合成生物学 ›› 2021, Vol. 2 ›› Issue (1): 59-90.DOI: 10.12211/2096-8280.2020-071
运动发酵单胞菌底盘细胞研究现状及展望
杨永富1, 耿碧男1, 宋皓月1, 何桥宁1, 何明雄2, 鲍杰3, 白凤武4, 杨世辉1
- 1.湖北大学生命科学学院,省部共建生物催化与酶工程国家重点实验室,湖北省环境微生物工程技术研究中心,湖北 武汉 430062
2.农业农村部沼气科学研究所,生物质能技术研究中心,四川 成都 610041
3.华东理工大学生物工程学院,生物反应器工程国家重点实验室,上海 200237
4.上海交通大学生命科学技术学院,微生物代谢国家重点实验室,上海 200240
-
收稿日期:2020-07-05修回日期:2020-10-01出版日期:2021-02-28发布日期:2021-03-12 -
通讯作者:白凤武,杨世辉 -
作者简介:杨永富(1994—),男,博士研究生,研究方向为微生物系统生物学与合成生物学。E-mail:yongfu.yang@stu.hubu.edu.cn
耿碧男(1995—),女,博士研究生,研究方向为微生物基因组优化及合成生物学。E-mail:binangeng@stu.hubu.edu.cn
白凤武(1964—),男,博士,教授,研究方向为生物质资源生物炼制及微生物代谢工程。E-mail:fwbai@sjtu.edu.cn
杨世辉(1971—),男,博士,教授,研究方向为微生物代谢工程与合成生物学。E-mail:Shihui.Yang@hubu.edu.cn -
基金资助:国家自然科学基金(21978071);国家重点研发计划(2018YFA0900300);浙江省引进培育领军型创新创业团队项目(2018R01014)
Progress and perspectives on developing Zymomonas mobilis as a chassis cell
YANG Yongfu1, GENG Binan1, SONG Haoyue1, HE Qiaoning1, HE Mingxiong2, BAO Jie3, BAI Fengwu4, YANG Shihui1
- 1.State Key Laboratory of Biocatalysis and Enzyme Engineering,Environmental Microbial Technology Center of Hubei Province,School of Life Sciences,Hubei University,Wuhan 430062,Hubei,China
2.Biomass Energy Technology Research Centre,Biogas Institute of Ministry of Agriculture,Chengdu 610041,Sichuan,China
3.State Key Laboratory of Bioreactor Engineering,School of Biotechnology,East China University of Science and Technology,Shanghai 200237,China
4.State Key Laboratory of Microbial Metabolism,School of Life Sciences and Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2020-07-05Revised:2020-10-01Online:2021-02-28Published:2021-03-12 -
Contact:BAI Fengwu, YANG Shihui
摘要:
运动发酵单胞菌(Zymomonas mobilis)是目前已知唯一能够在厌氧条件下利用Entner-Doudoroff(ED)途径代谢葡萄糖、果糖和蔗糖产乙醇的革兰氏阴性细菌,具有乙醇发酵速率高和对糖表观收率高、乙醇耐受性好及生物安全(generally regarded as safe,GRAS)等特点。基于合成生物学方法和代谢工程改造,可以作为纤维素乙醇及其他生物基产品生物炼制的细胞工厂。本文综述了运动发酵单胞菌独特的生理特点及其作为细胞工厂在不同领域的应用,重点介绍了构建运动发酵单胞菌作为底盘细胞,实现工业产品规模化经济生产涉及的系统生物学、合成生物学及代谢工程改造相关方法、技术与工具等方面的进展及瓶颈。同时探讨了持续开发、完善、应用高效精准的基因编辑技术、代谢途径精准时空调控方法及高通量自动筛选检测手段,在运动发酵单胞菌基因组精简优化以及生物固碳与固氮等方面取得的突破,推动合成生物学理论研究和实践应用的发展。
中图分类号:
引用本文
杨永富, 耿碧男, 宋皓月, 何桥宁, 何明雄, 鲍杰, 白凤武, 杨世辉. 运动发酵单胞菌底盘细胞研究现状及展望[J]. 合成生物学, 2021, 2(1): 59-90.
YANG Yongfu, GENG Binan, SONG Haoyue, HE Qiaoning, HE Mingxiong, BAO Jie, BAI Fengwu, YANG Shihui. Progress and perspectives on developing Zymomonas mobilis as a chassis cell[J]. Synthetic Biology Journal, 2021, 2(1): 59-90.

图1 运动发酵单胞菌的重要生理特性及其多样化的应用(绿色字体表示途径中的关键酶;蓝色字体表示可生产的平台化合物产品,加蓝色框线的为需引入外源基因的产物;灰色字体表示可能的产物)
Fig. 1 Physiological characteristics and diverse applications of Z. mobilis(The green font in the pathway represents the key enzymes. The blue font represents biochemicals produced by Z. mobilis, and those within the blue boxes are produced through metabolic engineering of heterologous pathways. Other potential bioproducts are presented with gray font)
| 化合物 | 产量 | 应用范围 | 文献 |
|---|---|---|---|
| 葡萄糖酸(gluconic acid) | 541 g·L-1 | 蛋白凝固剂和食品防腐剂 | [ |
| 聚羟基丁酸(PHB) | 0.7% DCW(DCW为细胞干重) | 可降解生物塑料 | [ |
| L-乳酸(L-lactate) | 10.8 g·L-1 | 医疗,食品,化妆品,化工原料等 | [ |
| D-乳酸(D-lactate) | 65.6 g·L-1 | 医疗,食品,化妆品,化工原料等 | [ |
| 果聚糖(levan) | 40.2 g·L-1 | 加工食品,常作为乳化稳定剂、气泡稳定剂 | [ |
| 丁二酸(succinate) | 2.8 g·L-1 | C4 平台化合物,有机化工原料及中间体 | [ |
| 乙醇(ethanol) | 136 g·L-1 | 生物能源 | [ |
| 山梨醇(sorbitol) | 135.7 g·L-1 | 保湿剂,软化剂,甜味剂 | [ |
| 2,3-丁二醇(2,3-butanediol) | 120 g·L-1 | 平台大宗化合物,液体燃料 | [ |
| 乙醛(acetaldehyde) | 4.0 g·L-1 | 化学工业平台化合物,重要的香味剂 | [ |
| 乙烯(ethylene) | 12.83 nmol/mL(OD600) | 基础化工原料,可用于合成橡胶、纤维、聚烯烃和聚氯乙烯塑料等 | [ |
| 乳糖酸(lactobionic acid) | 182.7 g·L-1 | 精细化学品,医药中间体,材料中间体等 | [ |
| 异丁醇(isobutanol) | 4 g·L-1 | 可再生生物能源,异丁烯前体物 | [ |
表1 运动发酵单胞菌可生产的不同化合物名单
Tab. 1 Biochemicals produced by Z. mobilis
| 化合物 | 产量 | 应用范围 | 文献 |
|---|---|---|---|
| 葡萄糖酸(gluconic acid) | 541 g·L-1 | 蛋白凝固剂和食品防腐剂 | [ |
| 聚羟基丁酸(PHB) | 0.7% DCW(DCW为细胞干重) | 可降解生物塑料 | [ |
| L-乳酸(L-lactate) | 10.8 g·L-1 | 医疗,食品,化妆品,化工原料等 | [ |
| D-乳酸(D-lactate) | 65.6 g·L-1 | 医疗,食品,化妆品,化工原料等 | [ |
| 果聚糖(levan) | 40.2 g·L-1 | 加工食品,常作为乳化稳定剂、气泡稳定剂 | [ |
| 丁二酸(succinate) | 2.8 g·L-1 | C4 平台化合物,有机化工原料及中间体 | [ |
| 乙醇(ethanol) | 136 g·L-1 | 生物能源 | [ |
| 山梨醇(sorbitol) | 135.7 g·L-1 | 保湿剂,软化剂,甜味剂 | [ |
| 2,3-丁二醇(2,3-butanediol) | 120 g·L-1 | 平台大宗化合物,液体燃料 | [ |
| 乙醛(acetaldehyde) | 4.0 g·L-1 | 化学工业平台化合物,重要的香味剂 | [ |
| 乙烯(ethylene) | 12.83 nmol/mL(OD600) | 基础化工原料,可用于合成橡胶、纤维、聚烯烃和聚氯乙烯塑料等 | [ |
| 乳糖酸(lactobionic acid) | 182.7 g·L-1 | 精细化学品,医药中间体,材料中间体等 | [ |
| 异丁醇(isobutanol) | 4 g·L-1 | 可再生生物能源,异丁烯前体物 | [ |

图2 运动发酵单胞菌系统与合成生物学研究进展(The omics research of genomics, transcriptomics, proteomics and metabolomics represents as G-moics, T-moics, P-omics and M-omics, respectively)
Fig. 2 Research progress of systems and synthetic biology in Z. mobilis
| 研究内容 | 年份 | 方法 | 数据序列号 | 参考文献 |
|---|---|---|---|---|
| ZM4基因组与比较基因组 | 2005 | 基因组(随机鸟枪测序法) 比较基因组(芯片技术) | AE008692;E-MEXP-217 | [ |
| 结合系统生物学数据更新ZM4基因组注释 | 2009 | 基因组(454焦磷酸测序) | SRP000908 | [ |
| 有氧与无氧条件下ZM4转录组与代谢组 | 2009 | 转录组(芯片技术); 代谢组(GC、GC-MS、HPLC) | GSE10302 | [ |
| AcR菌株基因型与表型关联及比较基因组 | 2010 | 基因组(454焦磷酸测序); 比较基因组(芯片技术);转录组(芯片技术) | GSE18106 | [ |
| 絮凝菌株ZM401重测序 | 2012 | 基因组(Illumina) | AMSR00000000 | [ |
| 絮凝菌株ZM401 转录组分析 | 2012 | 转录组(芯片技术) | GU560731-7 | [ |
| ZM4在5%(体积分数)乙醇压力下的转录组 | 2012 | 转录组(芯片技术) | GSE39558 | [ |
| ZM4在1 g/L糠醛压力下的转录组 | 2012 | 转录组(芯片技术) | GSE37848 | [ |
| ZM4在6%(体积分数)乙醇压力下的转录组、蛋白组和代谢组 | 2013 | 转录组(芯片技术); 蛋白组(鸟枪法蛋白质测序); 代谢组(GC-MS、HPLC) | GSE21165 | [ |
| ZM4 sRNA转录组 | 2014 | 转录组(sRNA-Seq) | GSE57773 | [ |
| 8b在不同碳源下外加乙酸的转录组 | 2014 | 转录组(芯片技术) | GSE57553 | [ |
| AcR菌株在10 g/L乙酸钠的转录组与蛋白组 | 2014 | 转录组(芯片技术);蛋白组(LC-MS/MS ) | GSE25443 | [ |
| ZM4基因转座子突变体文库的表型适应检测 | 2014 | 突变体文库适应检测 | GSE51870 | [ |
| ZM4五碳糖重组自然进化菌株(KLD1,KLD2)重测序 | 2015 | 基因组(Illumina) | NA | [ |
| ZM4添加5 mmol/L酚醛 抑制物的转录组 | 2015 | 转录组(芯片技术) | NA | [ |
| ZM4在220g/L葡萄糖添加山梨醇(10 mmol/L)的转录组 | 2015 | 转录组(芯片技术) | GSE49620 | [ |
| ZM4质粒序列更新及8b、2032基因组序列 | 2018 | 基因组(Illumina);转录组(RNA-Seq) | PRJNA391970 | [ |
| ZM4在多种抑制物环境下的蛋白组和代谢组 | 2018 | 蛋白组(RPLC-MS/MS);代谢组(GC-MS) | NA | [ |
| 耐低pH(pH 3.5)菌株(PH1 -29)重测序 | 2019 | 基因组(Illumina) | GDMCC60258;GDMCC60260 | [ |
| 乙酸及糠醛耐受菌株532与533重测序 | 2019 | 基因组(Illumina) | GDMCC60526;GDMCC60527 | [ |
| 2H和13C代谢组学研究ZM4 ED途径热力学 | 2019 | 代谢组(LC-MS、NMR) | NA | [ |
| ZM4在无氧、好氧和固氮条件的磷酸化蛋白质组 | 2019 | 蛋白组(LC-MS/MS) | PXD014065 | [ |
| 氧暴露对ZM4类异戊二烯生成的影响 | 2019 | 转录组(RNA-Seq);蛋白组(LC-MS/MS);代谢组(LC-MS、NMR) | GSE125123 | [ |
| 2032菌株利用合成水解液中葡萄糖与木糖 | 2019 | 转录组(RNA-Seq);蛋白组(LC-MS/MS);代谢组(IP-LC-MS、GC-MS、AEX-LC-MS、HILIC-MS) | GSE135718;PXD015007 | [ |
| ZM4葡萄糖与木糖共利用菌株AD50重测序 | 2020 | 基因组(Illumina) | NA | [ |
| ZM4耐低pH菌株3.6M和3.5M重测序及转录组 | 2020 | 基因组(Illumina);转录组(RNA-Seq) | PRJNA590883;PRJNA553033 | [ |
| 8b在3 g/L糠醛短期刺激或2 g/L糠醛长期胁迫下的转录组 | 2020 | 转录组(芯片技术、RNA-Seq) | GSE63540 | [ |
| ZM4过表达sRNA(Zms4与Zms6)的转录组 | 2020 | 转录组(RNA-Seq) | GSE107219 | [ |
| Mg2+对 ZM4细胞生长影响的转录组 | 2020 | 转录组(RNA-Seq) | PRJNA601020 | [ |
| 代谢活跃非生长Zm6菌株适应氧供应变化代谢组学 | 2020 | 代谢组(CIC-MS) | NA | [ |
表2 运动发酵单胞菌系统生物学研究进展总结
Tab. 2 Summary of systems biology research progress in Z. mobilis
| 研究内容 | 年份 | 方法 | 数据序列号 | 参考文献 |
|---|---|---|---|---|
| ZM4基因组与比较基因组 | 2005 | 基因组(随机鸟枪测序法) 比较基因组(芯片技术) | AE008692;E-MEXP-217 | [ |
| 结合系统生物学数据更新ZM4基因组注释 | 2009 | 基因组(454焦磷酸测序) | SRP000908 | [ |
| 有氧与无氧条件下ZM4转录组与代谢组 | 2009 | 转录组(芯片技术); 代谢组(GC、GC-MS、HPLC) | GSE10302 | [ |
| AcR菌株基因型与表型关联及比较基因组 | 2010 | 基因组(454焦磷酸测序); 比较基因组(芯片技术);转录组(芯片技术) | GSE18106 | [ |
| 絮凝菌株ZM401重测序 | 2012 | 基因组(Illumina) | AMSR00000000 | [ |
| 絮凝菌株ZM401 转录组分析 | 2012 | 转录组(芯片技术) | GU560731-7 | [ |
| ZM4在5%(体积分数)乙醇压力下的转录组 | 2012 | 转录组(芯片技术) | GSE39558 | [ |
| ZM4在1 g/L糠醛压力下的转录组 | 2012 | 转录组(芯片技术) | GSE37848 | [ |
| ZM4在6%(体积分数)乙醇压力下的转录组、蛋白组和代谢组 | 2013 | 转录组(芯片技术); 蛋白组(鸟枪法蛋白质测序); 代谢组(GC-MS、HPLC) | GSE21165 | [ |
| ZM4 sRNA转录组 | 2014 | 转录组(sRNA-Seq) | GSE57773 | [ |
| 8b在不同碳源下外加乙酸的转录组 | 2014 | 转录组(芯片技术) | GSE57553 | [ |
| AcR菌株在10 g/L乙酸钠的转录组与蛋白组 | 2014 | 转录组(芯片技术);蛋白组(LC-MS/MS ) | GSE25443 | [ |
| ZM4基因转座子突变体文库的表型适应检测 | 2014 | 突变体文库适应检测 | GSE51870 | [ |
| ZM4五碳糖重组自然进化菌株(KLD1,KLD2)重测序 | 2015 | 基因组(Illumina) | NA | [ |
| ZM4添加5 mmol/L酚醛 抑制物的转录组 | 2015 | 转录组(芯片技术) | NA | [ |
| ZM4在220g/L葡萄糖添加山梨醇(10 mmol/L)的转录组 | 2015 | 转录组(芯片技术) | GSE49620 | [ |
| ZM4质粒序列更新及8b、2032基因组序列 | 2018 | 基因组(Illumina);转录组(RNA-Seq) | PRJNA391970 | [ |
| ZM4在多种抑制物环境下的蛋白组和代谢组 | 2018 | 蛋白组(RPLC-MS/MS);代谢组(GC-MS) | NA | [ |
| 耐低pH(pH 3.5)菌株(PH1 -29)重测序 | 2019 | 基因组(Illumina) | GDMCC60258;GDMCC60260 | [ |
| 乙酸及糠醛耐受菌株532与533重测序 | 2019 | 基因组(Illumina) | GDMCC60526;GDMCC60527 | [ |
| 2H和13C代谢组学研究ZM4 ED途径热力学 | 2019 | 代谢组(LC-MS、NMR) | NA | [ |
| ZM4在无氧、好氧和固氮条件的磷酸化蛋白质组 | 2019 | 蛋白组(LC-MS/MS) | PXD014065 | [ |
| 氧暴露对ZM4类异戊二烯生成的影响 | 2019 | 转录组(RNA-Seq);蛋白组(LC-MS/MS);代谢组(LC-MS、NMR) | GSE125123 | [ |
| 2032菌株利用合成水解液中葡萄糖与木糖 | 2019 | 转录组(RNA-Seq);蛋白组(LC-MS/MS);代谢组(IP-LC-MS、GC-MS、AEX-LC-MS、HILIC-MS) | GSE135718;PXD015007 | [ |
| ZM4葡萄糖与木糖共利用菌株AD50重测序 | 2020 | 基因组(Illumina) | NA | [ |
| ZM4耐低pH菌株3.6M和3.5M重测序及转录组 | 2020 | 基因组(Illumina);转录组(RNA-Seq) | PRJNA590883;PRJNA553033 | [ |
| 8b在3 g/L糠醛短期刺激或2 g/L糠醛长期胁迫下的转录组 | 2020 | 转录组(芯片技术、RNA-Seq) | GSE63540 | [ |
| ZM4过表达sRNA(Zms4与Zms6)的转录组 | 2020 | 转录组(RNA-Seq) | GSE107219 | [ |
| Mg2+对 ZM4细胞生长影响的转录组 | 2020 | 转录组(RNA-Seq) | PRJNA601020 | [ |
| 代谢活跃非生长Zm6菌株适应氧供应变化代谢组学 | 2020 | 代谢组(CIC-MS) | NA | [ |
| 菌株 | 模型名称 | 模型尺度 | 参考文献 |
|---|---|---|---|
| Z. mobilis | 基于酶的动力学模型 | ED 和 PPP途径 | [ |
| Z. mobilis | 基于线性规划的计量学模型 | 79个反应与77个代谢物 | [ |
| Z. mobilis | 中心代谢的计量学模型 | 96 个反应 | [ |
| Z. mobilis ATCC29191 (Zm6) | ED 途径的动力学模型 | ED 途径 | [ |
| Z. mobilis | 多种方法的中心代谢模型 | 中等规模 | [ |
| Z. mobilis ATCC31821 (ZM4) | ZmoMBEL601代谢网络模型 | 基因组尺度:348个基因、601个反应和579个代谢物 | [ |
| Z. mobilis ATCC31821 (ZM4) | iZM363代谢网络模型 | 基因组尺度:363个基因、747个反应和704个代谢物 | [ |
| Z. mobilis ATCC10988 (ZM1) | iEM439代谢网络模型 | 基因组尺度:439个基因、692个反应和658个代谢物 | [ |
| Z. mobilis ATCC31821 (ZM4) | iHN446代谢网络模型 | 基因组尺度:446个基因、859个反应和894个代谢物 | [ |
表3 运动发酵单胞菌不同尺度代谢模型
Tab. 3 Metabolic models at different scales established for Z. mobilis
| 菌株 | 模型名称 | 模型尺度 | 参考文献 |
|---|---|---|---|
| Z. mobilis | 基于酶的动力学模型 | ED 和 PPP途径 | [ |
| Z. mobilis | 基于线性规划的计量学模型 | 79个反应与77个代谢物 | [ |
| Z. mobilis | 中心代谢的计量学模型 | 96 个反应 | [ |
| Z. mobilis ATCC29191 (Zm6) | ED 途径的动力学模型 | ED 途径 | [ |
| Z. mobilis | 多种方法的中心代谢模型 | 中等规模 | [ |
| Z. mobilis ATCC31821 (ZM4) | ZmoMBEL601代谢网络模型 | 基因组尺度:348个基因、601个反应和579个代谢物 | [ |
| Z. mobilis ATCC31821 (ZM4) | iZM363代谢网络模型 | 基因组尺度:363个基因、747个反应和704个代谢物 | [ |
| Z. mobilis ATCC10988 (ZM1) | iEM439代谢网络模型 | 基因组尺度:439个基因、692个反应和658个代谢物 | [ |
| Z. mobilis ATCC31821 (ZM4) | iHN446代谢网络模型 | 基因组尺度:446个基因、859个反应和894个代谢物 | [ |
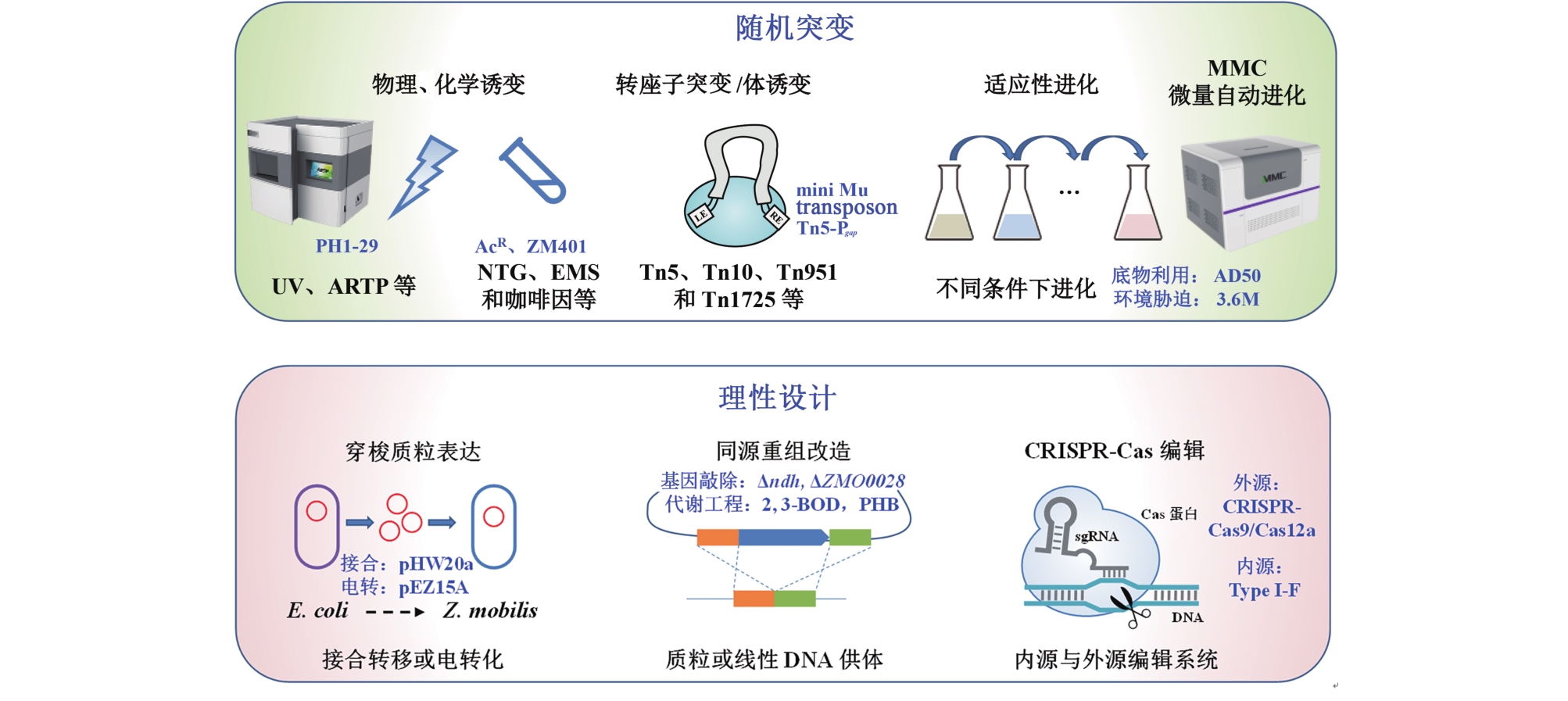
图3 运动发酵单胞菌中使用的遗传改造方法及代表性结果注:ARTP—atmospherie and room temperature plasma; NTG—nitrosoguanidine; EMS—ethyl methane sulfonate.
Fig. 3 Genetic engineering methods developed in Z. mobilis and representative examples(Random mutagenesis methods are shown at the top, and rational design methods are at the bottom)
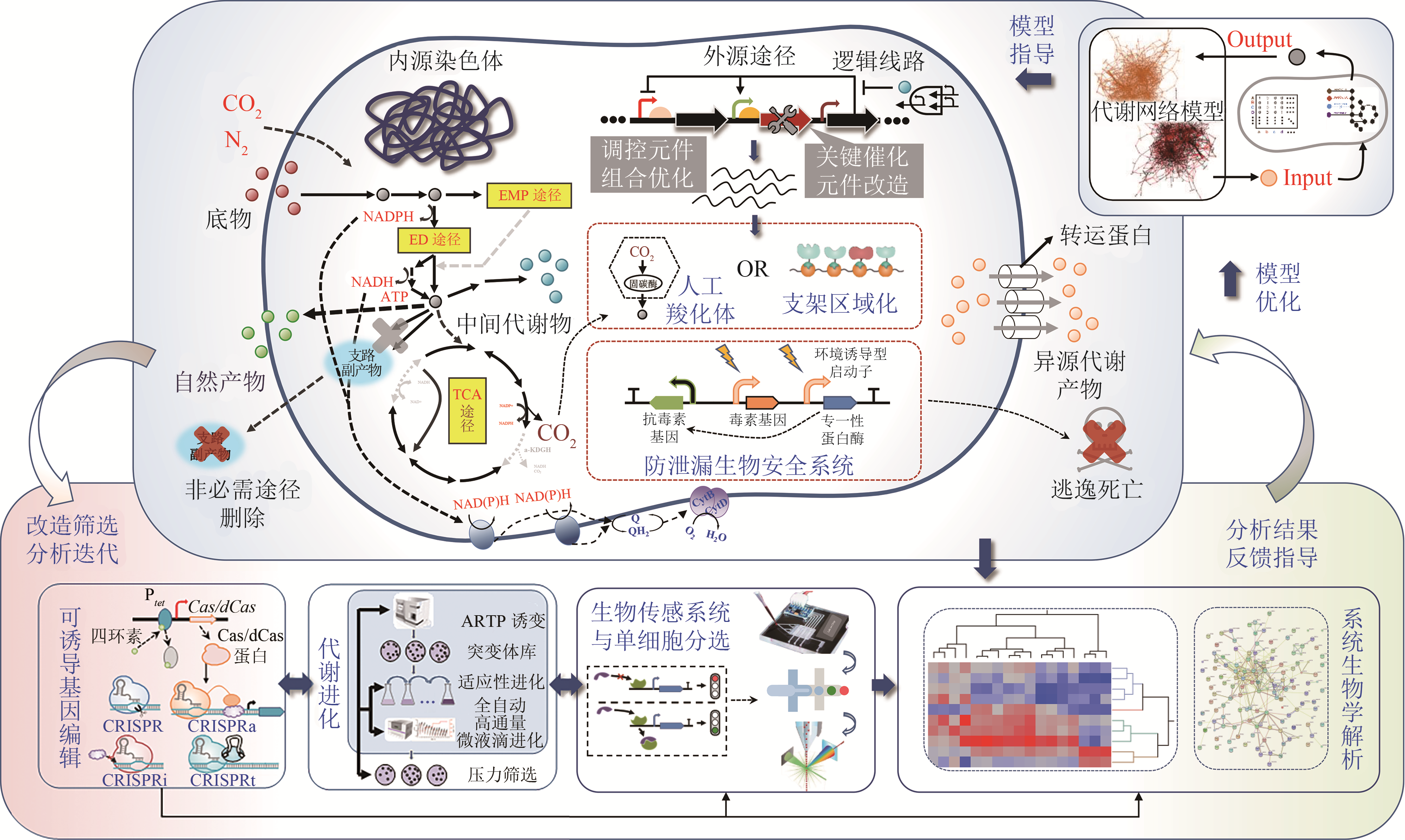
图4 运动发酵单胞菌底盘细胞构建展望(For developing Z.mobilis as an attractive microbial chasis to be able to fix CO2 and N2 for biochemical production, more effort is needed on reconstructing high-quality metabolic models, deploying efficient genome editing tools and strategies for fine-tuning metabolic and regulatory pathways spatiotemporally, as well as establishing the efficient iterative cycle of the design-build-test-learn)
Fig. 4 Perspectives on developing Z. mobilis as a chassis cell
| 1 | CAMPBELL K, XIA Jianye, NIELSEN J. The impact of systems biology on bioprocessing [J]. Trends in Biotechnology, 2017, 35(12): 1156-1168. |
| 2 | 赵国屏. 合成生物学: 开启生命科学"会聚"研究新时代[J]. 中国科学院院刊, 2018, 33(11): 1135-1149. |
| ZHAO Guoping. Synthetic biology: unsealing the convergence era of life science research [J]. Bulletin of the Chinese Academy of Sciences, 2018, 33(11): 1135-1149. | |
| 3 | 张先恩. 中国合成生物学发展回顾与展望[J]. 中国科学: 生命科学, 2019, 49(12): 1543-1572. |
| ZHANG Xian'en. Synthetic biology in China: review and prospects [J]. Science China Life Sciences, 2019, 49(12): 1543-1572. | |
| 4 | XIA Juan, YANG Yongfu, LIU Chenguang, et al. Engineering Zymomonas mobilis for robust cellulosic ethanol production [J]. Trends in Biotechnology, 2019, 37(9): 960-972. |
| 5 | WANG Xia, HE Qiaoning, YANG Yongfu, et al. Advances and prospects in metabolic engineering of Zymomonas mobilis [J]. Metabolic Engineering, 2018, 50: 57-73. |
| 6 | SWINGS J, DE LEY J. The biology of Zymomonas [J]. Bacteriological Reviews, 1977, 41(1): 1-46. |
| 7 | ROGERS P L, LEE K J, SKOTNICKI M L, et al. Ethanol production by Zymomonas mobilis [J]. Advances in Biochemical Engineering, 1982, 23: 37-84. |
| 8 | 蔺玉萍, 张木清, 陈柏铨. 产乙醇运动发酵单胞菌的研究进展[J]. 微生物学报, 2005, 45(3): 472-477. |
| LIN Yuping, ZHANG Muqing, CHEN Baiquan. Research progress of ethanologenic Zymomonas mobilis [J]. Acta Microbiologica Sinica, 2005, 45(3): 472-477. | |
| 9 | HE Mingxiong, WU Bo, QIN Han, et al. Zymomonas mobilis: a novel platform for future biorefineries [J]. Biotechnology for Biofuels, 2014, 7(101): 101. |
| 10 | YANG Shihui, FEI Qiang, ZHANG Yaoping, et al. Zymomonas mobilis as a model system for production of biofuels and biochemicals [J]. Microbial Biotechnology, 2016, 9(6): 699-717. |
| 11 | BOCHNER B, GOMEZ V, ZIMAN M, et al. Phenotype microarray profiling of Zymomonas mobilis ZM4 [J]. Applied Biochemistry and Biotechnology, 2010, 161: 116-123. |
| 12 | KREMER T A, LASARRE B, POSTO A L, et al. N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis [J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(7): 2222-2226. |
| 13 | PALAMAE S, CHOORIT W, CHATSUNGNOEN T, et al. Simultaneous nitrogen fixation and ethanol production by Zymomonas mobilis [J]. Journal of Biotechnology, 2020, ( 314/315): 41-52. |
| 14 | BRINGER S, HARTNER T, PORALLA K, et al. Influence of ethanol on the hopanoid content and the fatty acid pattern in batch and continuous cultures of Zymomonas mobilis [J]. Archives of Microbiology, 1985, 140: 312-316. |
| 15 | MOREAU R A, POWELL M J, FETT W F, et al. The effect of ethanol and oxygen on the growth of Zymomonas mobilis and the levels of hopanoids and other membrane lipids [J]. Current Microbiology, 1997, 35(2): 124-128. |
| 16 | VEGA M I, CUENCA M R, RODRIGUEZ E, et al. Identification of heptadecanoic and C19 cyclopropane fatty acids in the lipid fraction of Zymomonas mobilis [J]. Current Microbiology, 2000, 41(4): 305-306. |
| 17 | BRENAC L, BAIDOO E E K, KEASLING J D, et al. Distinct functional roles for hopanoid composition in the chemical tolerance of Zymomonas mobilis [J]. Molecular Microbiology, 2019, 112(5): 1564-1575. |
| 18 | YANG Qing, YANG Yongfu, TANG Ying, et al. Development and characterization of acidic-pH tolerant mutants of Zymomonas mobilis through adaptation and next generation sequencing based genome resequencing and RNA-Seq [J]. Biotechnology for Biofuels, 2020 13: 144. |
| 19 | AN Haejung, SCOPES R K, RODRIGUEZ M, et al. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein [J]. Journal of Bacteriology, 1991, 173(19): 5975-5982. |
| 20 | ALGAR E M, SCOPES R K. Studies on cell-free metabolism: ethanol production by extracts of Zymomonas mobilis [J]. Journal of Biotechnology, 1985, 2(5): 275-287. |
| 21 | YANG Yongfu, SHEN Wei, HUANG Ju, et al. Prediction and characterization of promoters and ribosomal binding sites of Zymomonas mobilis in system biology era [J]. Biotechnology for Biofuels, 2019, 12: 52. |
| 22 | KALNENIEKS U, GALININA N, TOMA M M, et al. Respiratory behaviour of a Zymomonas mobilis adhB::kanr mutant supports the hypothesis of two alcohol dehydrogenase isoenzymes catalysing opposite reactions [J]. FEBS Letters, 2006, 580(21): 5084-5088. |
| 23 | KALNENIEKS U, BALODITE E, RUTKIS R. Metabolic engineering of bacterial respiration: high vs. low P/O and the case of Zymomonas mobilis [J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 327. |
| 24 | KALNENIEKS U, GALININA N, STRAZDINA I, et al. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis [J]. Microbiology, 2008, 154(3): 989-994. |
| 25 | MARTIEN J I, HEBERT A S, STEVENSON D M, et al. Systems-level analysis of oxygen exposure in Zymomonas mobilis: implications for isoprenoid production [J]. mSystems, 2019, 4(1): 1-23. |
| 26 | KALNENIEKS U, DE GRAAF A A, BRINGER-MEYER S, et al. Oxidative phosphorylation in Zymomonas mobilis [J]. Archives of Microbiology, 1993, 160(1): 74-79. |
| 27 | CHEN R Ruizhen, AGRAWAL M, MAO Zichao. Impact of expression of EMP enzymes on glucose metabolism in Zymomonas mobilis [J]. Applied Biochemistry and Biotechnology, 2013, 170(4): 805-18.8 |
| 28 | FELCZAK M M, JACOBSON T B, ONG W K, et al. Expression of phosphofructokinase is not sufficient to enable Embden-Meyerhof-Parnas glycolysis in Zymomonas mobilis ZM4 [J]. Frontiers in Microbiology, 2019, 10: 2270. |
| 29 | ZHANG Min, EDDY C, DEANDA K, et al. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis [J]. Science, 1995, 267(5195): 240-243. |
| 30 | DEANDA K, ZHANG Min, EDDY C, et al. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering [J]. Applied and Environmental Microbiology, 1996, 62(12): 4465-4470. |
| 31 | SARKAR P, MUKHERJEE M, GOSWAMI G, et al. Adaptive laboratory evolution induced novel mutations in Zymomonas mobilis ATCC ZW658: a potential platform for co-utilization of glucose and xylose [J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(3): 329-341. |
| 32 | SCHELL D J, DOWE N, CHAPEAUX A, et al. Accounting for all sugars produced during integrated production of ethanol from lignocellulosic biomass [J]. Bioresource Technology, 2016, 205: 153-158. |
| 33 | HE Mingxiong, FENG Hong, BAI Fan, et al. Direct production of ethanol from raw sweet potato starch using genetically engineered Zymomonas mobilis [J]. African Journal of Microbiology Research, 2009, 3(11): 721-726. |
| 34 | SAHARKHIZ S, MAZAHERI D, SHOJAOSADATI S A. Evaluation of bioethanol production from carob pods by Zymomonas mobilis and Saccharomyces cerevisiae in solid submerged fermentation [J]. Preparative Biochemistry & Biotechnology, 2013, 43(5): 415-430. |
| 35 | NDABA B, CHIYANZU I, MARX S, et al. Effect of Saccharomyces cerevisiae and Zymomonas mobilis on the co-fermentation of sweet sorghum bagasse hydrolysates pretreated under varying conditions [J]. Biomass & Bioenergy, 2014, 71: 350-356. |
| 36 | SAVVIDES A L, KALLIMANIS A, VARSAKI A, et al. Simultaneous ethanol and bacterial ice nuclei production from sugar beet molasses by a Zymomonas mobilis CP4 mutant expressing the inaZ gene of Pseudomonas syringae in continuous culture [J]. Journal of Applied Microbiology, 2000, 89(6): 1002-1008. |
| 37 | SERATE J, XIE Dan, POHLMANN E, et al. Controlling microbial contamination during hydrolysis of AFEX-pretreated corn stover and switchgrass: effects on hydrolysate composition, microbial response and fermentation [J]. Biotechnology for Biofuels, 2015, 8: 180. |
| 38 | LUJAN-RHENALS D E, MORAWICKI R O, GBUR E E, et al. Fermentation of soybean meal hydrolyzates with Saccharomyces cerevisiae and Zymomonas mobilis for ethanol production [J]. Journal of Food Science, 2015, 80(7): E1512-E1518. |
| 39 | MA Kedong, RUAN Zhiyong, SHUI Zongxia, et al. Open fermentative production of fuel ethanol from food waste by an acid-tolerant mutant strain of Zymomonas mobilis [J]. Bioresource Technology, 2016, 203: 295-302. |
| 40 | GU Hanqi, ZHANG Jian, BAO Jie. High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue [J]. Biotechnology and Bioengineering, 2015, 112(9): 1770-1782. |
| 41 | TODHANAKASEM T, NARKMIT T, AREERAT K, et al. Fermentation of rice bran hydrolysate to ethanol using Zymomonas mobilis biofilm immobilization on DEAE-cellulose [J]. Electronic Journal of Biotechnology, 2015, 18(3): 196-201. |
| 42 | PERALTA-CONTRERAS M, AGUILAR-ZAMARRIPA E, PEREZ-CARRILLO E, et al. Ethanol production from extruded thermoplastic maize meal by high gravity fermentation with Zymomonas mobilis [J]. Biotechnology Research International, 2014, 2014: 654853. |
| 43 | BEHERA S, MOHANTY R C, RAY R C. Ethanol fermentation of sugarcane molasses by Zymomonas mobilis MTCC 92 immobilized in Luffa cylindrica L. sponge discs and Ca-alginate matrices [J]. Brazilian Journal of Microbiology, 2012, 43(4): 1499-1507. |
| 44 | ZHANG Jiayi, LYND L R. Ethanol production from paper sludge by simultaneous saccharification and co-fermentation using recombinant xylose-fermenting microorganisms [J]. Biotechnology and Bioengineering, 2010, 107(2): 235-244. |
| 45 | HE Mingxiong, LI Qing, LIU Xinying, et al. Bio-ethanol production from bamboo residues with lignocellulose fractionation technology (LFT) and separate hydrolysis fermentation (SHF) by Zymomonas mobilis [J]. Biomass & Bioenergy, 2013, 2(1): 15-24. |
| 46 | SULFAHRI, AMIN M, SUMITRO S B, et al. Bioethanol production from algae Spirogyra hyalina using Zymomonas mobilis [J]. Biofuels, 2016, 7(6): 621-626. |
| 47 | SOOTSUWAN K, THANONKEO P, KEERATIRAKHA N, et al. Sorbitol required for cell growth and ethanol production by Zymomonas mobilis under heat, ethanol, and osmotic stresses [J]. Biotechnology for Biofuels, 2013, 6(1): 180. |
| 48 | ZHANG Kun, SHAO Huanhuan, CAO Qinghua, et al. Transcriptional analysis of adaptation to high glucose concentrations in Zymomonas mobilis [J]. Applied Microbiology and Biotechnology, 2015, 99(4): 2009-2022. |
| 49 | ZHANG Linghua, LANG Yajun, WANG Chenxiang, et al. Promoting effect of compatible solute ectoine on the ethanol fermentation by Zymomonas mobilis CICC10232 [J]. Process Biochemistry, 2008, 43(6): 642-646. |
| 50 | LOOS H, KRAMER R, SAHM H, et al. Sorbitol promotes growth of Zymomonas mobilis in environments with high concentrations of sugar: evidence for a physiological function of glucose-fructose oxidoreductase in osmoprotection [J]. Journal of Bacteriology, 1994, 176(24): 7688-7693. |
| 51 | WU Bo, QIN Han, YANG Yiwei, et al. Engineered Zymomonas mobilis tolerant to acetic acid and low pH via multiplex atmospheric and room temperature plasma mutagenesis [J]. Biotechnology for Biofuels, 2019, 12: 10. |
| 52 | 李凯, 夏娟, 孜力汗, 等. 理化因素对运动发酵单胞菌絮凝的影响及絮凝机理 [J]. 中国科技论文, 2015, 10(24): 2880-2883. |
| LI Kai, XIA Juan, ZI Lihan, et al. Effect of physical and chemical factors on flocculation of Zymomonas mobilis and flocculation mechanism [J]. China Sciencepaper, 2015, 10(24): 2880-2883. | |
| 53 | JEON Young Jae, XUN Zhao, SU Ping, et al. Genome-wide transcriptomic analysis of a flocculent strain of Zymomonas mobilis [J]. Applied Microbiology and Biotechnology, 2012, 93(6): 2513-2518. |
| 54 | XIA Juan, LIU Chenguang, ZHAO Xinqing, et al. Contribution of cellulose synthesis, formation of fibrils and their entanglement to the self-flocculation of Zymomonas mobilis [J]. Biotechnology and Bioengineering, 2018, 115(11): 2714-2725. |
| 55 | 闻远, 夏娟, 戚良华, 等. 抗氧化基因过表达提高运动发酵单胞菌糠醛耐受性 [J]. 生物技术通报, 2019, 35(8): 85-94. |
| WEN Yuan, XIA Juan, QI Lianghua, et al. Enhanced furfural tolerance in Zymomonas mobilis by the overexpression of antioxidant genes [J]. Biotechnology Bulletin, 2019, 35(8): 85-94. | |
| 56 | GAO Xiaochuang, GAO Qiuqiang, BAO Jie. Improving cellulosic ethanol fermentability of Zymomonas mobilis by overexpression of sodium ion tolerance gene ZMO0119 [J]. Journal of Biotechnology, 2018, 282: 32-37. |
| 57 | YANG Shihui, LAND M L, KLINGEMAN D M, et al. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(23): 10395-10400. |
| 58 | WANG Weiting, WU Bo, QIN Han, et al. Genome shuffling enhances stress tolerance of Zymomonas mobilis to two inhibitors [J]. Biotechnology for Biofuels, 2019, 12(1): 288. |
| 59 | YANG Shihui, LINGER J, FRANDEN M A, et al. Biocatalysts with enhanced inhibitor tolerance: US9206445B2 [P]. 2015-12-08. |
| 60 | YANG Yongfu, HU Mimi, TANG Ying, et al. Progress and perspective on lignocellulosic hydrolysate inhibitor tolerance improvement in Zymomonas mobilis [J]. Bioresources and Bioprocessing, 2018, 5(1): 6. |
| 61 | CHEN Xiaowen, KUHN E, JENNINGS E W, et al. DMR (deacetylation and mechanical refining) processing of corn stover achieves high monomeric sugar concentrations (230 g·L-1) during enzymatic hydrolysis and high ethanol concentrations (>10% v/v) during fermentation without hydrolysate purification or concentration [J]. Energy & Environmental Science, 2016, 9(4): 1237-1245. |
| 62 | GENG Boyu, CAO Lianying, LI Feng, et al. Potential of Zymomonas mobilis as an electricity producer in ethanol production [J]. Biotechnology for Biofuels, 2019, 13: 36. |
| 63 | QIU Mengyue, SHEN W W, YAN Xiongyin, et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production [J]. Biotechnology for Biofuels, 2020, 13: 15. |
| 64 | CARRA S, RODRIGUES D C, BERALDO N M C, et al. High lactobionic acid production by immobilized Zymomonas mobilis cells: a great step for large-scale process [J]. Bioprocess and Biosystems Engineering, 2020, 43(7): 1265-1276. |
| 65 | ZHANG Min, CHOU Yat-Chen, FRANDEN Mary Ann, et al. Engineered Zymomonas for the production of 2,3 -butanediol: US 20190153483 A1 [P]. 2018-10-29. |
| 66 | KALNENIEKS U, BALODITE E, STRAHLER S, et al. Improvement of acetaldehyde production in Zymomonas mobilis by engineering of its aerobic metabolism [J]. Frontiers in Microbiology, 2019, 10: 2533. |
| 67 | HE Yan, WU Bo, XIA Wei, et al. Metabolic engineering of Zymomonas mobilis for ethylene production from straw hydrolysate[J].Applied Microbiology and Biotechnology, 2021.DOI:10.1007/s00253-021-11091-7 . |
| 68 | FOLLE A B, BASCHERA V M, VIVAN L T, et al. Assessment of different systems for the production of aldonic acids and sorbitol by calcium alginate-immobilized Zymomonas mobilis cells [J]. Bioprocess and Biosystems Engineering, 2018, 41(2): 185-194. |
| 69 | WANG Haoyong, CAO Shangzhi, WANG W Tianshuo, et al. Very high gravity ethanol and fatty acid production of Zymomonas mobilis without amino acid and vitamin [J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(6): 861-871. |
| 70 | KIM Jae-young, SHIN Sang-heum, CHONG Hyon-yong, et al. Transformant for production of lactic acid of high optical purity and method for producing lactic acid using the same: US9428775B2 [P]. 2016-08-30. |
| 71 | 税宗霞, 王景丽, 秦晗, 等. 产丁二酸运动发酵单胞菌的构建和发酵性能[J]. 应用与环境生物学报, 2015, 21(4): 657-664. |
| SHUI Zongxia, WANG Jingli, QIN Han, et al. Construction and preliminary fermentation of succinate-producing recombinant ethanologenic Zymomonas mobilis [J]. Chinese Journal of Applied & Environmental Biology, 2015, 21(4): 657-664. | |
| 72 | SILBIR S, DAGBAGLI S, YEGIN S, et al. Levan production by Zymomonas mobilis in batch and continuous fermentation systems [J]. Carbohydrate Polymers, 2014, 99: 454-461. |
| 73 | 姜坤妤, 苏喆, 王勇, 等. 产L-乳酸的运动发酵单胞菌代谢工程菌株的构建[J]. 生物技术通报, 2011(6): 170-174. |
| JIANG Kunyu, SU Zhe, WANG Yong, et al. Construction of metabolically-engineered Zymomonas mobilis strain for L-lactic acid production [J]. Biotechnology Bulletin, 2011(6): 170-174. | |
| 74 | 赖伟坚, 陈国强. 运动发酵单胞菌积累聚羟基丁酸提高乙醇产量[J]. 中国生物工程杂志, 2006, 26(8): 52-56. |
| LAI Weijian, CHEN Guoqiang. Polyhydroxybutyrate synthesis in recombinant Zymonomas mobilis affected ethanol production [J]. China Biotechnology, 2006, 26(8): 52-56. | |
| 75 | ERZINGER G S, VITOLO M. Zymomonas mobilis as catalyst for the biotechnological production of sorbitol and gluconic acid [J]. Applied Biochemistry and Biotechnology, 2006, 131(1/2/3): 787-794. |
| 76 | YANG Shihui, MOHAGHEGHI A, FRANDEN M A, et al. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars [J]. Biotechnology for Biofuels, 2016, 9(1): 189. |
| 77 | BRUNNER B, SCHEURER U, SEIBOLD F. Differences in yeast intolerance between patients with Crohn's disease and ulcerative colitis [J]. Diseases of the Colon & Rectum, 2007, 50(1): 83-88. |
| 78 | MUSATTI A, CAPPA C, MAPELLI C, et al. Zymomonas mobilis in bread dough: characterization of dough leavening performance in presence of sucrose [J]. Foods, 2020, 9(1): 89. |
| 79 | YUJI O, KENZO T. Dough-leavening ability by Zymomonas mobilis and its application to breadmaking [J]. Journal of Food Science, 1994, 59(1): 171-174. |
| 80 | MUSATTI A, MAPELLI C, ROLLINI M, et al. Can Zymomonas mobilis substitute Saccharomyces cerevisiae in cereal dough leavening? [J]. Foods, 2018, 7(4): 61. |
| 81 | MUSATTI A, MAPELLI C, FOSCHINO R, et al. Unconventional bacterial association for dough leavening [J]. International Journal of Food Microbiology, 2016, 237: 28-34. |
| 82 | NISSEN L, ROLLINI M, PICOZZI C, et al. Yeast-free doughs by Zymomonas mobilis: evaluation of technological and fermentation performances by using a metabolomic approach [J]. Microorganisms, 2020, 8(6): 792. |
| 83 | ZHOU Zhao, CHEN Xin, SHENG Huakang, et al. Engineering probiotics as living diagnostics and therapeutics for improving human health [J]. Microbial Cell Factories, 2020, 19(1): 56. |
| 84 | CALAZANS G M T, LIMA R C, DE FRANCA F P, et al. Molecular weight and antitumour activity of Zymomonas mobilis levans [J]. International Journal of Biological Macromolecules, 2000, 27(4): 245-247. |
| 85 | LIU Chenxi, KOLIDA S, CHARALAMPOPOULOS D, et al. An evaluation of the prebiotic potential of microbial levans from Erwinia sp. 10119 [J]. Journal of Functional Foods, 2020, 64: 103668. |
| 86 | ERDAL Ö, KAPLAN-TÜRKÖZ B, TAŞTAN O, et al. Levansucrase production by Zymomonas mobilis: optimization of process parameters and fructooligosaccharide production [J]. Journal of Food Biochemistry, 2017, 41(3): e12361. |
| 87 | GIBSON G R, SCOTT K P, RASTALL R A, et al. Dietary prebiotics: current status and new definition [J]. Food Science & Technology Bulletin: Functional Foods, 2010, 7(1): 1-19. |
| 88 | TAŞTAN O, SOZGEN G, BAYSAL T, et al. Production of prebiotic 6-kestose using Zymomonas mobilis levansucrase in carob molasses and its effect on 5-HMF levels during storage [J]. Food Chemistry, 2019, 297: 124897. |
| 89 | MARSH A J, O'SULLIVAN O, HILL C, et al. Sequence-based analysis of the microbial composition of water kefir from multiple sources [J]. FEMS Microbiology Letters, 2013, 348(1): 79-85. |
| 90 | CAO Chenxia, HOU Qiangchuan, HUI Wenyan, et al. Assessment of the microbial diversity of Chinese Tianshan tibicos by single molecule, real-time sequencing technology [J]. Food Science and Biotechnology, 2019, 28(1): 139-145. |
| 91 | DE AZERÊDO G A, STAMFORD T L M, DE SOUZA E L, et al. In vivo assessment of possible probiotic properties of Zymomonas mobilis in a Wistar rat model [J]. Electronic Journal of Biotechnology, 2010, 13(2). DOI: 10.2225/vol13-issue2-fulltext-4 . |
| 92 | DE AGUIAR SILVA A T, CAVALCANTI I D L, DE LIMA FERNANDES M A, et al. Effect of Zymomonas mobilis probiotic on cholesterol and its lipoprotein fractions and the intestinal regulation [J]. Clinical Nutrition, 2020, 39(12): 3750-3755. |
| 93 | MICHEL B, FLORES M J, VIGUERA E, et al. Rescue of arrested replication forks by homologous recombination [J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(15): 8181-8188. |
| 94 | JALES S T, SOARES-SOBRINHO J, NUNES L C, et al. Formulation technology of a probiotic (Zymomonas mobilis) in gelatinous capsules [J]. Latin American Journal of Pharmacy, 2007, 26(4): 553-557. |
| 95 | LUO Xiaozhou, REITER M A, D'ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast [J]. Nature, 2019, 567(7746): 123-126. |
| 96 | WEIR P M. The ecology of Zymomonas: a review [J]. Folia Microbiologica, 2016, 61(5): 385-392. |
| 97 | SEO J S, CHONG H, PARK H S, et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4 [J]. Nature Biotechnology, 2005, 23(1): 63-68. |
| 98 | YANG Shihui, PAPPAS K M, HAUSER L J, et al. Improved genome annotation for Zymomonas mobilis [J]. Nature Biotechnology, 2009, 27(10): 893-894. |
| 99 | YANG Shihui, VERA J M, GRASS J, et al. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose-utilizing derivatives 8b and 2032 [J]. Biotechnology for Biofuels, 2018, 11: 125. |
| 100 | CHEN Chen, WU Linfeng, CAO Qinghua, et al. Genome comparison of different Zymomonas mobilis strains provides insights on conservation of the evolution [J]. PLoS One, 2018, 13(4): e0195994. |
| 101 | YANG Shihui, TSCHAPLINSKI T J, ENGLE N L, et al. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations [J]. BMC Genomics, 2009, 10: 34. |
| 102 | HE Mingxiong, WU Bo, SHUI Zongxia, et al. Transcriptome profiling of Zymomonas mobilis under ethanol stress [J]. Biotechnology for Biofuels, 2012, 5(1): 75. |
| 103 | YANG Shihui, PAN Chongle, TSCHAPLINSKI T J, et al. Systems biology analysis of Zymomonas mobilis ZM4 ethanol stress responses [J]. PLoS One, 2013, 8(7): e68886. |
| 104 | HE Mingxiong, WU Bo, SHUI Zongxia, et al. Transcriptome profiling of Zymomonas mobilis under furfural stress [J]. Applied Microbiology and Biotechnology, 2012, 95(1): 189-199. |
| 105 | YANG Shihui, FRANDEN M A, WANG Xia, et al. Transcriptomic profiles of Zymomonas mobilis 8b to furfural acute and long-term stress in both glucose and xylose conditions [J]. Frontiers in Microbiology, 2020, 11: 13. |
| 106 | YANG Shihui, FRANDEN M A, BROWN S D, et al. Insights into acetate toxicity in Zymomonas mobilis 8b using different substrates [J]. Biotechnology for Biofuels, 2014, 7(1): 140. |
| 107 | YANG Shihui, PAN Chongle, HURST G B, et al. Elucidation of Zymomonas mobilis physiology and stress responses by quantitative proteomics and transcriptomics [J]. Frontiers in Microbiology, 2014, 5: 246. |
| 108 | YI Xia, GU Hanqi, GAO Qiuqiang, et al. Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment [J]. Biotechnology for Biofuels, 2015, 8: 153. |
| 109 | ZHAO Ning, BAI Yun, ZHAO Xinqing, et al. Draft genome sequence of the flocculating Zymomonas mobilis strain ZM401 (ATCC 31822) [J]. Journal of Bacteriology, 2012, 194(24): 7008-7009. |
| 110 | Seung Hee CHO, LEI R, HENNINGER T D, et al. Discovery of ethanol-responsive small RNAs in Zymomonas mobilis [J]. Applied and Environmental Microbiology, 2014, 80(14): 4189-4198. |
| 111 | DEUTSCHBAUER A, PRICE M N, WETMORE K M, et al. Towards an informative mutant phenotype for every bacterial gene [J]. Journal of Bacteriology, 2014, 196(20): 3643-3655. |
| 112 | DUNN K L, RAO C V. High-throughput sequencing reveals adaptation-induced mutations in pentose-fermenting strains of Zymomonas mobilis [J]. Biotechnology and Bioengineering, 2015, 112(11): 2228-2240. |
| 113 | CHANG Dongdong, YU Zhisheng, ISLAM Z UL, et al. Proteomic and metabolomic analysis of the cellular biomarkers related to inhibitors tolerance in Zymomonas mobilis ZM4 [J]. Biotechnology for Biofuels, 2018, 11: 283. |
| 114 | JACOBSON T B, ADAMCZYK P A, STEVENSON D M, et al. 2H and 13C metabolic flux analysis elucidates in vivo thermodynamics of the ED pathway in Zymomonas mobilis [J]. Metabolic Engineering, 2019, 54: 301-316. |
| 115 | TATLI M, HEBERT A S, COON J J, et al. Genome wide phosphoproteome analysis of Zymomonas mobilis under anaerobic, aerobic, and N2-fixing conditions [J]. Frontiers in Microbiology, 2019, 10: 1986. |
| 116 | ZHANG Yaoping, VERA J M, XIE Dan, et al. Multiomic fermentation using chemically defined synthetic hydrolyzates revealed multiple effects of lignocellulose-derived inhibitors on cell physiology and xylose utilization in Zymomonas mobilis [J]. Frontiers in Microbiology, 2019, 10: 2596. |
| 117 | HAN Runhua, HANING K, GONZALEZ-RIVERA J C, et al. Multiple small RNAs interact to co-regulate ethanol tolerance in Zymomonas mobilis [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 155. |
| 118 | LI Runxia, JIN Mingjie, DU Jun, et al. Underlying mechanism of uncoupled cell growth and ethanol fermentation of Zymomonas mobilis using different nitrogen sources [J]. Biotechnology for Biofuels, 2020. DOI: 10.21203/rs.2.21812/v1 |
| 119 | FUCHINO K, KALNENIEKS U, RUTKIS R, et al. Metabolic profiling of glucose-fed metabolically active resting Zymomonas mobilis strains [J]. Metabolites, 2020, 10(3): 1-12. |
| 120 | STARK R, GRZELAK M, HADFIELD J. RNA sequencing: the teenage years [J]. Nature Reviews Genetics, 2019, 20(11): 631-656. |
| 121 | SKERKER J M, LEON D, PRICE M N, et al. Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates [J]. Molecular Systems Biology, 2013, 9: 674. |
| 122 | CHAROENSUK K, SAKURADA T, TOKIYAMA A, et al. Thermotolerant genes essential for survival at a critical high temperature in thermotolerant ethanologenic Zymomonas mobilis TISTR 548 [J]. Biotechnology for Biofuels, 2017, 10: 204. |
| 123 | GU Changdai, KIM Gi Bae, KIM Won Jun, et al. Current status and applications of genome-scale metabolic models [J]. Genome Biology, 2019, 20(1): 121. |
| 124 | MONK J M, LLOYD C J, BRUNK E, et al. iML1515, a knowledgebase that computes Escherichia coli traits [J]. Nature Biotechnology, 2017, 35(10): 904-908. |
| 125 | LU Hongzhong, LI Feiran, SANCHEZ B J, et al. A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism [J]. Nature Communications, 2019, 10(1): 3586. |
| 126 | MOTAMEDIAN E, SAEIDI M, SHOJAOSADATI S A. Reconstruction of a charge balanced genome-scale metabolic model to study the energy-uncoupled growth of Zymomonas mobilis ZM1 [J]. Molecular BioSystems, 2016, 12(4): 1241-1249. |
| 127 | WIDIASTUTI H, KIM Jae Young, SELVARASU S, et al. Genome-scale modeling and in silico analysis of ethanologenic bacteria Zymomonas mobilis [J]. Biotechnology and Bioengineering, 2011, 108(3): 655-665. |
| 128 | Kyung Yun LEE, PARK Jong Myoung, KIM Tae Yong, et al. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies [J]. Microbial Cell Factories, 2010, 9: 94. |
| 129 | NOURI H, FOULADIHA H, MOGHIMI H, et al. A reconciliation of genome-scale metabolic network model of Zymomonas mobilis ZM4 [J]. Scientific Reports, 2020, 10(1): 7782. |
| 130 | ALTINTAS M M, EDDY C K, ZHANG Min, et al. Kinetic modeling to optimize pentose fermentation in Zymomonas mobilis [J]. Biotechnology and Bioengineering, 2006, 94(2): 273-295. |
| 131 | TSANTILI I C, KARIM M N, KLAPA M I. Quantifying the metabolic capabilities of engineered Zymomonas mobilis using linear programming analysis [J]. Microbial Cell Factories, 2007, 6: 8. |
| 132 | PENTJUSS A, ODZINA I, KOSTROMINS A, et al. Biotechnological potential of respiring Zymomonas mobilis: a stoichiometric analysis of its central metabolism [J]. Journal of Biotechnology, 2013, 165(1): 1-10. |
| 133 | RUTKIS R, KALNENIEKS U, STALIDZANS E, et al. Kinetic modelling of the Zymomonas mobilis Entner-Doudoroff pathway: insights into control and functionality [J]. Microbiology, 2013, 159: 2674-2689. |
| 134 | KALNENIEKS U, PENTJUSS A, RUTKIS R, et al. Modeling of Zymomonas mobilis central metabolism for novel metabolic engineering strategies [J]. Frontiers in Microbiology, 2014, 5: 42. |
| 135 | PINTO J P, DIAS O, LOURENÇO A, et al. Data integration issues in the reconstruction of the genome-scale metabolic model of Zymomonas mobillis [C]// 2nd International Workshop on Practical Applications of Computational Biology and Bioinformatics (IWPACBB 2008). Berlin: Springer, 2009: 92-101. |
| 136 | WIDIASTUTI H, Na-Rae LEE, KARIMI I, et al. Genome-scale in silico analysis for enhanced production of succinic acid in Zymomonas mobilis [J]. Processes, 2018, 6(4): 30. |
| 137 | Jae Sung CHO, GU Changdai, HAN Tae Hee, et al. Reconstruction of context-specific genome-scale metabolic models using multiomics data to study metabolic rewiring [J]. Current Opinion in Systems Biology, 2019, 15: 1-11. |
| 138 | XU Nan, LIU Liming. Computational inference of the transcriptional regulatory network of Candida glabrata [J]. FEMS Yeast Research, 2019, 19(4): foz036. DOI: 10.1093/femsyr/foz036 . |
| 139 | 赵欣, 杨雪, 毛志涛, 等. 基于酶约束的代谢网络模型研究进展及其应用[J]. 生物工程学报, 2020, 36(2): 1-11. |
| ZHAO Xin, YANG Xue, MAO Zhitao, et al. Progress and application of metabolic network model based on enzyme constraints [J]. Chinese Journal of Biotechnolog, 2020, 36(2): 1-11. | |
| 140 | THIELE I, PALSSON B O. A protocol for generating a high-quality genome-scale metabolic reconstruction [J]. Nature Protocols, 2010, 5(1): 93-121. |
| 141 | XAVIER J C, PATIL K R, ROCHA I. Integration of biomass formulations of genome-scale metabolic models with experimental data reveals universally essential cofactors in Prokaryotes [J]. Metabolic Engineering, 2017, 39: 200-208. |
| 142 | PAPPAS K M, GALANI I, TYPAS M A. Transposon mutagenesis and strain construction in Zymomonas mobilis [J]. Journal of Applied Microbiology, 1997, 82(3): 379-388. |
| 143 | GALEROS M, PAPPAS K M, BELETSIOTIS E, et al. ISZm1068: an IS5-like insertion element from Zymomonas mobilis [J]. Archives of Microbiology, 2001, 175(5): 323-333. |
| 144 | JOACHIMSTHAL E, HAGGETT K D, JANG Jin-Ho, et al. A mutant of Zymomonas mobilis ZM4 capable of ethanol production from glucose in the presence of high acetate concentrations [J]. Biotechnology Letters, 1998, 20(2): 137-142. |
| 145 | LEE J H, SKOTNICKI M L, ROGERS P L. Kinetic studies on a flocculent strain of Zymomonas mobilis [J]. Biotechnology Letters, 1982, 4(9): 615-620. |
| 146 | 杨依伟, 赵彩芳, 吴波, 等. 利用 Golden Gate "One-POT" 技术组装运动发酵单胞菌转录单元[J]. 应用与环境生物学报, 2019, 25(1): 170-175. |
| YANG Yiwei, ZHAO Caifang, WU Bo, et al. "One-POT" assembly of Zymomonas mobilis transcription unit via Golden Gate [J]. Chinese Journal of Applied & Environmental Biology, 2019, 25(1): 170-175. | |
| 147 | STRAZDINA I, BALODITE E, LASA Z, et al. Aerobic catabolism and respiratory lactate bypass in Ndh-negative Zymomonas mobilis [J]. Metabolic Engineering Communications, 2018, 7: e00081. |
| 148 | SENTHILKUMAR V, RAMESHKUMAR N, BUSBY S J, et al. Disruption of the Zymomonas mobilis extracellular sucrase gene (sacC) improves levan production [J]. Journal of Applied Microbiology, 2004, 96(4): 671-676. |
| 149 | 吴波, 何明雄, 冯红, 等. 运动发酵单胞菌限制-修饰系统缺失突变株的构建及其性质分析[J]. 应用与环境生物学报, 2013, 19(2): 189-197. |
| WU Bo, HE Mingxiong, FENG Hong, et al. Construction and characterization of restriction-modification deficient mutants in Zymomonas mobilis ZM4 [J]. Chinese Journal of Applied & Environmental Biology, 2013, 19(2): 189-197. | |
| 150 | KERR A L, JEON Y J, SVENSON C J, et al. DNA restriction-modification systems in the ethanologen, Zymomonas mobilis ZM4 [J]. Applied Microbiology and Biotechnology, 2011, 89(3): 761-769. |
| 151 | ZOU Shaolan, HONG Jiefang, WANG Cui, et al. Construction of an unmarked Zymomonas mobilis mutant using a site-specific FLP recombinase [J]. Food Technology and Biotechnology, 2012, 50(4): 406-411. |
| 152 | LAL P B, WELLS F M, Yucai LÜ, et al. A markerless method for genome engineering in Zymomonas mobilis ZM4 [J]. Frontiers in Microbiology, 2019, 10: 2216. |
| 153 | CAO Qinghua, SHAO Huanhuan, QIU Hui, et al. Using the CRISPR/Cas9 system to eliminate native plasmids of Zymomonas mobilis ZM4 [J]. Bioscience, Biotechnology, and Biochemistry, 2017, 81(3): 453-459. |
| 154 | SHEN Wei, ZHANG Jun, GENG Binan, et al. Establishment and application of a CRISPR-Cas12a assisted genome-editing system in Zymomonas mobilis [J]. Microbial Cell Factories, 2019, 18(1): 162. |
| 155 | ZHENG Yanli, HAN Jiamei, WANG Baiyang, et al. Characterization and repurposing of the endogenous Type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering [J]. Nucleic Acids Research, 2019, 47(21): 11461-11475. |
| 156 | ZHENG Yanli, LI Jie, WANG Baiyang, et al. Endogenous type I CRISPR-Cas: from foreign DNA defense to prokaryotic engineering [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 62. |
| 157 | 卫玮, 李晓玲, 袁文常. 金黄色葡萄球菌基因敲除质粒的构建及应用[J]. 中国组织工程研究, 2015, 19(5): 799-804. |
| WEI Wei, LI Xiaoling, YUAN Wenchang. Construction and application of Staphylococcus aureus gene knockout plasmid [J]. Chinese Journal of Tissue Engineering Research, 2015, 19(5): 799-804. | |
| 158 | LIU Dingyu, HUANG Can, GUO Jiaxin, et al. Development and characterization of a CRISPR/Cas9n-based multiplex genome editing system for Bacillus subtilis [J]. Biotechnology for Biofuels, 2019, 12: 197. |
| 159 | YANG Yongfu, RONG Ziyue, SONG Haoyue, et al. Identification and characterization of ethanol-inducible promoters of Zymomonas mobilis based on omics data and dual reporter-gene system [J]. Biotechnology and Applied Biochemistry, 2020, 67(1): 158-165. |
| 160 | ZHENG Yang, MENG Fankang, ZHU Zihui, et al. A tight cold-inducible switch built by coupling thermosensitive transcriptional and proteolytic regulatory parts [J]. Nucleic Acids Research, 2019, 47(21): e137. |
| 161 | NORA L C, WESTMANN C A, GUAZZARONI M E, et al. Recent advances in plasmid-based tools for establishing novel microbial chassis [J]. Biotechnology Advances, 2019: 107433. |
| 162 | Lok Yan SO, CHEN Wenyang, LACAP-BUGLER D C, et al. pZMO7-Derived shuttle vectors for heterologous protein expression and proteomic applications in the ethanol-producing bacterium Zymomonas mobilis [J]. BMC Microbiology, 2014, 14: 68. |
| 163 | CAO Qinghua, LI Tao, SHAO Huanhuan, et al. Three new shuttle vectors for heterologous expression in Zymomonas mobilis [J]. Electronic Journal of Biotechnology, 2016, 19: 33-40. |
| 164 | YANG Shihui, PELLETIER D A, LU Tse-Yuan, et al. The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors [J]. BMC Microbiology, 2010, 10(1): 135. |
| 165 | 邹少兰, 张鲲, 井欣, 等. 运动发酵单胞菌ZM4、CP4菌株基因工程选择标记的研究 [J]. 工业微生物, 2012, 42(3): 72-77. |
| ZOU Shaolan, ZHANG Kun, JING Xin, et al. Studies on selectable marker for genetic engineering of Zymomonas mobilis ZM4 and CP4 strain [J]. Industrial Microbiology, 2012, 42(3): 72-77. | |
| 166 | GLIESSMAN J R, KREMER T A, SANGANI A A, et al. Pantothenate auxotrophy in Zymomonas mobilis ZM4 is due to a lack of aspartate decarboxylase activity [J]. FEMS Microbiology Letters, 2017, 364(13). DOI: 10.1093/femsle/fnx136 . |
| 167 | Dong-Wuk CHO, PETER R L, DELANEY S F. Construction of a shuttle vector for Zymomonas mobilis [J]. Applied Microbiology and Biotechnology, 1989, 32: 50-53. |
| 168 | BYUN O K, KAPER J B, INGRAM L O. Construction of a new vector for the expression of foreign genes in Zymomonas mobilis [J]. Journal of Industrial Microbiology, 1986, 1(1): 9-15. |
| 169 | BUCHHOLZ S E, EVELEIGH D E. Genetic modification of Zymomonas mobilis [J]. Biotechnology Advances, 1990, 8(3): 547-581. |
| 170 | JEON Y J, SVENSON C J, ROGERS P L. Over-expression of xylulokinase in a xylose-metabolising recombinant strain of Zymomonas mobilis [J]. FEMS Microbiology Letters, 2005, 244(1): 85-92. |
| 171 | TATE W P, MANSELL J B, MANNERING S A, et al. UGA: a dual signal for 'stop' and for recoding in protein synthesis [J]. Biochemistry (Moscow), 1999, 64(12): 1342-1353. |
| 172 | CAREY V C, WALIA S K, INGRAM L O. Expression of a lactose transposon (Tn951) in Zymomonas mobilis [J]. Applied and Environmental Microbiology, 1983, 46(5): 1163-1168. |
| 173 | BAUMLER D J, HUNG K F, BOSE J L, et al. Enhancement of acid tolerance in Zymomonas mobilis by a proton-buffering peptide [J]. Applied Biochemistry and Biotechnology, 2006, 134(1): 15-26. |
| 174 | DENG Mingde, COLEMAN J R. Ethanol synthesis by genetic engineering in cyanobacteria [J]. Applied and Environmental Microbiology, 1999, 65(2): 523-528. |
| 175 | SMITH C J, ROLLINS L A, PARKER A C. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143 [J]. Plasmid, 1995, 34(3): 211-222. |
| 176 | DELGADO O D, ABATE C M, SINERIZ F. Construction of an integrative shuttle vector for Zymomonas mobilis [J]. FEMS Microbiology Letters, 1995, 132(1/2): 23-26. |
| 177 | OKAMOTO T, YAMANO S, IKEAGA H, et al. Cloning of the Acetobacter xylinum cellulase gene and its expression in Escherichia coli and Zymomonas mobilis [J]. Applied Microbiology and Biotechnology, 1994, 42(4): 563-568. |
| 178 | DANILEVICH V N, DUZHII D E, BRAGA E A. Design of recombinant plasmids for effective Zymomonas mobilis pyruvate decarboxylase (pdk) gene expression in Bacillus subtilis cells [J]. Molecular Biology, 1994, 28(1): 158-166. |
| 179 | KANAGASUNDARAM V, SCOPES R K. Cloning, sequence analysis, and expression of the structural gene encoding glucose-fructose oxidoreductase from Zymomonas mobilis [J]. Journal of Bacteriology, 1992, 174(5): 1439-1447. |
| 180 | SPRENGER G A, TYPAS M A, DRAINAS C. Genetics and genetic engineering of Zymomonas mobilis [J]. World Journal of Microbiology and Biotechnology, 1993, 9(1): 17-24. |
| 181 | SKOTNICKI M L, TRIBE D E, ROGERS P L. R-plasmid transfer in Zymomonas mobilis [J]. Applied and Environmental Microbiology, 1980, 40(1): 7-12. |
| 182 | DONG Hongwei, BAO Jie, RYU D D, et al. Design and construction of improved new vectors for Zymomonas mobilis recombinants [J]. Biotechnology and Bioengineering, 2011, 108(7): 1616-1627. |
| 183 | OKAMOTO T, NAKAMURA K. Simple and highly efficient transformation method for Zymomonas mobilis: electroporation [J]. Bioscience, Biotechnology, and Biochemistry, 1992, 56(5): 833. |
| 184 | JEON Young Jae, SVENSON C J, JOACHIMSTHAL E L, et al. Kinetic analysis of ethanol production by an acetate-resistant strain of recombinant Zymomonas mobilis [J]. Biotechnology Letters, 2002, 24: 819-824. |
| 185 | TU Qiang, YIN Jia, FU Jun, et al. Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency [J]. Scientific Reports, 2016, 6: 24648. |
| 186 | MORALES-RUIZ E, LOPEZ-CEBALLOS A, MALDONADO-MENDOZA I E. Transformation of the rhizospheric Bacillus cereus sensu lato B25 strain using a room-temperature electrocompetent cells preparation protocol [J]. Plasmid, 2019, 105: 102435. |
| 187 | DU Xinjun, WANG Fei, GE Yue, et al. Improvement of electrotransformation efficiency by lysozyme treatment in Cronobacter sakazakii [C]// Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012). Berlin: Springer, 2014. |
| 188 | ERSHOVA A S, RUSINOV I S, SPIRIN S A, et al. Role of restriction-modification systems in prokaryotic evolution and ecology [J]. Biochemistry (Moscow), 2015, 80(10): 1373-1386. |
| 189 | GEORG J, HESS W R. Regulatory RNAs in cyanobacteria: developmental decisions, stress responses and a plethora of chromosomally encoded cis-antisense RNAs [J]. Biological Chemistry, 2011, 392(4): 291-297. |
| 190 | HANING K, ENGELS S M, WILLIAMS P, et al. Applying a new REFINE approach in Zymomonas mobilis identifies novel sRNAs that confer improved stress tolerance phenotypes [J]. Frontiers in Microbiology, 2019, 10: 2987. |
| 191 | IVAIN L, BORDEAU V, EYRAUD A, et al. An in vivo reporter assay for sRNA-directed gene control in Gram-positive bacteria: identifying a novel sRNA target in Staphylococcus aureus [J]. Nucleic Acids Research, 2017, 45(8): 4994-5007. |
| 192 | Seung Hee CHO, HANING K, SHEN Wei, et al. Identification and characterization of 5' untranslated regions (5'UTRs) in Zymomonas mobilis as regulatory biological parts [J]. Frontiers in Microbiology, 2017, 8: 2432. |
| 193 | BARNELL W O, LIU J, HESMAN T L, et al. The Zymomonas mobilis glf, zwf, edd, and glk genes form an operon: localization of the promoter and identification of a conserved sequence in the regulatory region [J]. Journal of Bacteriology, 1992, 174(9): 2816-2823. |
| 194 | VERA J M, GHOSH I N, ZHANG Yaoping, et al. Genome-scale transcription-translation mapping reveals features of Zymomonas mobilis transcription units and promoters [J]. mSystems, 2020, 5(4): e00250-20. |
| 195 | 罗利. 植物固氮细胞器的合成生物学研究[J]. 生物技术通报, 2019, 35(10): 1-6. |
| LUO Li. Synthetic biology research of plant nitrogen-fixation organelle [J]. Biotechnology Bulletin, 2019, 35(10): 1-6. | |
| 196 | LE QUÉRÉ C, ANDREW R M, FRIEDLINGSTEIN P, et al. Global warming will happen faster than we think [J]. Nature, 2018, 10(4): 2141-2194. |
| 197 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263. |
| 198 | SUTTER M, LAUGHLIN T G, SLOAN N B, et al. Structure of a synthetic beta-carboxysome shell [J]. Plant Physiology, 2019, 181(3): 1050-1058. |
| 199 | CAI Fei, BERNSTEIN S L, WILSON S C, et al. Production and characterization of synthetic carboxysome shells with incorporated luminal proteins [J]. Plant Physiology, 2016, 170(3): 1868-1877. |
| 200 | RAE B D, LONG B M, BADGER M R, et al. Structural determinants of the outer shell of beta-carboxysomes in Synechococcus elongatus PCC 7942: roles for CcmK2, K3-K4, CcmO, and CcmL [J]. PLoS One, 2012, 7(8): e43871. |
| 201 | LIU Zihe, WANG Kai, CHEN Yun, et al. Third-generation biorefineries as the means to produce fuels and chemicals from CO2 [J]. Nature Catalysis, 2020, 3: 274-288. |
| 202 | GARST A D, BASSALO M C, PINES G, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering [J]. Nature Biotechnology, 2017, 35(1): 48-55. |
| 203 | CHEN Cong, CHOUDHURY A, ZHANG Shuanghong, et al. Integrating CRISPR-enabled trackable genome engineering and transcriptomic analysis of global regulators for antibiotic resistance selection and identification in Escherichia coli [J]. mSystems, 2020, 5(2): e00232-20. |
| 204 | WALTON R T, CHRISTIE K A, WHITTAKER M N, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants [J]. Science, 2020, 368(6488): 290-296. |
| 205 | STRECKER J, LADHA A, GARDNER Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases [J]. Science, 2019, 365(6448): 48-53. |
| 206 | KLOMPE S E, VO P L H, HALPIN-HEALY T S, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration [J]. Nature, 2019, 571(7764): 219-225. |
| 207 | LU Zhenghui, YANG Shihui, YUAN Xin, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 47(7): e40. |
| 208 | LEE H H, OSTROV N, WONG B G, et al. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi [J]. Nature Microbiology, 2019, 4(7): 1105-1113. |
| 209 | WANG Tianmin, GUAN Changge, GUO Jiahui, et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance [J]. Nature Communications, 2018, 9(1): 2475. |
| 210 | RISHI H S, TORO E, LIU Honglei, et al. Systematic genome-wide querying of coding and non-coding functional elements in E . coli using CRISPRi [EB/OL]. [2020-03-05]. . |
| 211 | FANG Lixia, FAN Jie, WANG Congya, et al. Genome-wide targets identification by CRISPRi-Omics for high-titer production of free fatty acids in Escherichia coli [EB/OL]. [2020-03-04]. . |
| 212 | FRANDEN M A, PIENKOS P T, ZHANG Min. Development of a high-throughput method to evaluate the impact of inhibitory compounds from lignocellulosic hydrolysates on the growth of Zymomonas mobilis [J]. Journal of Biotechnology, 2009, 144(4): 259-267. |
| 213 | JIAN Xingjin, GUO Xiaojie, WANG Jia, et al. Microbial microdroplet culture system (MMC): an integrated platform for automated, high-throughput microbial cultivation and adaptive evolution [J]. Biotechnology and Bioengineering, 2020, 117(6): 1724-1737. |
| 214 | LI Sijin, SI Tong, WANG Meng, et al. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening [J]. ACS Synthetic Biology, 2015, 4(12): 1308-1315. |
| 215 | QIU Xueliang, XU Peng, ZHAO Xinrui, et al. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica [J]. Metabolic Engineering, 2020, 60: 66-76. |
| 216 | TAN Yumeng, ZHANG Yong, HAN Yunbin, et al. Directed evolution of an α1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method [J]. Science Advances, 2019, 5(10): eaaw8451. |
| 217 | BJORK S M, JOENSSON H N. Microfluidics for cell factory and bioprocess development [J]. Current Opinion in Biotechnology, 2019, 55: 95-102. |
| 218 | MA Xiaoyan, HUO Yixin. The application of microfluidic-based technologies in the cycle of metabolic engineering [J]. Synthetic and Systems Biotechnology, 2016, 1(3): 137-142. |
| 219 | WANG Xixian, REN Lihui, SU Yetian, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells [J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 220 | WANG Xixian, XIN Yi, REN Lihui, et al. Positive dielectrophoresis based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo [J].Science Advances, 2020, 6(32): eabb3521. DOI: 10. 1126/sciadv. abb3521 . |
| 221 | XU Teng, GONG Yanhai, SU Xiaolu, et al. Phenome-genome profiling of single bacterial cell by raman-activated gravity-driven encapsulation and sequencing [J]. Small, 2020, 16(30): e2001172. |
| 222 | MOCCIARO A, ROTH T L, BENNETT H M, et al. Light-activated cell identification and sorting (LACIS) for selection of edited clones on a nanofluidic device [J]. Communications Biology, 2018, 1: 41. |
| 223 | TIAN Rongzhen, LIU Yanfeng, CHEN Junrong, et al. Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering, 2019(55): 131-141. |
| 224 | WU Zaiqiang, ZHAO Dongdong, LI Siwei, et al. Combinatorial modulation of initial codons for improved zeaxanthin synthetic pathway efficiency in Escherichia coli [J]. Microbiologyopen, 2019, 8(12): e930. |
| 225 | 王晨, 赵雨佳, 李春, 等. 动态转录调控微生物代谢途径研究进展[J]. 化工进展, 2019, 38(9): 4238-4246. |
| WANG Chen, ZHAO Yujia, LI Chun, et al. Advances in dynamic transcriptional regulation of microbial metabolic pathways [J]. Chemical Industry and Engineering Progress, 2019, 38(9): 4238-4246. | |
| 226 | XU Ning, WEI Liang, LIU Jun. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis [J]. World Journal of Microbiology and Biotechnology, 2019, 35(2): 33. |
| 227 | LIU Min, CAO Zhijun. Regulation of NADH oxidase expression via a thermo-regulated genetic switch for pyruvate production in Escherichia coli [J]. Biotechnology and Bioprocess Engineering, 2018, 23(1): 93-99. |
| 228 | XIONG Zhiqiang, WEI Yunying, KONG Linghui, et al. Short communication: an inducible CRISPR/dCas9 gene repression system in Lactococcus lactis [J]. Journal of Dairy Science, 2020, 103(1): 161-165. |
| 229 | DONG Xiaomin, LI Nan, LIU Zhenmin, et al. CRISPRi-guided multiplexed fine-tuning of metabolic flux for enhanced lacto-N-neotetraose production in Bacillus subtilis [J]. Journal of Agricultural and Food Chemistry, 2020, 68(8): 2477-2484. |
| 230 | MOROZ-OMORI E V, SATYAPERTIWI D, RAMEL M C, et al. Photoswitchable gRNAs for spatiotemporally controlled CRISPR-Cas-based genomic regulation [J]. ACS Central Science, 2020, 6(5): 695-703. |
| 231 | LIU Yang, ZOU R S, HE Shuaixin, et al. Very fast CRISPR on demand [J]. Science, 2020, 368(6496): 1265-1269. |
| 232 | KOBAYASHI S, KAWAGUCHI H, SHIRAI T, et al. Automatic redirection of carbon flux between glycolysis and pentose phosphate pathway using an oxygen-responsive metabolic switch in Corynebacterium glutamicum [J]. ACS Synthetic Biology, 2020, 9(4): 814-826. |
| 233 | SHEN Yuping, FONG Lai San, YAN Zhibo, et al. Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli [J]. Biotechnology for Biofuels, 2019, 12: 94. |
| 234 | LIU Yang, GHOSH I N, MARTIEN J, et al. Regulated redirection of central carbon flux enhances anaerobic production of bioproducts in Zymomonas mobilis [J]. Metabolic Engineering, 2020, 61: 261-274. DOI: 10.1016/j.ymben.2020.06.005 . |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 陈盈盈, 刘扬, 史俊杰, 马俊英, 鞠建华. CRISPR/Cas基因编辑及其新兴技术在丝状真菌研究中的系统应用[J]. 合成生物学, 2024, 5(3): 672-693. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
