38(5): 2402-2412. |
| YANG Z M, WANG Y, YAO M D, et al. High-throughput screening technology in strain evolution[J]. Chemical Industry and Engineering Progress, 2019, 38(5): 2402-2412. | |
| 10 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 11 | FU L H, GUO E P, ZHANG J Z, et al. Towards one sample per second for mass spectrometric screening of engineered microbial strains[J]. Current Opinion in Biotechnology, 2022, 76: 102725. |
| 12 | MARCELLIN E, NIELSEN L K. Advances in analytical tools for high throughput strain engineering[J]. Current Opinion in Biotechnology, 2018, 54: 33-40. |
| 13 | PILLAI-KASTOORI L, SCHUTZ-GESCHWENDER A R, HARFORD J A. A systematic approach to quantitative Western blot analysis[J]. Analytical Biochemistry, 2020, 593: 113608. |
| 14 | SIMARD J R, LEE L D, VIEUX E, et al. High-throughput quantitative assay technologies for accelerating the discovery and optimization of targeted protein degradation therapeutics[J]. SLAS Discovery, 2021, 26(4): 503-517. |
| 15 | AARTHY M, GEORGE A, AYYADURAI N. Beyond protein tagging: rewiring the genetic code of fluorescent proteins - a review[J]. International Journal of Biological Macromolecules, 2021, 191: 840-851. |
| 16 | REICHE M A, AARON J S, BOEHM U, et al. When light meets biology-how the specimen affects quantitative microscopy[J]. Journal of Cell Science, 2022, 135(6): jcs259656. |
| 17 | HIRANO M, ANDO R, SHIMOZONO S, et al. A highly photostable and bright green fluorescent protein[J]. Nature Biotechnology, 2022, 40(7): 1132-1142. |
| 18 | YE D Y, LI X W, SHEN J Z, et al. Microbial metabolomics: from novel technologies to diversified applications[J]. TrAC Trends in Analytical Chemistry, 2022, 148: 116540. |
| 19 | AEBERSOLD R, MANN M. Mass spectrometry-based proteomics[J]. Nature, 2003, 422(6928): 198-207. |
| 20 | AEBERSOLD R, MANN M. Mass-spectrometric exploration of proteome structure and function[J]. Nature, 2016, 537(7620): 347-355. |
| 21 | DING J, FENG Y Q. Mass spectrometry-based metabolomics for clinical study: recent progresses and applications[J]. TrAC Trends in Analytical Chemistry, 2023, 158: 116896. |
| 22 | SI T, LI B, COMI T J, et al. Profiling of microbial colonies for high-throughput engineering of multistep enzymatic reactions via optically guided matrix-assisted laser desorption/ionization mass spectrometry[J]. Journal of the American Chemical Society, 2017, 139(36): 12466-12473. |
| 23 | PLUCHINSKY A J, WACKELIN D J, HUANG X Y, et al. High throughput screening with SAMDI mass spectrometry for directed evolution[J]. Journal of the American Chemical Society, 2020, 142(47): 19804-19808. |
| 24 | CHENG X L, HIRAS J, DENG K, et al. High throughput nanostructure-initiator mass spectrometry screening of microbial growth conditions for maximal β-glucosidase production[J]. Frontiers in Microbiology, 2013, 4: 365. |
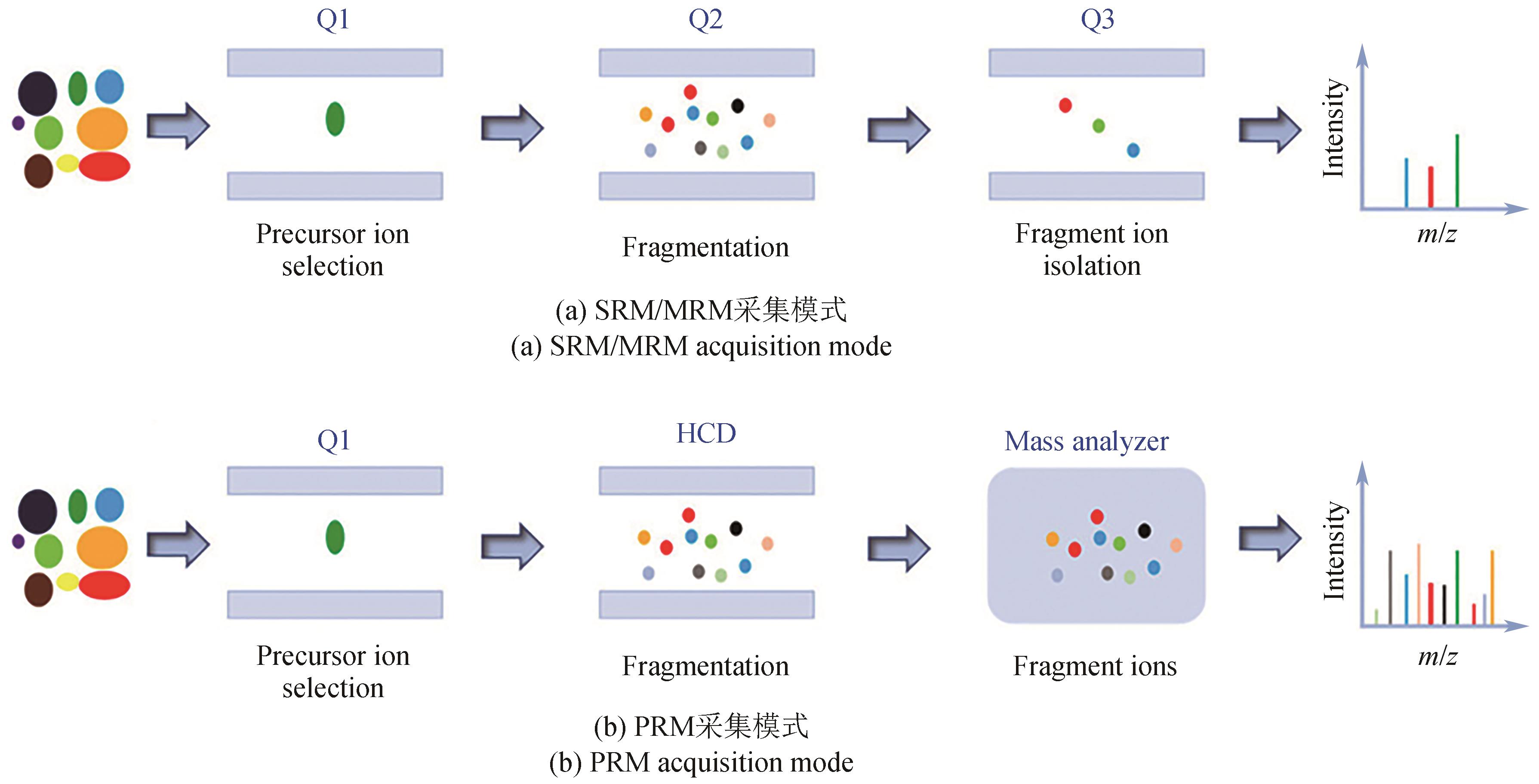
| 25 | DE ROND T, GAO J, ZARGAR A, et al. A high-throughput mass spectrometric enzyme activity assay enabling the discovery of cytochrome P450 biocatalysts[J]. Angewandte Chemie International Edition, 2019, 58(30): 10114-10119. |
| 26 | GOWERS G O F, CHEE S M, BELL D, et al. Improved betulinic acid biosynthesis using synthetic yeast chromosome recombination and semi-automated rapid LC-MS screening[J]. Nature Communications, 2020, 11: 868. |
| 27 | SAWYER W S, SRIKUMAR N, CARVER J, et al. High-throughput antibody screening from complex matrices using intact protein electrospray mass spectrometry[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(18): 9851-9856. |
| 28 | ZHANG H, LIU C, HUA W Y, et al. Acoustic ejection mass spectrometry for high-throughput analysis[J]. Analytical Chemistry, 2021, 93(31): 10850-10861. |
| 29 | SINCLAIR I, BACHMAN M, ADDISON D, et al. Acoustic mist ionization platform for direct and contactless ultrahigh-throughput mass spectrometry analysis of liquid samples[J]. Analytical Chemistry, 2019, 91(6): 3790-3794. |
| 30 | WEN X J, LIU C, GHISLAIN L, et al. Direct analysis from phase-separated liquid samples using ADE-OPI-MS: applicability to high-throughput screening for inhibitors of diacylglycerol acyltransferase 2[J]. Analytical Chemistry, 2021, 93(15): 6071-6079. |
| 31 | HÄBE T T, LIU C, COVEY T R, et al. Ultrahigh-throughput ESI-MS: sampling pushed to six samples per second by acoustic ejection mass spectrometry[J]. Analytical Chemistry, 2020, 92(18): 12242-12249. |
| 32 | BRETSCHNEIDER T, OZBAL C, HOLSTEIN M, et al. RapidFire BLAZE-mode is boosting ESI-MS toward high-throughput-screening[J]. SLAS Technology, 2019, 24(4): 386-393. |
| 33 | SINCLAIR I, STEARNS R, PRINGLE S, et al. Novel acoustic loading of a mass spectrometer: toward next-generation high-throughput MS screening[J]. Journal of Laboratory Automation, 2016, 21(1): 19-26. |
| 34 | WAGNER A, ZHANG J, LIU C, et al. Ultrahigh-throughput and chromatography-free bioanalysis of polar analytes with acoustic ejection mass spectrometry[J]. Analytical Chemistry, 2020, 92(19): 13525-13531. |
| 35 | ZHANG J, ZHANG Y, LIU C, et al. Acoustic ejection/full-scan mass spectrometry analysis for high-throughput compound quality control[J]. SLAS Technology, 2021, 26(2): 178-188. |
| 36 | SCHIRMER M, WINK K, OHLA S, et al. Conversion efficiencies of a few living microbial cells detected at a high throughput by droplet-based ESI-MS[J]. Analytical Chemistry, 2020, 92(15): 10700-10708. |
| 37 | WEI X H, HAO Y Y, HUANG X Y, et al. Automated solid phase extraction and electrospray chip based on programmatic pneumatic micro-valves[J]. Talanta, 2019, 198: 404-411. |
| 38 | WELLS S S, KENNEDY R T. High-throughput liquid-liquid extractions with nanoliter volumes[J]. Analytical Chemistry, 2020, 92(4): 3189-3197. |
| 39 | CHEN Y J, GONG T, YU C L, et al. Microfluidic flow-through SPME chip for online separation and MS detection of multiple analyses in complex matrix[J]. Micromachines, 2020, 11(2): 120. |
| 40 | ELLIS B M, FISCHER C N, MARTIN L B, et al. Spatiochemically profiling microbial interactions with membrane scaffolded desorption electrospray ionization-ion mobility-imaging mass spectrometry and unsupervised segmentation[J]. Analytical Chemistry, 2019, 91(21): 13703-13711. |
| 41 | DREISEWERD K. The desorption process in MALDI[J]. Chemical Reviews, 2003, 103(2): 395-426. |
| 42 | LI D D, YI J, HAN G B, et al. MALDI-TOF mass spectrometry in clinical analysis and research[J]. ACS Measurement Science Au, 2022, 2(5): 385-404. |
| 43 | HOU T Y, CHIANG-NI C, TENG S H. Current status of MALDI-TOF mass spectrometry in clinical microbiology[J]. Journal of Food and Drug Analysis, 2019, 27(2): 404-414. |
| 44 | TORRES-SANGIAO E, LEAL RODRIGUEZ C, GARCÍA-RIESTRA C. Application and perspectives of MALDI-TOF mass spectrometry in clinical microbiology laboratories[J]. Microorganisms, 2021, 9(7): 1539. |
| 45 | XUE P, SI T, MISHRA S, et al. A mass spectrometry-based high-throughput screening method for engineering fatty acid synthases with improved production of medium-chain fatty acids[J]. Biotechnology and Bioengineering, 2020, 117(7): 2131-2138. |
| 46 | CHOE K, XUE P, ZHAO H M, et al. macroMS: image-guided analysis of random objects by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry[J]. Journal of the American Society for Mass Spectrometry, 2021, 32(5): 1180-1188. |
| 47 | ZHANG S Y, ZHU J, FAN S, et al. Directed evolution of a cyclodipeptide synthase with new activities via label-free mass spectrometric screening[J]. Chemical Science, 2022, 13(25): 7581-7586. |
| 48 | GUO E P, FU L H, FANG X T, et al. Robotic construction and screening of lanthipeptide variant libraries in Escherichia coli [J]. ACS Synthetic Biology, 2022, 11(12): 3900-3911. |
| 49 | KRENKEL H, HARTMANE E, PIRAS C, et al. Advancing liquid atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry toward ultrahigh-throughput analysis[J]. Analytical Chemistry, 2020, 92(4): 2931-2936. |
| 50 | HAIDAS D, BACHLER S, KÖHLER M, et al. Microfluidic platform for multimodal analysis of enzyme secretion in nanoliter droplet arrays[J]. Analytical Chemistry, 2019, 91(3): 2066-2073. |
| 51 | HAIDAS D, NAPIORKOWSKA M, SCHMITT S, et al. Parallel sampling of nanoliter droplet arrays for noninvasive protein analysis in discrete yeast cultivations by MALDI-MS[J]. Analytical Chemistry, 2020, 92(5): 3810-3818. |
| 52 | SU J, MRKSICH M. Using mass spectrometry to characterize self-assembled monolayers presenting peptides, proteins, and carbohydrates[J]. Angewandte Chemie International Edition, 2002, 41(24): 4715-4718. |
| 53 | KIGHTLINGER W, LIN L, ROSZTOCZY M, et al. Design of glycosylation sites by rapid synthesis and analysis of glycosyltransferases[J]. Nature Chemical Biology, 2018, 14(6): 627-635. |
| 54 | GRANT J, GOUDARZI S H, MRKSICH M. High-throughput enzyme kinetics with 3D microfluidics and imaging SAMDI mass spectrometry[J]. Analytical Chemistry, 2018, 90(21): 13096-13103. |
| 55 | ANDERSON S E, FAHEY N S, PARK J, et al. A high-throughput SAMDI-mass spectrometry assay for isocitrate dehydrogenase 1[J]. The Analyst, 2020, 145(11): 3899-3908. |
| 56 | SCHOLLE M D, GURARD-LEVIN Z A. Development of a novel label-free and high-throughput arginase-1 assay using self-assembled monolayer desorption ionization mass spectrometry[J]. SLAS Discovery, 2021, 26(6): 775-782. |
| 57 | PATINO C A, MUKHERJEE P, BERNS E J, et al. High-throughput microfluidics platform for intracellular delivery and sampling of biomolecules from live cells[J]. ACS Nano, 2022, 16(5): 7937-7946. |
| 58 | NORTHEN T R, YANES O, NORTHEN M T, et al. Clathrate nanostructures for mass spectrometry[J]. Nature, 2007, 449(7165): 1033-1036. |
| 59 | WOO H K, NORTHEN T R, YANES O, et al. Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis[J]. Nature Protocols, 2008, 3(8): 1341-1349. |
| 60 | CECH N B, ENKE C G. Practical implications of some recent studies in electrospray ionization fundamentals[J]. Mass Spectrometry Reviews, 2001, 20(6): 362-387. |
| 61 | ZHOU S Z, FATMA Z, XUE P, et al. Mass spectrometry-based high-throughput quantification of bioproducts in liquid culture[J]. Analytical Chemistry, 2023, 95(8): 4067-4076. |
| 62 | DUSNY C, LOHSE M, REEMTSMA T, et al. Quantifying a biocatalytic product from a few living microbial cells using microfluidic cultivation coupled to FT-ICR-MS[J]. Analytical Chemistry, 2019, 91(11): 7012-7018. |
| 63 | TAKÁTS Z, WISEMAN J M, GOLOGAN B, et al. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization[J]. Science, 2004, 306(5695): 471-473. |
| 64 | YAN C Y, PARMEGGIANI F, JONES E A, et al. Real-time screening of biocatalysts in live bacterial colonies[J]. Journal of the American Chemical Society, 2017, 139(4): 1408-1411. |
| 65 | ELLIS B M, BABELE P K, MAY J C, et al. Accelerating strain phenotyping with desorption electrospray ionization-imaging mass spectrometry and untargeted analysis of intact microbial colonies[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(49): e2109633118. |
| 66 | SCHÄFER K C, DÉNES J, ALBRECHT K, et al. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry[J]. Angewandte Chemie International Edtion, 2009, 48(44): 8240-8242. |
| 67 | GOWERS G O F, CAMERON S J S, PERDONES-MONTERO A, et al. Off-colony screening of biosynthetic libraries by rapid laser-enabled mass spectrometry[J]. ACS Synthetic Biology, 2019, 8(11): 2566-2575. |
| 68 | CAMERON S J S, PERDONES-MONTERO A, VAN MEULEBROEK L, et al. Sample preparation free mass spectrometry using laser-assisted rapid evaporative ionization mass spectrometry: applications to microbiology, metabolic biofluid phenotyping, and food authenticity[J]. Journal of the American Society for Mass Spectrometry, 2021, 32(6): 1393-1401. |
| 69 | PU F, RADOSEVICH A J, SAWICKI J W, et al. High-throughput label-free biochemical assays using infrared matrix-assisted desorption electrospray ionization mass spectrometry[J]. Analytical Chemistry, 2021, 93(17): 6792-6800. |
| 70 | RADOSEVICH A J, PU F, CHANG-YEN D, et al. Ultra-high-throughput ambient MS: direct analysis at 22 samples per second by infrared matrix-assisted laser desorption electrospray ionization mass spectrometry[J]. Analytical Chemistry, 2022, 94(12): 4913-4918. |
| 71 | RACE A M, PALMER A D, DEXTER A, et al. SpectralAnalysis: software for the masses[J]. Analytical Chemistry, 2016, 88(19): 9451-9458. |
| 72 | RÜBEL O, GREINER A, CHOLIA S, et al. OpenMSI: a high-performance web-based platform for mass spectrometry imaging[J]. Analytical Chemistry, 2013, 85(21): 10354-10361. |
| 73 | DE RAAD M, DE ROND T, RÜBEL O, et al. OpenMSI arrayed analysis toolkit: analyzing spatially defined samples using mass spectrometry imaging[J]. Analytical Chemistry, 2017, 89(11): 5818-5823. |
| 74 | TORRES-ACOSTA M A, LYE G J, DIKICIOGLU D. Automated liquid-handling operations for robust, resilient, and efficient bio-based laboratory practices[J]. Biochemical Engineering Journal, 2022, 188: 108713. |
| 75 | KO S C, CHO M, LEE H J, et al. Biofoundry palette: planning-assistant software for liquid handler-based experimentation and operation in the biofoundry workflow[J]. ACS Synthetic Biology, 2022, 11(10): 3538-3543. |
| 76 | CHEN Y, KAPLAN LEASE N, GIN J W, et al. Modular automated bottom-up proteomic sample preparation for high-throughput applications[J]. PLoS One, 2022, 17(2): e0264467. |
| 77 | HUGHES C S, FOEHR S, GARFIELD D A, et al. Ultrasensitive proteome analysis using paramagnetic bead technology[J]. Molecular Systems Biology, 2014, 10(10): 757. |
| 78 | HUGHES C S, MOGGRIDGE S, MÜLLER T, et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments[J]. Nature Protocols, 2019, 14(1): 68-85. |
| 79 | HAYOUN K, GAILLARD J C, PIBLE O, et al. High-throughput proteotyping of bacterial isolates by double barrel chromatography-tandem mass spectrometry based on microplate paramagnetic beads and phylopeptidomics[J]. Journal of Proteomics, 2020, 226: 103887. |
| 80 | LIU X Y, GYGI S P, PAULO J A. A semiautomated paramagnetic bead-based platform for isobaric tag sample preparation[J]. Journal of the American Society for Mass Spectrometry, 2021, 32(6): 1519-1529. |
| 81 | QU Y, WU S, ZHAO R, et al. Automated immobilized metal affinity chromatography system for enrichment of Escherichia coli phosphoproteome[J]. Electrophoresis, 2013, 34(11): 1619-1626. |
| 82 | LEUTERT M, RODRÍGUEZ-MIAS R A, FUKUDA N K, et al. R2-P2 rapid-robotic phosphoproteomics enables multidimensional cell signaling studies[J]. Molecular Systems Biology, 2019, 15(12): e9021. |
| 83 | SCHAEFER U, BOOS W, TAKORS R, et al. Automated sampling device for monitoring intracellular metabolite dynamics[J]. Analytical Biochemistry, 1999, 270(1): 88-96. |
| 84 | VISSER D, VAN ZUYLEN G A, VAN DAM J C, et al. Rapid sampling for analysis of in vivo kinetics using the BioScope: a system for continuous-pulse experiments[J]. Biotechnology and Bioengineering, 2002, 79(6): 674-681. |
| 85 | MASHEGO M R, VAN GULIK W M, VINKE J L, et al. In vivo kinetics with rapid perturbation experiments in Saccharomyces cerevisiae using a second-generation BioScope[J]. Metabolic Engineering, 2006, 8(4): 370-383. |
| 86 | ROCKENBACH A, SUDARSAN S, BERENS J, et al. Microfluidic irreversible electroporation—a versatile tool to extract intracellular contents of bacteria and yeast[J]. Metabolites, 2019, 9(10): 211. |
| 87 | CAUSON T J, HANN S. Review of sample preparation strategies for MS-based metabolomic studies in industrial biotechnology[J]. Analytica Chimica Acta, 2016, 938: 18-32. |
| 88 | WIDYA M, PASUTTI W D, SACHDEVA M, et al. Development and optimization of a higher-throughput bacterial compound accumulation assay[J]. ACS Infectious Diseases, 2019, 5(3): 394-405. |
| 89 | REYES-GARCÉS N, GIONFRIDDO E, GÓMEZ-RÍOS G A, et al. Advances in solid phase microextraction and perspective on future directions[J]. Analytical Chemistry, 2018, 90(1): 302-360. |
| 90 | MOUSAVI F, BOJKO B, PAWLISZYN J. Development of high throughput 96-blade solid phase microextraction-liquid chromatrography-mass spectrometry protocol for metabolomics[J]. Analytica Chimica Acta, 2015, 892: 95-104. |
| 91 | ACQUARO V R JR, RODRIGUES J P, ALBERTO BERALDO MORAES L. Solid phase microextraction as a powerful alternative for screening of secondary metabolites in actinomycetes[J]. Journal of Mass Spectrometry, 2019, 54(10): 823-833. |
| 92 | MOUSAVI F, BOJKO B, PAWLISZYN J. High-throughput solid-phase microextraction-liquid chromatography-mass spectrometry for microbial untargeted metabolomics[M/OL]//Methods in molecular biology: microbial metabolomics. New York: Springer New York, 2019: 133-152 [2023-03-01]. . |
| 93 | MOUSAVI F, BOJKO B, BESSONNEAU V, et al. Cinnamaldehyde characterization as an antibacterial agent toward E. coli metabolic profile using 96-blade solid-phase microextraction coupled to liquid chromatography-mass spectrometry[J]. Journal of Proteome Research, 2016, 15(3): 963-975. |
| 94 | YUNUS I S, LEE T S. Applications of targeted proteomics in metabolic engineering: advances and opportunities[J]. Current Opinion in Biotechnology, 2022, 75: 102709. |
| 95 | BIAN Y Y, GAO C L, KUSTER B. On the potential of micro-flow LC-MS/MS in proteomics[J]. Expert Review of Proteomics, 2022, 19(3): 153-164. |
| 96 | CHEN Y, GUENTHER J M, GIN J W, et al. Automated "cells-to-peptides" sample preparation workflow for high-throughput, quantitative proteomic assays of microbes[J]. Journal of Proteome Research, 2019, 18(10): 3752-3761. |
| 97 | MUENZNER J, TRÉBULLE P, AGOSTINI F, et al. The natural diversity of the yeast proteome reveals chromosome-wide dosage compensation in aneuploids[EB/OL]. bioRxiv, 2022, DOI: 10.1101/2022.04.06.487392[2023-03-01]. . |
| 98 | PAGLIA G, ASTARITA G. A high-throughput HILIC-MS-based metabolomic assay for the analysis of polar metabolites[M/OL]//Plant Metabolic Engineering. New York: Springer US, 2021: 137-159 [2023-03-01]. . |
| 99 | JEUCKEN A, MOLENAAR M R, VAN DE LEST C H A, et al. A comprehensive functional characterization of Escherichia coli lipid genes[J]. Cell Reports, 2019, 27(5): 1597-1606.e2. |
| 100 | BACHE N, GEYER P E, BEKKER-JENSEN D B, et al. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics[J]. Molecular & Cellular Proteomics, 2018, 17(11): 2284-2296. |
| 101 | BEKKER-JENSEN D B, MARTÍNEZ-VAL A, STEIGERWALD S, et al. A compact quadrupole-orbitrap mass spectrometer with FAIMS interface improves proteome coverage in short LC gradients[J]. Molecular & Cellular Proteomics, 2020, 19(4): 716-729. |
| 102 | MEIER F, BRUNNER A D, FRANK M, et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition[J]. Nature Methods, 2020, 17(12): 1229-1236. |
| 103 | ORTON D J, WALL M J, DOUCETTE A A. Dual LC-MS platform for high-throughput proteome analysis[J]. Journal of Proteome Research, 2013, 12(12): 5963-5970. |
| 104 | HOSP F, SCHELTEMA R A, EBERL H C, et al. A double-barrel liquid chromatography-tandem mass spectrometry (LC-MS/MS) system to quantify 96 interactomes per day[J]. Molecular & Cellular Proteomics, 2015, 14(7): 2030-2041. |
| 105 | CARDOZO K H M, LEBKUCHEN A, OKAI G G, et al. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts[J]. Nature Communications, 2020, 11: 6201. |
| 106 | BONDARENKO P V, CHELIUS D, SHALER T A. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry[J]. Analytical Chemistry, 2002, 74(18): 4741-4749. |
| 107 | ZHANG R J, REGNIER F E. Minimizing resolution of isotopically coded peptides in comparative proteomics[J]. Journal of Proteome Research, 2002, 1(2): 139-147. |
| 108 | ROSS P L, HUANG Y N, MARCHESE J N, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents[J]. Molecular & Cellular Proteomics, 2004, 3(12): 1154-1169. |
| 109 | DAYON L, HAINARD A, LICKER V, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags[J]. Analytical Chemistry, 2008, 80(8): 2921-2931. |
| 110 | KREY J F, WILMARTH P A, SHIN J B, et al. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers[J]. Journal of Proteome Research, 2014, 13(2): 1034-1044. |
| 111 | GUO J, HUAN T. Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography-mass spectrometry based untargeted metabolomics[J]. Analytical Chemistry, 2020, 92(12): 8072-8080. |
| 112 | KRASNY L, HUANG P H. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology[J]. Molecular Omics, 2021, 17(1): 29-42. |
| 113 | COLLINS B C, HUNTER C L, LIU Y S, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry[J]. Nature Communications, 2017, 8: 291. |
| 114 | LUDWIG C, GILLET L, ROSENBERGER G, et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial[J]. Molecular Systems Biology, 2018, 14(8): e8126. |
| 115 | KARAYEL O, MICHAELIS A C, MANN M, et al. DIA-based systems biology approach unveils E3 ubiquitin ligase-dependent responses to a metabolic shift[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(51): 32806-32815. |
| 116 | MESSNER C B, DEMICHEV V, BLOOMFIELD N, et al. Ultra-fast proteomics with scanning SWATH[J]. Nature Biotechnology, 2021, 39(7): 846-854. |
| 117 | MAY J C, MCLEAN J A. Ion mobility-mass spectrometry: time-dispersive instrumentation[J]. Analytical Chemistry, 2015, 87(3): 1422-1436. |
| 118 | PIČMANOVÁ M, MOSES T, CORTADA-GARCIA J, et al. Rapid HILIC-Z ion mobility mass spectrometry (RHIMMS) method for untargeted metabolomics of complex biological samples[J]. Metabolomics, 2022, 18(3): 16. |
| 119 | CHEN W W, FREINKMAN E, SABATINI D M. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites[J]. Nature Protocols, 2017, 12(10): 2215-2231. |
| 120 | KIRKPATRICK D S, GERBER S A, GYGI S P. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications[J]. Methods, 2005, 35(3): 265-273. |
| 121 | PRATT J M, SIMPSON D M, DOHERTY M K, et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes[J]. Nature Protocols, 2006, 1(2): 1029-1043. |
| 122 | BRUN V, DUPUIS A, ADRAIT A, et al. Isotope-labeled protein standards[J]. Molecular & Cellular Proteomics, 2007, 6(12): 2139-2149. |
| 123 | PICOTTI P, AEBERSOLD R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions[J]. Nature Methods, 2012, 9(6): 555-566. |
| 124 | BATTH T S, SINGH P, RAMAKRISHNAN V R, et al. A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins[J]. Metabolic Engineering, 2014, 26: 48-56. |
| 125 | GAO Y Q, FILLMORE T L, MUNOZ N, et al. High-throughput large-scale targeted proteomics assays for quantifying pathway proteins in Pseudomonas putida KT2440[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 603488. |
| 126 | MCCLOSKEY D, XU J, SCHRÜBBERS L, et al. RapidRIP quantifies the intracellular metabolome of 7 industrial strains of E. coli [J]. Metabolic Engineering, 2018, 47: 383-392. |
| 127 | SALEH S, STAES A, DEBORGGRAEVE S, et al. Targeted proteomics for studying pathogenic bacteria[J]. PROTEOMICS, 2019, 19(16): 1800435. |
| 128 | SCHIFFMANN C, HANSEN R, BAUMANN S, et al. Comparison of targeted peptide quantification assays for reductive dehalogenases by selective reaction monitoring (SRM) and precursor reaction monitoring (PRM)[J]. Analytical and Bioanalytical Chemistry, 2014, 406(1): 283-291. |
| 129 | LI Z C, LI Y J, CHEN W J, et al. Integrating MS1 and MS2 scans in high-resolution parallel reaction monitoring assays for targeted metabolite quantification and dynamic 13C-labeling metabolism analysis[J]. Analytical Chemistry, 2017, 89(1): 877-885. |
| 130 | LU Y, HU X X, PANG J, et al. Parallel reaction monitoring mass spectrometry for rapid and accurate identification of β-lactamases produced by Enterobacteriaceae [J]. Frontiers in Microbiology, 2022, 13: 784628. |
| 131 | MIGGIELS P, WOUTERS B, VAN WESTEN G J P, et al. Novel technologies for metabolomics: more for less[J]. TrAC Trends in Analytical Chemistry, 2019, 120: 115323. |
| 132 | AOSHIMA K, TAKAHASHI K, IKAWA M, et al. A simple peak detection and label-free quantitation algorithm for chromatography-mass spectrometry[J].BMC Bioinformatics, 2014, 15(1): 376. |
| 133 | PLUSKAL T, CASTILLO S, VILLAR-BRIONES A, et al. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data[J]. BMC Bioinformatics, 2010, 11: 395. |
| 134 | STURM M, BERTSCH A, GRÖPL C, et al. OpenMS-an open-source software framework for mass spectrometry[J]. BMC Bioinformatics, 2008, 9: 163. |
| 135 | PALAGI P M, WALTHER D, QUADRONI M, et al. MSight: an image analysis software for liquid chromatography-mass spectrometry[J]. Proteomics, 2005, 5(9): 2381-2384. |
| 136 | MUELLER L N, RINNER O, SCHMIDT A, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling[J]. Proteomics, 2007, 7(19): 3470-3480. |
| 137 | KIEFER P, SCHMITT U, MÜLLER J E N, et al. DynaMet: a fully automated pipeline for dynamic LC-MS data[J]. Analytical Chemistry, 2015, 87(19): 9679-9686. |
| 138 | LIAO X P, MA H W, TANG Y J. Artificial intelligence: a solution to involution of design-build-test-learn cycle[J]. Current Opinion in Biotechnology, 2022, 75: 102712. |
| 139 | PATRA P, DISHA B R, KUNDU P, et al. Recent advances in machine learning applications in metabolic engineering[J]. Biotechnology Advances, 2023, 62: 108069. |
| 140 | DEMICHEV V, MESSNER C B, VERNARDIS S I, et al. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput[J]. Nature Methods, 2020, 17(1): 41-44. |
| 141 | RÖST H L, LIU Y S, D'AGOSTINO G, et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics[J]. Nature Methods, 2016, 13(9): 777-783. |
| 142 | CAI Y, WENG K, GUO Y, et al. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing[J]. Metabolomics, 2015, 11(6): 1575-1586. |
| 143 | REITER A, ASGARI J, WIECHERT W, et al. Metabolic footprinting of microbial systems based on comprehensive in silico predictions of MS/MS relevant data[J]. Metabolites, 2022, 12(3): 257. |
| 144 | KANG K, LI J, LIM B L, et al. MESSI: metabolic engineering target selection and best strain identification tool[J]. Database, 2015, 2015: bav076. |
| 145 | PALEY S, BILLINGTON R, HERSON J, et al. Pathway tools visualization of organism-scale metabolic networks[J]. Metabolites, 2021, 11(2): 64-77. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||