合成生物学 ›› 2020, Vol. 1 ›› Issue (4): 454-469.DOI: 10.12211/2096-8280.2020-026
典型模式微生物基因表达精细调控工具的研究进展
田荣臻1,2, 刘延峰1,2, 李江华2, 刘龙1,2, 堵国成1,2
- 1.江南大学糖化学与生物技术教育部重点实验室,江苏 无锡 214122
2.江南大学工业生物技术教育部重点实验室,江苏 无锡 214122
-
收稿日期:2020-03-15修回日期:2020-06-21出版日期:2020-08-31发布日期:2020-10-09 -
通讯作者:堵国成 -
作者简介:田荣臻(1995—),男,博士研究生,研究方向为发酵工程。E-mail:rz_tian@stu.jiangnan.edu.cn
作者简介:堵国成(1965—),男,博士生导师,教授,研究方向为发酵工程与酶工程。E-mail:gcdu@jiangnan.edu.cn -
基金资助:国家重点研发计划(2018YFA0900300);国家自然科学基金(31972854)
Progress in the regulatory tools of gene expression for model microorganisms
TIAN Rongzhen1,2, LIU Yanfeng1,2, LI Jianghua2, LIU Long1,2, DU Guocheng1,2
- 1.Key Laboratory of Carbohydrate Chemistry and Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
2.Key Laboratory of Industrial Biotechnology,Ministry of Education,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2020-03-15Revised:2020-06-21Online:2020-08-31Published:2020-10-09 -
Contact:DU Guocheng
摘要:
以大肠杆菌、枯草芽孢杆菌和酿酒酵母等为代表的典型模式微生物是合成生物学研究中的重要底盘细胞。典型模型微生物的新型基因表达调控工具开发与应用实现了细胞代谢途径的精确工程设计和新型遗传回路的设计,极大促进了合成生物学和代谢工程的发展。本文针对典型模式微生物基因表达精细调控工具,特别是人工基因表达调控元件和精确调控的工具进行了系统总结和讨论。首先总结了经典的基因表达调控元件,然后介绍和讨论了基于中心法则创建的新型基因表达调控元件、基于全局调控蛋白的基因表达全局调控工具,以及响应特定信号的基因表达调控工具三个方面的最新研究进展,最后展望了如何通过新型天然基因表达调控元件的发现和响应代谢压力等细胞生理特性的基因表达调控系统的开发,进一步提升基因表达精细调控的范围和精度。通过将系统生物学数据与生物信息学相结合,能够进一步促进基因表达调控元件的标准化和多元化,提升基因表达精细调控的效率。
中图分类号:
引用本文
田荣臻, 刘延峰, 李江华, 刘龙, 堵国成. 典型模式微生物基因表达精细调控工具的研究进展[J]. 合成生物学, 2020, 1(4): 454-469.
TIAN Rongzhen, LIU Yanfeng, LI Jianghua, LIU Long, DU Guocheng. Progress in the regulatory tools of gene expression for model microorganisms[J]. Synthetic Biology Journal, 2020, 1(4): 454-469.
| 调控工具 | 调控水平 | 调控机理 | 典型实例 | 参考 文献 |
|---|---|---|---|---|
| 合成启动子 | 转录起始 | 实现天然启动子难以达到的特性,如更高的强度、更短的序列等 | 枯草芽孢杆菌混合启动子文库的构建 | [ |
| 广谱启动子 | 转录起始 | 实现启动子在所有模式微生物中的通用性 | 大肠杆菌、枯草芽孢杆菌、酿酒酵母广谱启动子 | [ |
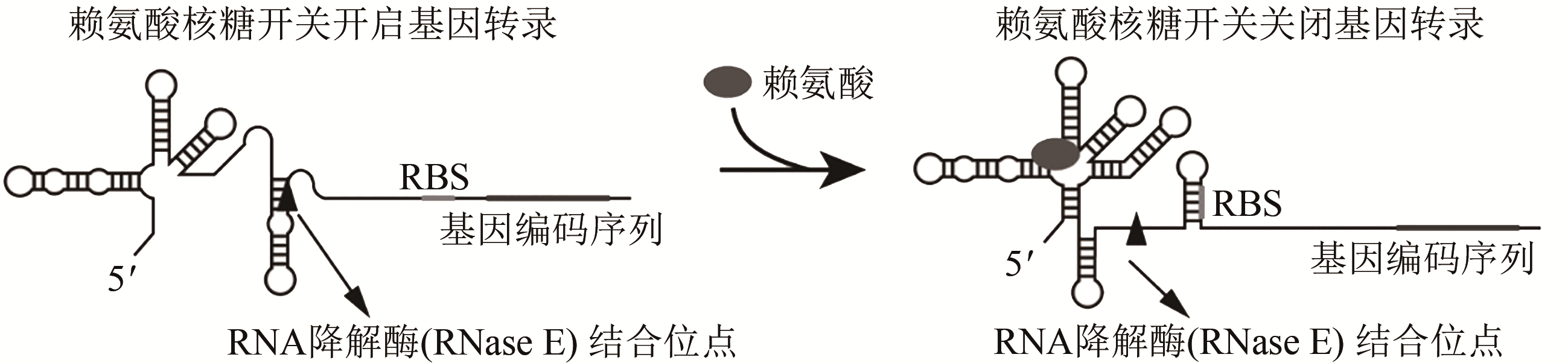
| 核糖开关 | 转录终止, 翻译起始, mRNA丰度 | 通过响应特定代谢物的结合而改变自身结构,可以调节转录终止、翻译起始和mRNA降解速率 | 大肠杆菌赖氨酸核糖开关、枯草芽孢杆菌glmS核糖开关 | [ |
| 核酸适配体 | 转录起始, 翻译起始 | 通过特异性结合响应核糖配体可以调节基因的转录起始或翻译起始 | 在枯草芽孢杆菌中使用凝血酶配体-适配体构建双功能动态代谢调控回路提高2′-岩藻糖基产量 | [ |
| sRNA | 转录延伸, 翻译起始 与延伸 | 在基因组上的反义转录可以造成目的基因转录中止,同时通过与特定mRNA进行碱基配对可以影响核糖体的结合与延伸 | 通过合成asRNA调节丙二酰辅酶A代谢,增强4-羟基香豆素、白藜芦醇和柚皮素的生物合成 | [ |
| 转录因子 | 转录起始 | 充当基因转录的阻遏物或激活物 | 构建CodY和CcpA突变文库调控枯草芽孢杆菌内碳氮代谢网络以提高β-半乳糖苷酶、木聚糖酶和肽酶产量 | [ |
| σ因子 | 转录起始 | 通过偏好性引导RNA聚合酶转录不同基因实现全局基因转录调控 | σ70的编码基因rpoD随机突变获得高性能的目标大肠杆菌 | [ |
| 基因表观 遗传修饰 | 转录起始 | 通过在宿主基因组中引入DNA修饰以实现基因转录的调控 | 通过调控DNA甲基转移酶表达提高大肠杆菌环境抗逆性 | [ |
| CRISPRa/i | 转录起始, 翻译起始 | 将dCas9蛋白融合强激活域或强抑制域靶向特定DNA或mRNA序列调控基因转录或翻译 | 在酿酒酵母中使用CRISPR/dCsd9系统同时实现对基因表达的激活、抑制与沉默 | [ |
| 群体响应 | 转录起始 | 响应细胞密度信号并将其转化为基因转录状态的变化 | 在大肠杆菌基于LuxR群体相应系统实现基因表达状态自动切换,提高了异丙醇产量 | [ |
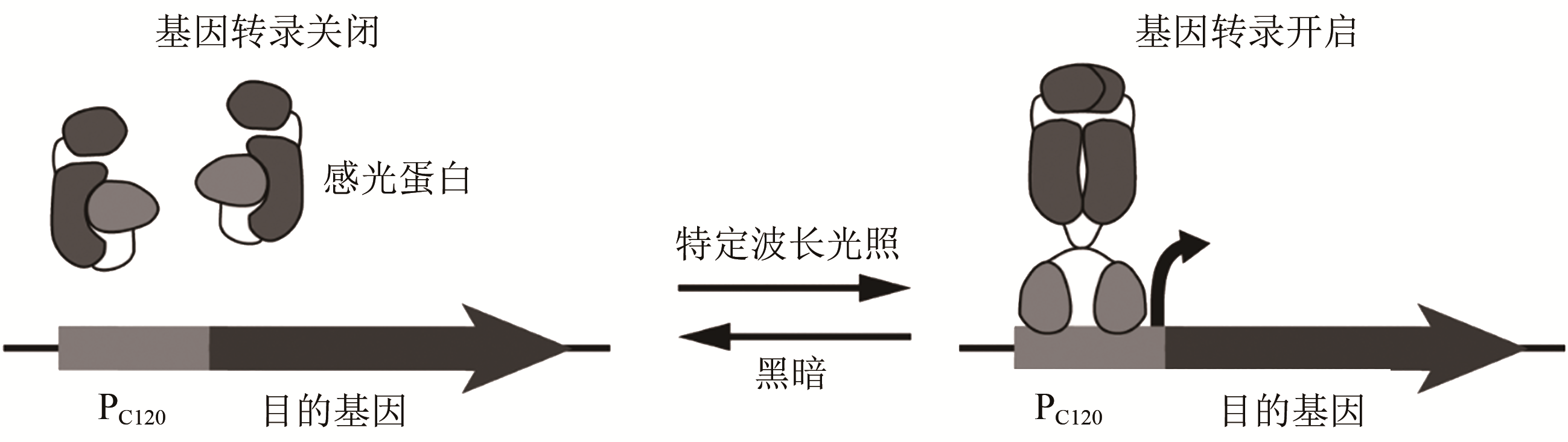
| 光遗传学调控 | 转录起始 | 响应特定波长光信号并将其转化为基因转录状态的变化 | 在酵母中实现蓝光依赖的基因表达激活与抑制以提高异丁醇和2-甲基-1-丁醇产量 | [ |
| 生物传感器 | 转录起始 | 响应特定代谢产物浓度并将其转化为基因转录状态的变化 | 在大肠杆菌中使用丙二酰-CoA生物传感器自动调控脂肪酸合成关键基因的表达,实现细胞生长和产物和成间的平衡 | [ |
| N端编码序列 | 翻译延伸 | 影响核糖体在mRNA上的延伸速率或脱落率 | 构建枯草芽孢杆菌N端编码序列文库调控NeuAc合成途径关键基因表达水平与动态模式以提高NeuAc产量 | [ |
| 蛋白降解标签 | 蛋白丰度 | 通过对目的蛋白的降解调控目的蛋白的丰度 | 将蛋白降解标签用于构建大肠杆菌正交基因回路,实现基因表达水平高强度激活与抑制 | [ |
表1 模式微生物新型基因表达调控工具
Tab. 1 Regulatory tools of gene expression for model microorganisms
| 调控工具 | 调控水平 | 调控机理 | 典型实例 | 参考 文献 |
|---|---|---|---|---|
| 合成启动子 | 转录起始 | 实现天然启动子难以达到的特性,如更高的强度、更短的序列等 | 枯草芽孢杆菌混合启动子文库的构建 | [ |
| 广谱启动子 | 转录起始 | 实现启动子在所有模式微生物中的通用性 | 大肠杆菌、枯草芽孢杆菌、酿酒酵母广谱启动子 | [ |
| 核糖开关 | 转录终止, 翻译起始, mRNA丰度 | 通过响应特定代谢物的结合而改变自身结构,可以调节转录终止、翻译起始和mRNA降解速率 | 大肠杆菌赖氨酸核糖开关、枯草芽孢杆菌glmS核糖开关 | [ |
| 核酸适配体 | 转录起始, 翻译起始 | 通过特异性结合响应核糖配体可以调节基因的转录起始或翻译起始 | 在枯草芽孢杆菌中使用凝血酶配体-适配体构建双功能动态代谢调控回路提高2′-岩藻糖基产量 | [ |
| sRNA | 转录延伸, 翻译起始 与延伸 | 在基因组上的反义转录可以造成目的基因转录中止,同时通过与特定mRNA进行碱基配对可以影响核糖体的结合与延伸 | 通过合成asRNA调节丙二酰辅酶A代谢,增强4-羟基香豆素、白藜芦醇和柚皮素的生物合成 | [ |
| 转录因子 | 转录起始 | 充当基因转录的阻遏物或激活物 | 构建CodY和CcpA突变文库调控枯草芽孢杆菌内碳氮代谢网络以提高β-半乳糖苷酶、木聚糖酶和肽酶产量 | [ |
| σ因子 | 转录起始 | 通过偏好性引导RNA聚合酶转录不同基因实现全局基因转录调控 | σ70的编码基因rpoD随机突变获得高性能的目标大肠杆菌 | [ |
| 基因表观 遗传修饰 | 转录起始 | 通过在宿主基因组中引入DNA修饰以实现基因转录的调控 | 通过调控DNA甲基转移酶表达提高大肠杆菌环境抗逆性 | [ |
| CRISPRa/i | 转录起始, 翻译起始 | 将dCas9蛋白融合强激活域或强抑制域靶向特定DNA或mRNA序列调控基因转录或翻译 | 在酿酒酵母中使用CRISPR/dCsd9系统同时实现对基因表达的激活、抑制与沉默 | [ |
| 群体响应 | 转录起始 | 响应细胞密度信号并将其转化为基因转录状态的变化 | 在大肠杆菌基于LuxR群体相应系统实现基因表达状态自动切换,提高了异丙醇产量 | [ |
| 光遗传学调控 | 转录起始 | 响应特定波长光信号并将其转化为基因转录状态的变化 | 在酵母中实现蓝光依赖的基因表达激活与抑制以提高异丁醇和2-甲基-1-丁醇产量 | [ |
| 生物传感器 | 转录起始 | 响应特定代谢产物浓度并将其转化为基因转录状态的变化 | 在大肠杆菌中使用丙二酰-CoA生物传感器自动调控脂肪酸合成关键基因的表达,实现细胞生长和产物和成间的平衡 | [ |
| N端编码序列 | 翻译延伸 | 影响核糖体在mRNA上的延伸速率或脱落率 | 构建枯草芽孢杆菌N端编码序列文库调控NeuAc合成途径关键基因表达水平与动态模式以提高NeuAc产量 | [ |
| 蛋白降解标签 | 蛋白丰度 | 通过对目的蛋白的降解调控目的蛋白的丰度 | 将蛋白降解标签用于构建大肠杆菌正交基因回路,实现基因表达水平高强度激活与抑制 | [ |
| 1 | KENT R, DIXON N. Contemporary tools for regulating gene expression in bacteria [J]. Trends in Biotechnology, 2020, 38(3): 316-333. DOI: 10.1016/j.tibtech.2019.09.007 . |
| 2 | TYO K E, ALPER H S, STEPHANOPOULOS G N. Expanding the metabolic engineering toolbox: more options to engineer cells [J]. Trends in Biotechnology, 2007, 25(3): 132-137. DOI: 10.1016/j.tibtech.2007.01.003 . |
| 3 | YANG Sen, DU Guocheng, CHEN Jian, et al. Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis [J]. Applied Microbiology and Biotechnology, 2017, 101(10): 4151-4161. DOI: 10.1007/s00253-017-8142-7 . |
| 4 | ALPER H, FISCHER C, NEVOIGT E, et al. Tuning genetic control through promoter engineering [J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(36): 12678. DOI: 10.1073/pnas.0504604102 . |
| 5 | REZNIKOFF W S. The lactose operon-controlling elements: a complex paradigm [J]. Molecular Microbiology, 1992, 6(17): 2419-2422. DOI: 10.1111/j.1365-2958.1992.tb01416.x . |
| 6 | GUIZIOU S, SAUVEPLANE V, CHANG Hung-Ju, et al. A part toolbox to tune genetic expression in Bacillus subtilis [J]. Nucleic Acids Research, 2016, 44(15): 7495-7508. DOI: 10.1093/nar/gkw624 . |
| 7 | SALIS H M, MIRSKY E A, VOIGT C A. Automated design of synthetic ribosome binding sites to control protein expression [J]. Nature Biotechnology, 2009, 27(10): 946-950. DOI: 10.1038/nbt.1568 . |
| 8 | SINUMVAYO J P, 杨森, 陈坚, 等. 枯草芽孢杆菌168新型转录终止子的构建与表征[J]. 生物工程学报, 2017, 33(7): 1091-1100. DOI: 10.13345/j.cjb.160484 . |
| SINUMVAYO J P, YANG S, CHEN J, et al. Engineering and characterization of new intrinsic transcriptional terminators in Bacillus subtilis 168 [J]. Chinese Journal of Biotechnology, 2017, 33(7): 1091-1100. DOI: 10.13345/j.cjb.160484 . | |
| 9 | POPE S D, MEDZHITOV R. Emerging principles of gene expression programs and their regulation [J]. Molecular Cell, 2018, 71(3): 389-397. DOI: 10.1016/j.molcel.2018.07.017 . |
| 10 | LU Zhenghui, YANG Shihui, YUAN Xin, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic acids research, 2019, 47(7): e40. DOI: 10.1093/nar/gkz072 . |
| 11 | YANG Sen, LIU Qingtao, ZHANG Yunfeng, et al. Construction and characterization of broad-spectrum promoters for synthetic biology [J]. ACS Synthetic Biology, 2018, 7(1): 287-291. DOI: 10.1021/acssynbio.7b00258 . |
| 12 | PAPENFORT K, VANDERPOOL C K. Target activation by regulatory RNAs in bacteria [J]. FEMS Microbiology Reviews, 2015, 39(3): 362-378. DOI: 10.1093/femsre/fuv016 . |
| 13 | BREAKER R R. Riboswitches and translation control [J]. Cold Spring Harbor Perspectives in Biology, 2018, 10(11). DOI: 10.1101/cshperspect.a032797 . |
| 14 | MANDAL M, BREAKER R R. Gene regulation by riboswitches [J]. Nature Reviews Molecular Cell Biology, 2004, 5(6): 451-463. DOI: 10.1038/nrm1403 . |
| 15 | CARON M P, BASTET L, LUSSIER A, et al. Dual-acting riboswitch control of translation initiation and mRNA decay [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(50): E3444-E3453. DOI: 10.1073/pnas.1214024109 . |
| 16 | JANG Sungho, JUNG Gyoo Yeol. Systematic optimization of L-tryptophan riboswitches for efficient monitoring of the metabolite in Escherichia coli [J]. Biotechnology and Bioengineering, 2018, 115(1): 266-271. DOI: 10.1002/bit.26448 . |
| 17 | JANG Sungho, JANG Sungyeon, XIU Yu, et al. Development of artificial riboswitches for monitoring of naringenin in vivo [J]. ACS Synthetic Biology, 2017, 6(11): 2077-2085. DOI: 10.1021/acssynbio.7b00128 . |
| 18 | XIU Yu, JANG Sungho, JONES J A, et al. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures [J]. Biotechnology and Bioengineering, 2017, 114(10): 2235-2244. DOI: 10.1002/bit.26340 . |
| 19 | CAI Yao, HU Huasi, PAN Ziheng, et al. Metaheuristic optimization in shielding design for neutrons and gamma rays reducing dose equivalent as much as possible [J]. Annals of Nuclear Energy, 2018, 120: 27-34. DOI: 10.1016/j.anucene.2018.05.038 . |
| 20 | BURKE-AGUERO D H. Methods in enzymology : riboswitches as targets and tools [J]. Methods in Enzymology, 2015. DOI: 10.1016/S0076-6879(15)00012-9 . |
| 21 | SEDLYAROVA N, SHAMOVSKY I, BHARATI B K, et al. sRNA-mediated control of transcription termination in E. coli [J]. Cell, 2016, 167(1): 111-121. e13. DOI: 10.1016/j.cell.2016.09.004. |
| 22 | Young Je LEE, HOYNES-O'CONNOR A, LEONG M C, et al. Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system [J]. Nucleic Acids Research, 2016, 44(5): 2462-2473. DOI: 10.1093/nar/gkw056 . |
| 23 | YANG Yaping, LIN Yuheng, LI Lingyun, et al. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products [J]. Metabolic Engineering, 2015, 29: 217-226. DOI: 10.1016/j.ymben.2015.03.018 . |
| 24 | NAKASHIMA N, TAMURA T, GOOD L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli [J]. Nucleic Acids Research, 2006, 34(20): e138-e138. DOI: 10.1093/nar/gkl697 . |
| 25 | SHERWOOD A V, HENKIN T M. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses [J]. Annual Review of Microbiology, 2016, 70(1): 361-374. DOI: 10.1146/annurev-micro-091014-104306 . |
| 26 | RÖTHLISBERGER P, HOLLENSTEIN M. Aptamer chemistry [J]. Advanced Drug Delivery Reviews, 2018, 134: 3-21. DOI: 10.1016/j.addr.2018.04.007 . |
| 27 | ABOUL-ELA F, HUANG Wei, ELRAHMAN M A, et al. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design [J]. Wiley Interdisciplinary Reviews. RNA, 2015, 6(6): 631-650. DOI: 10.1002/wrna.1300 . |
| 28 | WANG j, YANG Le, CUI Xun, et al. A DNA bubble-mediated gene regulation system based on thrombin-bound DNA aptamers [J]. ACS Synthetic Biology, 2017, 6(5): 758-765. DOI: 10.1021/acssynbio.6b00391 . |
| 29 | DENG Jieying, CHEN Chunmei, GU Yang, et al. Creating an in vivo bifunctional gene expression circuit through an aptamer-based regulatory mechanism for dynamic metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering, 2019, 55: 179-190. DOI: 10.1016/j.ymben.2019.07.008 . |
| 30 | GOODMAN D B, CHURCH G M, KOSURI S. Causes and effects of N-terminal codon bias in bacterial genes [J]. Science, 2013, 342(6157): 475. DOI: 10.1126/science.1241934 . |
| 31 | KUDLA G, MURRAY A W, TOLLERVEY D, et al. Coding-sequence determinants of gene expression in Escherichia coli [J]. Science, 2009, 324(5924): 255. DOI: 10.1126/science.1170160 . |
| 32 | SAUER C, THEMAAT E V L VAN, BOENDER L G M, et al. Exploring the nonconserved sequence space of synthetic expression modules in Bacillus subtilis [J]. ACS Synthetic Biology, 2018, 7(7): 1773-1784. DOI: 10.1021/acssynbio.8b00110 . |
| 33 | ESPAH BORUJENI A, SALIS H M. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism [J]. Journal of the American Chemical Society, 2016, 138(22): 7016-7023. DOI: 10.1021/jacs.6b01453 . |
| 34 | BORUJENI A E, CETNAR D, FARASAT I, et al. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences [J]. Nucleic Acids Research, 2017, 45(9): 5437-5448. DOI: 10.1093/nar/gkx061 . |
| 35 | DOUGAN D A, TRUSCOTT K N, ZETH K. The bacterial N-end rule pathway: expect the unexpected [J]. Molecular Microbiology, 2010, 76(3): 545-558. DOI: 10.1111/j.1365-2958.2010 .07120.x. |
| 36 | LU Jianli, DEUTSCH C. Electrostatics in the ribosomal tunnel modulate chain elongation rates [J]. Journal of Molecular Biology, 2008, 384(1): 73-86. DOI: 10.1016/j.jmb.2008.08.089 . |
| 37 | SHARMA A K, BUKAU B, O'BRIEN E P. Physical origins of codon positions that strongly influence cotranslational folding: a framework for controlling nascent-protein folding [J]. Journal of the American Chemical Society, 2016, 138(4): 1180-1195. DOI: 10.1021/jacs.5b08145 . |
| 38 | TOBIAS J W, SHRADER T E, ROCAP G, et al. The N-end rule in bacteria [J]. Science, 1991, 254(5036): 1374-1377. |
| 39 | ZADEH J N, STEENBERG C D, BOIS J S, et al. NUPACK: analysis and design of nucleic acid systems [J]. Journal of Computational Chemistry, 2011, 32(1): 170-173. DOI: 10.1002/jcc.21596 . |
| 40 | CAMBRAY G, GUIMARAES J C, ARKIN A P. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli [J]. Nature Biotechnology, 2018, 36(10): 1005-1015. DOI: 10.1038/nbt.4238 . |
| 41 | TIAN Rongzhen, LIU Yanfeng, CHEN Junrong, et al. Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering, 2019, 55: 131-141. DOI: 10.1016/j.ymben.2019 .07.001. |
| 42 | XU Peng. Production of chemicals using dynamic control of metabolic fluxes [J]. Current Opinion in Biotechnology, 2018, 53: 12-19. DOI: 10.1016/j.copbio.2017.10.009 . |
| 43 | HOLTZ W J, KEASLING J D. Engineering static and dynamic control of synthetic pathways [J]. Cell, 2010, 140(1): 19-23. DOI: 10.1016/j.cell.2009.12.029 . |
| 44 | ALPER H, STEPHANOPOULOS G. Global transcription machinery engineering: a new approach for improving cellular phenotype [J]. Metabolic Engineering, 2007, 9(3): 258-267. DOI: 10.1016/j.ymben.2006.12.002 . |
| 45 | MY L, ACHKAR N G, VIALA J P, et al. Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: global positive control by the dual functional regulator FadR [J]. Journal of Bacteriology, 2015, 197(11): 1862-1872. DOI: 10.1128/JB.00064-15 . |
| 46 | KURODA K, UEDA M. Engineering of global regulators and cell surface properties toward enhancing stress tolerance in Saccharomyces cerevisiae [J]. Journal of Bioscience and Bioengineering, 2017, 124(6): 599-605. DOI: 10.1016/j.jbiosc.2017 .06.010. |
| 47 | NGUYEN-VO T P, LIANG Yunxiao, SANKARANARAYANAN M, et al. Development of 3-hydroxypropionic-acid-tolerant strain of Escherichia coli W and role of minor global regulator yieP [J]. Metabolic Engineering, 2019, 53: 48-58. DOI: 10.1016/j.ymben.2019.02.001 . |
| 48 | BRINSMADE S R, ALEXANDER E L, LIVNY J, et al. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(22): 8227. DOI: 10.1073/pnas.1321308111 . |
| 49 | CAO Haojie, KUIPERS O P. Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in Bacillus subtilis [J]. Microbial Cell Factories, 2018, 17(1): 134. DOI: 10.1186/s12934-018-0985-9 . |
| 50 | ZHU Liying, GAO Shan, ZHANG Hongman, et al. Improvement of lead tolerance of Saccharomyces cerevisiae by random mutagenesis of transcription regulator SPT3 [J]. Applied Biochemistry and Biotechnology, 2018, 184(1): 155-167. DOI: 10.1007/s12010-017-2531-3 . |
| 51 | PARK Kyung-Soon, Dong-ki LEE, Horim LEE, et al. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors [J]. Nature Biotechnology, 2003, 21(10): 1208-1214. DOI: 10.1038/nbt868 . |
| 52 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production [J]. Science, 2006, 314(5805): 1565. DOI: 10.1126/science.1131969 . |
| 53 | BURGESS R R, ANTHONY L. How sigma docks to RNA polymerase and what sigma does [J]. Current Opinion in Microbiology, 2001, 4(2): 126-131. DOI: 10.1016/S1369-5274(00)00177-6 . |
| 54 | GAO Xi, JIANG Ling, ZHU Liying, et al. Tailoring of global transcription sigma D factor by random mutagenesis to improve Escherichia coli tolerance towards low-pHs [J]. Journal of Biotechnology, 2016, 224: 55-63. DOI: 10.1016/j.jbiotec.2016.03.012 . |
| 55 | ADHIKARI S, CURTIS P D. DNA methyltransferases and epigenetic regulation in bacteria [J]. FEMS Microbiology Reviews, 2016, 40(5): 575-591. DOI: 10.1093/femsre/fuw023 . |
| 56 | KANG Jeong Gu, PARK Jin Suk, Jeong-Heosn KO, et al. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system [J]. Scientific Reports, 2019, 9(1): 11960. DOI: 10.1038/s41598-019-48130-3 . |
| 57 | GRUNSTEIN M, GASSER S M. Epigenetics in Saccharomyces cerevisiae [J]. Cold Spring Harbor Perspectives in Biology, 2013, 5(7): a017491. DOI: 10.1101/cshperspect.a017491 . |
| 58 | CHEN Chao, WANG Lianrong, CHEN Si, et al. Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes [J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(17): 4501-4506. DOI: 10.1073/pnas.1702450114 . |
| 59 | LAL A, KRISHNA S, SESHASAYEE A S N. Regulation of global transcription in Escherichia coli by Rsd and 6S RNA [J]. G3: Genes, Genomes, Genetics, 2018, 8(6): 2079-2089. DOI: 10.1534/g3.118.200265 . |
| 60 | WASSARMAN K M, STORZ G. 6S RNA Regulates E. coli RNA polymerase activity [J]. Cell, 2000, 101(6): 613-623. DOI: 10.1016/S0092-8674(00)80873-9 . |
| 61 | WASSARMAN K M. 6S RNA, a global regulator of transcription [J]. Microbiology Spectrum, 2018, 6(3): 10.1128/microbiolspec. RWR-0019-2018. DOI: 10.1128/microbiolspec.RWR-0019-2018 . |
| 62 | CAVANAGH A T, WASSARMAN K M. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond [J]. Annual Review of Microbiology, 2014, 68(1): 45-60. DOI: 10.1146/annurev-micro-092611-150135 . |
| 63 | QI Lei S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression [J]. Cell, 2013, 152(5): 1173-1183. DOI: 10.1016/j.cell.2013.02.022 . |
| 64 | RAN F A, HSU P D, WRIGHT J, et al. Genome engineering using the CRISPR-Cas9 system [J]. Nature Protocols, 2013, 8(11): 2281-2308. DOI: 10.1038/nprot.2013.143 . |
| 65 | HSU P D, LANDER E S, ZHANG Feng. Development and applications of CRISPR-Cas9 for genome engineering [J]. Cell, 2014, 157(6): 1262-1278. DOI: 10.1016/j.cell.2014.05.010 . |
| 66 | LIU Yang, WAN Xinyi, WANG Baojun. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria [J]. Nature Communications, 2019, 10(1): 3693. DOI: 10.1038/s41467-019-11479-0 . |
| 67 | LIAN Jiazhang, HAMEDIRAD M, HU Sumeng, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system [J]. Nature Communications, 2017, 8(1): 1688. DOI: 10.1038/s41467-017-01695-x . |
| 68 | WU Yaokang, CHEN Taichi, LIU Yanfeng, et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 48(2): 996-1009. DOI: 10.1093/nar/gkz1123 . |
| 69 | FENNO L, YIZHAR O, DEISSEROTH K. The development and application of optogenetics [J]. Annual Review of Neuroscience, 2011, 34: 389-412. DOI: 10.1146/annurev-neuro-061010-113817 . |
| 70 | BACCHUS W, FUSSENEGGER M. The use of light for engineered control and reprogramming of cellular functions [J]. Current Opinion in Biotechnology, 2012, 23(5): 695-702. DOI: 10.1016/j.copbio.2011.12.004 . |
| 71 | OLSON E J, TABOR J J. Optogenetic characterization methods overcome key challenges in synthetic and systems biology [J]. Nature Chemical Biology, 2014, 10(7): 502-511. DOI: 10.1038/nchembio.1559 . |
| 72 | PATHAK G P, VRANA J D, TUCKER C L. Optogenetic control of cell function using engineered photoreceptors [J]. Biology of the Cell, 2013, 105(2): 59-72. DOI: 10.1111/boc.201200056 . |
| 73 | ZHAO E M, ZHANG Yanfei, MEHL J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production [J]. Nature, 2018, 555(7698): 683-687. DOI: 10.1038/nature26141 . |
| 74 | SHIMIZU-SATO S, HUQ E, TEPPERMAN J M, et al. A light-switchable gene promoter system [J]. Nature Biotechnology, 2002, 20(10): 1041-1044. DOI: 10.1038/nbt734 . |
| 75 | MOTTA-MENA L B, READE A, MALLORY M J, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics [J]. Nature Chemical Biology, 2014, 10(3): 196-202. DOI: 10.1038/nchembio.1430 . |
| 76 | SHIN Yongdae, BERRY J, PANNUCCI N, et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets [J]. Cell, 2017, 168(1/2): 159-171.e 14. DOI: 10.1016/j.cell.2016.11.054 . |
| 77 | TABOR J J, LEVSKAYA A, VOIGT C A. Multichromatic control of gene expression in Escherichia coli [J]. Journal of Molecular Biology, 2011, 405(2): 315-324. DOI: 10.1016/j.jmb.2010.10.038 . |
| 78 | MILIAS-ARGEITIS A, RULLAN M, AOKI S K, et al. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth [J]. Nature Communications, 2016, 7: 12546. DOI: 10.1038/ncomms12546 . |
| 79 | CASTILLO-HAIR S M, BAERMAN E A, FUJITA M, et al. Optogenetic control of Bacillus subtilis gene expression [J]. Nature Communications, 2019, 10(1): 3099. DOI: 10.1038/s41467-019-10906-6 . |
| 80 | GAO Cong, HOU Jianshen, XU Peng, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli [J]. Nature Communications, 2019, 10(1): 3751. DOI: 10.1038/s41467-019-11793-7 . |
| 81 | CAMERON D E, COLLINS J J. Tunable protein degradation in bacteria [J]. Nature Biotechnology, 2014, 32(12): 1276-1281. DOI: 10.1038/nbt.3053 . |
| 82 | CHUNG Hokyung K, JACOBS C L, HUO Yunwen, et al. Tunable and reversible drug control of protein production via a self-excising degron [J]. Nature Chemical Biology, 2015, 11(9): 713-720. DOI: 10.1038/nchembio.1869 . |
| 83 | DOONG S J, GUPTA A, PRATHER K L J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): 2964-2969. DOI: 10.1073/pnas.1716920115 . |
| 84 | MARTÍNEZ V, LAURITSEN I, HOBEL T, et al. CRISPR/Cas9-based genome editing for simultaneous interference with gene expression and protein stability [J]. Nucleic Acids Research, 2017, 45(20): e171-e171. DOI: 10.1093/nar/gkx797 . |
| 85 | FERNANDEZ-RODRIGUEZ J, VOIGT C A. Post-translational control of genetic circuits using Potyvirus proteases [J]. Nucleic Acids Research, 2016, 44(13): 6493-6502. DOI: 10.1093/nar/gkw537 . |
| 86 | TAN S Z, PRATHER K L J. Dynamic pathway regulation:recent advances and methods of construction [J]. Current Opinion in Chemical Biology, 2017, 41: 28-35. DOI: 10.1016/j.cbpa.2017.10.004 . |
| 87 | WHITELEY M, DIGGLE S P, GREENBERG E P. Progress in and promise of bacterial quorum sensing research [J]. Nature, 2017, 551(7680): 313-320. DOI: 10.1038/nature24624 . |
| 88 | SOMA Y, HANAI T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production [J]. Metabolic Engineering, 2015, 30: 7-15. DOI: 10.1016/j.ymben.2015.04.005 . |
| 89 | GUPTA A, REIZMAN I M B, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit [J]. Nature biotechnology, 2017, 35(3): 273-279. DOI: 10.1038/nbt.3796 . |
| 90 | CUI Shixiu, Xueqin LÜ, WU Yaokang, et al. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis [J]. ACS Synthetic Biology, 2019, 8(8): 1826-1837. DOI: 10.1021/acssynbio.9b00140 . |
| 91 | WILLIAMS T C, AVERESCH N, WINTER G, et al. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 29: 124-134. DOI: 10.1016/j.ymben.2015.03.008 . |
| 92 | XU Peng, LI Lingyun, ZHANG Fuming, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. DOI: 10.1073/pnas.1406401111 . |
| 93 | RUGBJERG P, SARUP-LYTZEN K, NAGY M, et al. Synthetic addiction extends the productive life time of engineered Escherichia coli populations [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2347. DOI: 10.1073/pnas.1718622115 . |
| 94 | SANDBERG T E, SALAZAR M J, WENG L L, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology [J]. Metabolic Engineering, 2019, 56: 1-16. DOI: 10.1016/j.ymben.2019.08.004 . |
| 95 | BUERGER J, GRONENBERG L S, GENEE H J, et al. Wiring cell growth to product formation [J]. Current Opinion in Biotechnology, 2019, 59: 85-92. DOI: 10.1016/j.copbio.2019.02.014 . |
| 96 | CHOU H H, KEASLING J D. Programming adaptive control to evolve increased metabolite production [J]. Nature Communications, 2013, 4(1): 2595. DOI: 10.1038/ncomms3595 . |
| 97 | LEAVITT J M, WAGNER J M, TU C C, et al. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae [J]. Biotechnology Journal, 2017, 12(10): 1600687. DOI: 10.1002/biot.201600687 . |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [11] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [12] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [13] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [14] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [15] | 禹伟, 高教琪, 周雍进. 一碳生物转化合成有机酸的研究进展[J]. 合成生物学, 2024, 5(5): 1169-1188. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||