合成生物学 ›› 2025, Vol. 6 ›› Issue (3): 685-700.DOI: 10.12211/2096-8280.2024-079
合成生物学视角下的基因功能探索与酵母工程菌株文库构建
章益蜻1,2, 刘高雯1
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,深圳合成基因组学重点实验室,广东省合成基因组学重点实验室,定量合成生物学重点实验室,广东 深圳 518055
2.中国科学院大学,北京 100049
-
收稿日期:2024-11-11修回日期:2025-02-20出版日期:2025-06-30发布日期:2025-06-27 -
通讯作者:刘高雯 -
作者简介:章益蜻 (2001—),女,硕士研究生。研究方向为合成生物学与系统基因组学。E-mail:yq.zhang3@siat.ac.cn刘高雯 (1987—),女,副研究员,博士生导师。研究方向为酵母系统基因组学与适应性进化。E-mail:gaowen.liu@siat.ac.cn -
基金资助:广东省合成基因组学重点实验室资助项目(2023B1212060054);深圳合成基因组学重点实验室资助项目(ZDSYS201802061806209)
Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective
ZHANG Yiqing1,2, LIU Gaowen1
- 1.Shenzhen Institute of Synthetic Biology,Shenzhen Key Laboratory of Synthetic Genomics,Guangdong Provincial Key Laboratory of Synthetic Genomics,Key Laboratory of Quantitative Synthetic Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2024-11-11Revised:2025-02-20Online:2025-06-30Published:2025-06-27 -
Contact:LIU Gaowen
摘要:
合成生物学作为一门通过设计、构建和改造生物系统来实现其特定功能的学科,被广泛应用于生物制造、环境保护和药物合成等领域。基因功能的系统性探索和工程菌株文库的构建是推动合成生物学发展的重要手段。本文重点介绍了不同酵母文库在合成生物学中的构建方法及其应用前景。随着基因组测序和高通量技术的快速进展,酿酒酵母和裂殖酵母等微生物文库在系统性研究中发挥了关键作用。基因缺失文库、过表达文库、转座子插入文库等多种类型的酵母文库为基因组合优化和代谢路径设计提供了重要工具,促进了代谢工程和合成生物学的创新应用。这些文库在工业生产中支持高产菌株的构建,如用于生物燃料和化学品的高效生产;在环境领域,通过基因改造筛选,生成具备污染物降解能力的菌株,为生态修复提供解决方案;在药物合成方面,文库帮助构建高效合成药用化合物的菌株,推动生物制药的发展。然而,当前文库构建和应用仍面临诸如构建成本、基因组编辑的精确度及筛选效率等技术瓶颈。未来,自动化、数字化和新型筛选技术的进步有望突破这些瓶颈,推动酵母文库的快速构建和高效筛选,从而加速合成生物学在可持续发展和生态工程中的应用。
中图分类号:
引用本文
章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700.
ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective[J]. Synthetic Biology Journal, 2025, 6(3): 685-700.
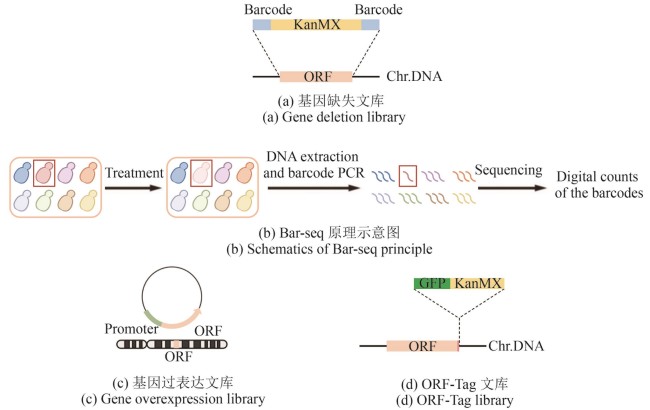
图1 经典文库构建方法(本图由biorender绘制)[(a)基因缺失文库中,目的基因ORF(橘色片段)被替换成卡那霉素抗性筛选标签KanMX(黄色片段),两侧伴有“分子条形码”barcode(蓝色片段);Chr.DNA:染色体DNA。(b)Bar-seq原理示意图。在特定条件处理下,目标菌株(红色)的生长量相对低(淡粉色),表现为测序时相应barcode的读数量相对低(红色线条比其他颜色少)。(c)基因过表达文库中,目的基因ORF(橘色片段)除了在基因组上正常表达之外,还额外在质粒上由通用型启动子(淡绿色片段)驱动表达。(d)ORF-Tag文库中,在删除终止密码子的目的基因ORF(橘色片段)C端插入荧光蛋白GFP(深绿色片段)和筛选标签KanMX(黄色片段)]
Fig. 1 Schematics for constructing classical libraries (created by biorender)[(a) In the gene deletion library, the ORF (orange fragment) of target gene is replaced by the KanMX (kanamycin resistance selection marker, yellow fragment), flanked by unique barcodes (blue fragments). Chr.DNA: chromosomal DNA.(b) Schematic diagram of the Bar-seq principle. Under specific treatment conditions, the target strain (red) shows reduced growth (light pink), which is reflected by a lower read count of its corresponding barcode during sequencing (fewer red lines compared to others).(c) In the gene overexpression library, the ORF (orange fragment) of target gene is not only expressed from its endogenous locus but is also additionally expressed from a plasmid, driven by a constitutive promoter (light green fragment).(d) In the ORF-Tag library, the C-terminus of the target gene ORF (orange fragment), with its stop codon removed, is fused to the fluorescent protein GFP (dark green fragment) and the selection marker KanMX (yellow fragment).]
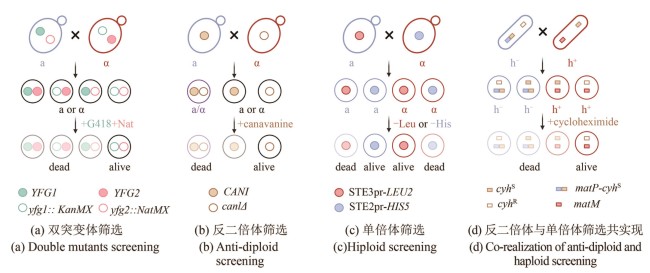
图2 SGA与PEM方法原理(本图由biorender绘制)[(a)双突变体筛选策略。两种交配型的细胞分别在各自的突变位点携带不同抗性标签KanMX(绿色空心圆点)和NatMX(粉色空心圆点)用于筛选双突变体子代(黑线框细胞);YFG (your favourite gene):目的基因。(b)反二倍体筛选策略。含有野生型CAN1基因的单倍体细胞(蓝色细胞,a型)或二倍体细胞(紫色细胞)会摄入有毒的刀豆氨酸(canavanine),从而被杀死,而can1Δ突变体无法将刀豆氨酸运转入体内,因此能够存活(黑线框细胞)。(c) 单倍体筛选策略。在某一细胞型的母本菌株中,构建另一细胞型特异性启动子与营养缺陷筛选标签的表达盒,用于选择任一性别的单倍体子代细胞。例如在a型细胞(蓝色)中,携带只能在α型细胞中表达的STE3pr-LEU2基因线路(红色实心圆点),只有细胞交配使STE3pr-LEU2基因线路存在于α型细胞中时,该细胞存活(含有红色实心圆点的红线框细胞)。(d)反二倍体与单倍体筛选共实现。通过将一个显性致死的抗性基因cyhS(棕色实心方块)“镶嵌”在裂殖酵母交配位点mat1(在蓝色的h-细胞中为蓝色方块表示的matP)附近,使得某一单倍体表型与抗性基因的表达偶联(matP-cyhS)]
Fig. 2 Schematics for SGA and PEM methods (created by biorender)[(a) Double mutant selection strategy. Two mating-type cells each carry different resistance markers at their respective mutation sites: KanMX (green hollow circle) and NatMX (pink hollow circle). These allow for the selection of double mutant progeny (outlined in black). YFG (your favorite gene): target gene of interest.(b) Counter-diploid selection strategy. Haploid cells carrying a wild-type CAN1 gene (blue, a-type) and diploid cells (purple) can uptake the toxic analog canavanine and are killed, while can1Δ mutants cannot transport canavanine and thus survive (outlined in black).(c) Haploid selection strategy. A mating-type-specific promoter is used to drive the expression of a nutritional selection marker in the opposite mating type, allowing selection of haploid progeny of a specific mating type. For example, when an a-type parent cell (blue), harboring a gene cassette expressing STE3pr-LEU2 (red solid circle) — which is only active in α-type cell — mates with another cell, only the α-type progeny that inherit this construct (outlined in red with a red solid circle) will survive.(d) Combined counter-diploid and haploid selection strategy. A dominant-lethal resistance gene, cyhS (brown solid square), is inserted near the mat1 mating-type locus to link its expression with a specific haploid phenotype. For example, insertion near matP (blue square) in h- cells (blue) leads to the death of cyhS -containing h- haploids and diploids, enabling selection of h+ haploids.]
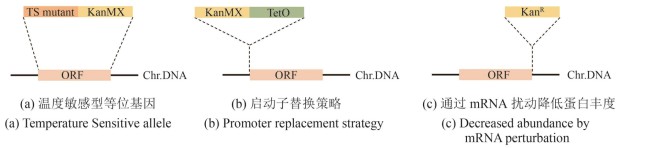
图3 条件等位基因文库(本图由biorender绘制)[(a)温度敏感型(temperature sensitive,TS)等位基因中,目的基因ORF(橘色片段)被替换成相应的温敏突变基因TS mutant(深橘色片段)并携带一个抗性筛选标签KanMX(黄色片段)。Chr.DNA:染色体DNA。(b)启动子替换策略中,携带抗性筛选标签KanMX(黄色片段)的诱导性启动子TetO(绿色片段)被插入目的基因ORF(橘色片段)的起始密码子上游。Chr.DNA:染色体DNA。(c)通过mRNA扰动降低蛋白丰度的DAmP策略中,目的基因ORF(橘色片段)的3′UTR区被插入一个抗性标签KanR(黄色片段)。Chr.DNA:染色体DNA]
Fig. 3 Construction of conditional allele libraries (created by biorender)[(a) In the temperature-sensitive (TS) allele strategy, the ORF (orange fragment) of target gene is replaced with a corresponding TS mutant (dark orange fragment) and tagged with a KanMX resistance marker (yellow fragment). Chr.DNA: chromosomal DNA.(b) In the promoter replacement strategy, an inducible promoter (green fragment) and a KanMX resistance marker (yellow fragment) is inserted upstream of the start codon of the target gene ORF (orange fragment). Chr.DNA: chromosomal DNA.(c) In the DAmP (Decreased Abundance by mRNA Perturbation) strategy, the 3′UTR of the target gene ORF (orange fragment) is disrupted by insertion of a KanR resistance cassette (yellow fragment) to reduce transcript stability and protein abundance. Chr.DNA: chromosomal DNA.]
| 1 | PRZYBYLA L, GILBERT L A. A new era in functional genomics screens[J]. Nature Reviews Genetics, 2022, 23(2): 89-103. |
| 2 | GALANIE S, THODEY K, TRENCHARD I J, et al. Complete biosynthesis of opioids in yeast[J]. Science, 2015, 349(6252): 1095-1100. |
| 3 | RUNGUPHAN W, KEASLING J D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals[J]. Metabolic Engineering, 2014, 21: 103-113. |
| 4 | SHENG J Y, FENG X Y. Metabolic engineering of yeast to produce fatty acid-derived biofuels: bottlenecks and solutions[J]. Frontiers in Microbiology, 2015, 6: 554. |
| 5 | SAMAL S K, PREETAM S. Synthetic biology: refining human health[M/OL]//SUAR M, MISRA N, DASH C. Microbial engineering for therapeutics. Singapore: Springer Nature Singapore, 2022: 57-70 [2024-11-01]. . |
| 6 | TSAI C S, KWAK S, TURNER T L, et al. Yeast synthetic biology toolbox and applications for biofuel production[J]. FEMS Yeast Research, 2015, 15(1): 1-15. |
| 7 | GOFFEAU A, BARRELL B G, BUSSEY H, et al. Life with 6000 genes[J]. Science, 1996, 274(5287): 546-567. |
| 8 | ARITA Y, KIM G, LI Z J, et al. A genome-scale yeast library with inducible expression of individual genes[J]. Molecular Systems Biology, 2021, 17(6): e10207. |
| 9 | WOOD V, GWILLIAM R, RAJANDREAM M A, et al. The genome sequence of Schizosaccharomyces pombe [J]. Nature, 2002, 415(6874): 871-880. |
| 10 | GREWAL S I S, JIA S T. Heterochromatin revisited[J]. Nature Reviews Genetics, 2007, 8(1): 35-46. |
| 11 | ROGUEV A, RYAN C J, HARTSUIKER E, et al. High-throughput quantitative genetic interaction mapping in the fission yeast Schizosaccharomyces pombe [J]. Cold Spring Harbor Protocols, 2018, 2018(2): pdb.top079905. |
| 12 | ZHANG W, GENG A L. Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method[J]. Biotechnology for Biofuels, 2012, 5(1): 46. |
| 13 | BOTSTEIN D, FINK G R. Yeast: an experimental organism for 21st century biology[J]. Genetics, 2011, 189(3): 695-704. |
| 14 | RODRIGUEZ A, STRUCKO T, STAHLHUT S G, et al. Metabolic engineering of yeast for fermentative production of flavonoids[J]. Bioresource Technology, 2017, 245: 1645-1654. |
| 15 | MYBURGH M W, FAVARO L, VAN ZYL W H, et al. Engineered yeast for the efficient hydrolysis of polylactic acid[J]. Bioresource Technology, 2023, 378: 129008. |
| 16 | CHEN J S, BECKLEY J R, MCDONALD N A, et al. Identification of new players in cell division, DNA damage response, and morphogenesis through construction of Schizosaccharomyces pombe deletion strains[J]. G3 Genes|Genomes|Genetics, 2015, 5(3): 361-370. |
| 17 | TURCO G, CHANG C, WANG R Y, et al. Global analysis of the yeast knockout phenome[J]. Science Advances, 2023, 9(21): eadg5702. |
| 18 | KIM D U, HAYLES J, KIM D, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe [J]. Nature Biotechnology, 2010, 28(6): 617-623. |
| 19 | SCHERENS B, GOFFEAU A. The uses of genome-wide yeast mutant collections[J]. Genome Biology, 2004, 5(7): 229. |
| 20 | GIAEVER G, NISLOW C. The yeast deletion collection: a decade of functional genomics[J]. Genetics, 2014, 197(2): 451-465. |
| 21 | SOPKO R, HUANG D Q, PRESTON N, et al. Mapping pathways and phenotypes by systematic gene overexpression[J]. Molecular Cell, 2006, 21(3): 319-330. |
| 22 | MATSUYAMA A, ARAI R, YASHIRODA Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe [J]. Nature Biotechnology, 2006, 24(7): 841-847. |
| 23 | TONG A H, EVANGELISTA M, PARSONS A B, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants[J]. Science, 2001, 294(5550): 2364-2368. |
| 24 | ROGUEV A, WIREN M, WEISSMAN J S, et al. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe [J]. Nature Methods, 2007, 4(10): 861-866. |
| 25 | GIAEVER G, CHU A M, NI L, et al. Functional profiling of the Saccharomyces cerevisiae genome[J]. Nature, 2002, 418(6896): 387-391. |
| 26 | HWANG Y C, LIN C C, CHANG J Y, et al. Predicting essential genes based on network and sequence analysis[J]. Molecular BioSystems, 2009, 5(12): 1672-1678. |
| 27 | JEONG H, MASON S P, BARABÁSI A L, et al. Lethality and centrality in protein networks[J]. Nature, 2001, 411(6833): 41-42. |
| 28 | LI Z J, VIZEACOUMAR F J, BAHR S, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants[J]. Nature Biotechnology, 2011, 29(4): 361-367. |
| 29 | SHORTLE D, NOVICK P, BOTSTEIN D. Construction and genetic characterization of temperature-sensitive mutant alleles of the yeast actin gene[J]. Proceedings of the National Academy of Sciences of the United States of America, 1984, 81(15): 4889-4893. |
| 30 | BEN-AROYA S, COOMBES C, KWOK T, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae [J]. Molecular Cell, 2008, 30(2): 248-258. |
| 31 | POULTNEY C S, BUTTERFOSS G L, GUTWEIN M R, et al. Rational design of temperature-sensitive alleles using computationalstructure prediction[J]. PLoS One, 2011, 6(9): e23947. |
| 32 | CHAKSHUSMATHI G, MONDAL K, LAKSHMI G S, et al. Design of temperature-sensitive mutants solely from amino acid sequence[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(21): 7925-7930. |
| 33 | TAN G H, CHEN M, FOOTE C, et al. Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges[J]. Genetics, 2009, 183(1): 13-22. |
| 34 | WIDLUND P O, DAVIS T N. A high-efficiency method to replace essential genes with mutant alleles in yeast[J]. Yeast, 2005, 22(10): 769-774. |
| 35 | KOFOED M, MILBURY K L, CHIANG J H, et al. An updated collection of sequence barcoded temperature-sensitive alleles of yeast essential genes[J]. G3 Genes|Genomes|Genetics, 2015, 5(9): 1879-1887. |
| 36 | MNAIMNEH S, DAVIERWALA A P, HAYNES J, et al. Exploration of essential gene functions via titratable promoter alleles[J]. Cell, 2004, 118(1): 31-44. |
| 37 | BRESLOW D K, CAMERON D M, COLLINS S R, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome[J]. Nature Methods, 2008, 5(8): 711-718. |
| 38 | SCHULDINER M, COLLINS S R, THOMPSON N J, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile[J]. Cell, 2005, 123(3): 507-519. |
| 39 | LIU G W, YONG M Y J, YURIEVA M, et al. Gene essentiality is a quantitative property linked to cellular evolvability[J]. Cell, 2015, 163(6): 1388-1399. |
| 40 | GUO Y B, PARK J M, CUI B W, et al. Integration profiling of gene function with dense maps of transposon integration[J]. Genetics, 2013, 195(2): 599-609. |
| 41 | EVERTTS A G, PLYMIRE C, CRAIG N L, et al. The Hermes transposon of Musca domestica is an efficient tool for the mutagenesis of Schizosaccharomyces pombe [J]. Genetics, 2007, 177(4): 2519-2523. |
| 42 | PARK J M, EVERTTS A G, LEVIN H L. The Hermes transposon of Musca domestica and its use as a mutagen of Schizosaccharomyces pombe [J]. Methods, 2009, 49(3): 243-247. |
| 43 | GANGADHARAN S, MULARONI L, FAIN-THORNTON J, et al. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(51): 21966-21972. |
| 44 | CAIN A K, BARQUIST L, GOODMAN A L, et al. A decade of advances in transposon-insertion sequencing[J]. Nature Reviews Genetics, 2020, 21(9): 526-540. |
| 45 | MICHEL A H, HATAKEYAMA R, KIMMIG P, et al. Functional mapping of yeast genomes by saturated transposition[J]. eLife, 2017, 6: e23570. |
| 46 | BLAKE BILLMYRE R, EICKBUSH M T, CRAIG C J, et al. Genome-wide quantification of contributions to sexual fitness identifies genes required for spore viability and health in fission yeast[J]. PLoS Genetics, 2022, 18(10): e1010462. |
| 47 | ADAMES N R, GALLEGOS J E, PECCOUD J. Yeast genetic interaction screens in the age of CRISPR/Cas[J]. Current Genetics, 2019, 65(2): 307-327. |
| 48 | LI L, LIU X C, WEI K K, et al. Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems[J]. Biotechnology Advances, 2019, 37(5): 730-745. |
| 49 | GASIUNAS G, BARRANGOU R, HORVATH P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(39): E2579-E2586. |
| 50 | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2021, 184: 844. |
| 51 | LIAN J Z, HAMEDIRAD M, HU S M, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nature Communications, 2017, 8(1): 1688. |
| 52 | GUO X G, CHAVEZ A, TUNG A, et al. High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR-Cas9 in yeast[J]. Nature Biotechnology, 2018, 36(6): 540-546. |
| 53 | SI T, CHAO R, MIN Y H, et al. Automated multiplex genome-scale engineering in yeast[J]. Nature Communications, 2017, 8: 15187. |
| 54 | SAKAI A, SHIMIZU Y, HISHINUMA F. Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty[J]. Applied Microbiology and Biotechnology, 1990, 33(3): 302-306. |
| 55 | DICARLO J E, CONLEY A J, PENTTILÄ M, et al. Yeast oligo-mediated genome engineering (YOGE)[J]. ACS Synthetic Biology, 2013, 2(12): 741-749. |
| 56 | BARBIERI E M, MUIR P, AKHUETIE-ONI B O, et al. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes[J]. Cell, 2017, 171(6): 1453-1467.e13. |
| 57 | JAKOČIŪNAS T, RAJKUMAR A S, ZHANG J, et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(11): 1226-1234. |
| 58 | LIU R M, LIANG L Y, CHOUDHURY A, et al. Multiplex navigation of global regulatory networks (MINR) in yeast for improved ethanol tolerance and production[J]. Metabolic Engineering, 2019, 51: 50-58. |
| 59 | KUZMIN E, VANDERSLUIS B, NGUYEN BA A N, et al. Exploring whole-genome duplicate gene retention with complex genetic interaction analysis[J]. Science, 2020, 368(6498): eaaz5667. |
| 60 | PENG J R. Gene redundancy and gene compensation: an updated view[J]. Journal of Genetics and Genomics, 2019, 46(7): 329-333. |
| 61 | MERZ S, WESTERMANN B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae [J]. Genome Biology, 2009, 10(9): R95. |
| 62 | LOUCA S, POLZ M F, MAZEL F, et al. Function and functional redundancy in microbial systems[J]. Nature Ecology & Evolution, 2018, 2(6): 936-943. |
| 63 | BIDLINGMAIER S, LIU B. Construction of yeast surface-displayed cDNA libraries[J]. Methods in Molecular Biology, 2011, 729: 199-210. |
| 64 | LIU Z H, TYO K E J, MARTÍNEZ J L, et al. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2012, 109(5): 1259-1268. |
| 65 | SMITH V, BOTSTEIN D, BROWN P O. Genetic footprinting: a genomic strategy for determining a gene’s function given its sequence[J]. Proceedings of the National Academy of Sciences of the United States of America, 1995, 92(14): 6479-6483. |
| 66 | SMITH V, CHOU K N, LASHKARI D, et al. Functional analysis of the genes of yeast chromosome Ⅴ by genetic footprinting[J]. Science, 1996, 274(5295): 2069-2074. |
| 67 | HAN T X, XU X Y, ZHANG M J, et al. Global fitness profiling of fission yeast deletion strains by barcode sequencing[J]. Genome Biology, 2010, 11(6): R60. |
| 68 | STEINMETZ L M, SCHARFE C, DEUTSCHBAUER A M, et al. Systematic screen for human disease genes in yeast[J]. Nature Genetics, 2002, 31(4): 400-404. |
| 69 | WARRINGER J, ERICSON E, FERNANDEZ L, et al. High-resolution yeast phenomics resolves different physiological features in the saline response[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15724-15729. |
| 70 | BIRRELL G W, BROWN J A, IRENE WU H, et al. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(13): 8778-8783. |
| 71 | CHANG M, BELLAOUI M, BOONE C, et al. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(26): 16934-16939. |
| 72 | PARSONS A B, BROST R L, DING H M, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways[J]. Nature Biotechnology, 2004, 22(1): 62-69. |
| 73 | PARSONS A B, LOPEZ A, GIVONI I E, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast[J]. Cell, 2006, 126(3): 611-625. |
| 74 | ENYENIHI A H, SAUNDERS W S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae [J]. Genetics, 2003, 163(1): 47-54. |
| 75 | MATECIC M, SMITH D L, PAN X W, et al. A microarray-based genetic screen for yeast chronological aging factors[J]. PLoS Genetics, 2010, 6(4): e1000921. |
| 76 | ROMILA C A, TOWNSEND S, MALECKI M, et al. Barcode sequencing and a high-throughput assay for chronological lifespan uncover ageing-associated genes in fission yeast[J]. Microbial Cell, 2021, 8(7): 146-160. |
| 77 | FERRARI S, BERETTA S, JACOB A, et al. BAR-Seq clonal tracking of gene-edited cells[J]. Nature Protocols, 2021, 16(6): 2991-3025. |
| 78 | RALLIS C, LÓPEZ-MAURY L, GEORGESCU T, et al. Systematic screen for mutants resistant to TORC1 inhibition in fission yeast reveals genes involved in cellular ageing and growth[J]. Biology Open, 2014, 3(2): 161-171. |
| 79 | KENNEDY P J, VASHISHT A A, HOE K L, et al. A genome-wide screen of genes involved in cadmium tolerance in Schizosaccharomyces pombe [J]. Toxicological Sciences, 2008, 106(1): 124-139. |
| 80 | RODRÍGUEZ-LÓPEZ M, BORDIN N, LEES J, et al. Broad functional profiling of fission yeast proteins using phenomics and machine learning[J]. eLife, 2023, 12: RP88229. |
| 81 | NI L, SNYDER M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae [J]. Molecular Biology of the Cell, 2001, 12(7): 2147-2170. |
| 82 | KELLY F D, NURSE P. Spatial control of Cdc42 activation determines cell width in fission yeast[J]. Molecular Biology of the Cell, 2011, 22(20): 3801-3811. |
| 83 | NAVARRO F J, NURSE P. A systematic screen reveals new elements acting at the G2/M cell cycle control[J]. Genome Biology, 2012, 13(5): R36. |
| 84 | BLYTH J, MAKRANTONI V, BARTON R E, et al. Genes important for Schizosaccharomyces pombe meiosis identified through a functional genomics screen[J]. Genetics, 2018, 208(2): 589-603. |
| 85 | DESHPANDE G P, HAYLES J, HOE K L, et al. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance[J]. DNA Repair, 2009, 8(5): 672-679. |
| 86 | PAN X, LEI B K, ZHOU N, et al. Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library[J]. BMC Genomics, 2012, 13: 662. |
| 87 | COSTANZO M, VANDERSLUIS B, KOCH E N, et al. A global genetic interaction network maps a wiring diagram of cellular function[J]. Science, 2016, 353(6306): aaf1420. |
| 88 | ROSS-MACDONALD P, COELHO P S, ROEMER T, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption[J]. Nature, 1999, 402(6760): 413-418. |
| 89 | WHITE W H, JOHNSON D I. Characterization of synthetic-lethal mutants reveals a role for the Saccharomyces cerevisiae guanine-nucleotide exchange factor Cdc24p in vacuole function and Na+ tolerance[J]. Genetics, 1997, 147(1): 43-55. |
| 90 | HUH W K, FALVO J V, GERKE L C, et al. Global analysis of protein localization in budding yeast[J]. Nature, 2003, 425(6959): 686-691. |
| 91 | RAZDAIBIEDINA A, BRECHALOV A, FRIESEN H, et al. PIFiA: self-supervised approach for protein functional annotation from single-cell imaging data[J]. Molecular Systems Biology, 2024, 20(5): 521-548. |
| 92 | CHONG Y T, KOH J L Y, FRIESEN H, et al. Yeast proteome dynamics from single cell imaging and automated analysis[J]. Cell, 2015, 161: 1413-1424 |
| 93 | HAYASHI A, DING D Q, TSUTSUMI C, et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe [J]. Genes to Cells, 2009, 14(2): 217-225. |
| 94 | JIA B, WU Y, LI B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9(1): 1933. |
| 95 | SI T, LUO Y Z, BAO Z H, et al. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering[J]. ACS Synthetic Biology, 2015, 4(3): 283-291. |
| 96 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 97 | RUGBJERG P, SOMMER M O A. Overcoming genetic heterogeneity in industrial fermentations[J]. Nature Biotechnology, 2019, 37(8): 869-876. |
| 98 | WEHRS M, TANJORE D, ENG T, et al. Engineering robust production microbes for large-scale cultivation[J]. Trends in Microbiology, 2019, 27(6): 524-537. |
| 99 | JAKOČIŪNAS T, BONDE I, HERRGÅRD M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 28: 213-222. |
| 100 | LI Y H, MOLYNEAUX N, ZHANG H T, et al. A multiplexed, three-dimensional pooling and next-generation sequencing strategy for creating barcoded mutant arrays: construction of a Schizosaccharomyces pombe transposon insertion library[J]. Nucleic Acids Research, 2022, 50(17): e102. |
| 101 | COOPE R J N, MATIC N, PANDOH P K, et al. Automated library construction and analysis for high-throughput nanopore sequencing of SARS-CoV-2[J]. The Journal of Applied Laboratory Medicine, 2022, 7(5): 1025-1036. |
| 102 | SANTACRUZ D, ENANE F O, FUNDEL-CLEMENS K, et al. Automation of high-throughput mRNA-seq library preparation: a robust, hands-free and time efficient methodology[J]. SLAS Discovery, 2022, 27(2): 140-147. |
| 103 | VAN DEVENTER J A, WITTRUP K D. Yeast surface display for antibody isolation: library construction, library screening, and affinity maturation[J]. Methods in Molecular Biology, 2014, 1131: 151-181. |
| 104 | YOFE I, WEILL U, MEURER M, et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-tag strategy[J]. Nature Methods, 2016, 13(4): 371-378. |
| 105 | COSTANZO M, HOU J, MESSIER V, et al. Environmental robustness of the global yeast genetic interaction network[J]. Science, 2021, 372(6542): eabf8424. |
| 106 | KUZMIN E, VANDERSLUIS B, WANG W, et al. Systematic analysis of complex genetic interactions[J]. Science, 2018, 360(6386): eaao1729. |
| 107 | PENNISI E. Building the ultimate yeast genome[J]. Science, 2014, 343(6178): 1426-1429. |
| 108 | ZHAO Y, COELHO C, HUGHES A L, et al. Debugging and consolidating multiple synthetic chromosomes reveals combinatorial genetic interactions[J]. Cell, 2023, 186(24): 5220-5236.e16. |
| 109 | SCHINDLER D, WALKER R S K, JIANG S Y, et al. Design, construction, and functional characterization of a tRNA neochromosome in yeast[J]. Cell, 2023, 186(24): 5237-5253.e22. |
| 110 | ZHANG W M, LAZAR-STEFANITA L, YAMASHITA H, et al. Manipulating the 3D organization of the largest synthetic yeast chromosome[J]. Molecular Cell, 2023, 83(23): 4424-4437.e5. |
| 111 | DAI J B, BOEKE J D, LUO Z Q, et al. Sc3.0: revamping and minimizing the yeast genome[J]. Genome Biology, 2020, 21(1): 205. |
| 112 | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 113 | WANG P, LIN Y, ZOU C J, et al. Construction and screening of a glycosylphosphatidylinositol protein deletion library in Pichia pastoris [J]. BMC Microbiology, 2020, 20(1): 262. |
| [1] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [2] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [3] | 李永珠, 陈禹. 酵母基因组规模模型进展及发展趋势[J]. 合成生物学, 2025, 6(3): 585-602. |
| [4] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [5] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [6] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| [7] | 盛周煌, 陈智仙, 张彦. 酵母甘露糖蛋白的研究进展[J]. 合成生物学, 2025, 6(2): 408-421. |
| [8] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [9] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [10] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [11] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [12] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [13] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [14] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [15] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
