合成生物学 ›› 2022, Vol. 3 ›› Issue (4): 626-637.DOI: 10.12211/2096-8280.2021-087
细菌生物被膜的软物质特性及其工程化应用
朱润涛, 钟超, 戴卓君
- 中国科学院深圳先进技术研究院合成生物学研究所,广东 深圳 518055
-
收稿日期:2021-08-27修回日期:2021-12-22出版日期:2022-08-31发布日期:2022-09-08 -
通讯作者:钟超,戴卓君 -
作者简介:朱润涛 (1993—),男,硕士。研究方向为基于工程细菌及工程菌群的活体材料。E-mail:rt.zhu@siat.ac.cn钟超 (1979—),男,博士,研究员。研究方向为利用合成生物学技术发展新材料,包括活体功能材料和蛋白水下黏合材料。E-mail:chao.zhong@siat.ac.cn戴卓君 (1987—),女,博士,副研究员。研究方向是编辑合成功能菌群并结合高分子化学与物理的手段实现生物制剂及活体材料的智能制造。E-mail:zj.dai@siat.ac.cn -
基金资助:国家重点研发计划(2018YFA0903000);国家自然科学基金(32071427);深圳市科技计划(KQTD20180413181837372)
Biofilm matrixes-from soft matters to engineered materials
ZHU Runtao, ZHONG Chao, DAI Zhuojun
- Institute of Synthetic Biology,Shenzhen Institude of Advanced Technology,Chinese Acdamey of Science,Shenzhen 518055,Guangdong,China
-
Received:2021-08-27Revised:2021-12-22Online:2022-08-31Published:2022-09-08 -
Contact:ZHONG Chao, DAI Zhuojun
摘要:
2014年,利用工程化生物被膜大肠杆菌淀粉样蛋白纤维(curli)组装活材料工作的发表,正式拉开了活体功能材料这一新兴领域的序幕。截至目前,围绕活体材料主题,针对curli系统的编辑、组装、性能拓展以及应用等各个方向的相关研究层出不穷,也有一系列的综述对相关工作进行了详细的梳理,并对活体功能材料以及材料合成生物学新方向及新学科进行归纳及定义。然而,当研究人员试着去构建整幅活体材料领域发生及发展的拼图时,其中有一些关键信息缺失。在众多的生物体系中,为什么活体材料新方向会从生物被膜开启?另外,在生物被膜的繁杂组分中,是如何剥离出curli核心系统,并成为整个活体功能材料工程化的中心?围绕着这些疑问,本篇综述从生物被膜的软物质特性以及curli生物起源及调控的研究开始挖掘。从高分子物理及合成生物学的观点解读工程化生物被膜从体系选择、去粗取精、工程化设计、系统构建以及性能推广及优化中,由繁至简,再由简至繁的全过程。作者希望借这篇综述回顾工程化生物被膜curli从发掘到发展的历程,并进一步思考相关领域背后发展及推动的知识积累、设计思维以及发展理念,并期待这些思考将对未来活体材料研究的新体系与新范式带来启示、借鉴以及推动。
中图分类号:
引用本文
朱润涛, 钟超, 戴卓君. 细菌生物被膜的软物质特性及其工程化应用[J]. 合成生物学, 2022, 3(4): 626-637.
ZHU Runtao, ZHONG Chao, DAI Zhuojun. Biofilm matrixes-from soft matters to engineered materials[J]. Synthetic Biology Journal, 2022, 3(4): 626-637.
| 材料 | 弹性 /GPa | 黏度 /mPa·s | 参考文献 |
|---|---|---|---|
| 钛 | 106~108 | [ | |
| 铝 | 68~70 | [ | |
| 透明质酸基地的组织工程化骨架 | 10-4 | 107 | [ |
| 皮肤 | 0.015~0.15 | [ | |
| 人皮质骨 | 15~30 | [ | |
| 牙釉质 | 80 | [ | |
| 毛发 | 7 | [ | |
| 水 | — | 1 | |
| 唾液 | — | 1.3~2.0 | [ |
| 血液 | 3~4 | [ | |
| 尿液 | 0.8 | [ | |
| Pseudomonas生物外膜(剪切模式) | 10-10 | [ | |
| Pseudomonas全生物膜(剪切模式) | 10-5 | [ | |
| Miscellaneous生物膜(剪切模式) | 10-10~10-4 | 103~1013 | [ |
| 环境与工业的生物膜(拉伸模式) | 10-8 | [ | |
| 口腔生物膜(压缩模式) | 10-8~10-7 | [ |
表1 不同生物被膜在室温条件下的弹性模量、黏度
Tab. 1 Viscoelasticity of different biological and synthetic materials at room temperature
| 材料 | 弹性 /GPa | 黏度 /mPa·s | 参考文献 |
|---|---|---|---|
| 钛 | 106~108 | [ | |
| 铝 | 68~70 | [ | |
| 透明质酸基地的组织工程化骨架 | 10-4 | 107 | [ |
| 皮肤 | 0.015~0.15 | [ | |
| 人皮质骨 | 15~30 | [ | |
| 牙釉质 | 80 | [ | |
| 毛发 | 7 | [ | |
| 水 | — | 1 | |
| 唾液 | — | 1.3~2.0 | [ |
| 血液 | 3~4 | [ | |
| 尿液 | 0.8 | [ | |
| Pseudomonas生物外膜(剪切模式) | 10-10 | [ | |
| Pseudomonas全生物膜(剪切模式) | 10-5 | [ | |
| Miscellaneous生物膜(剪切模式) | 10-10~10-4 | 103~1013 | [ |
| 环境与工业的生物膜(拉伸模式) | 10-8 | [ | |
| 口腔生物膜(压缩模式) | 10-8~10-7 | [ |
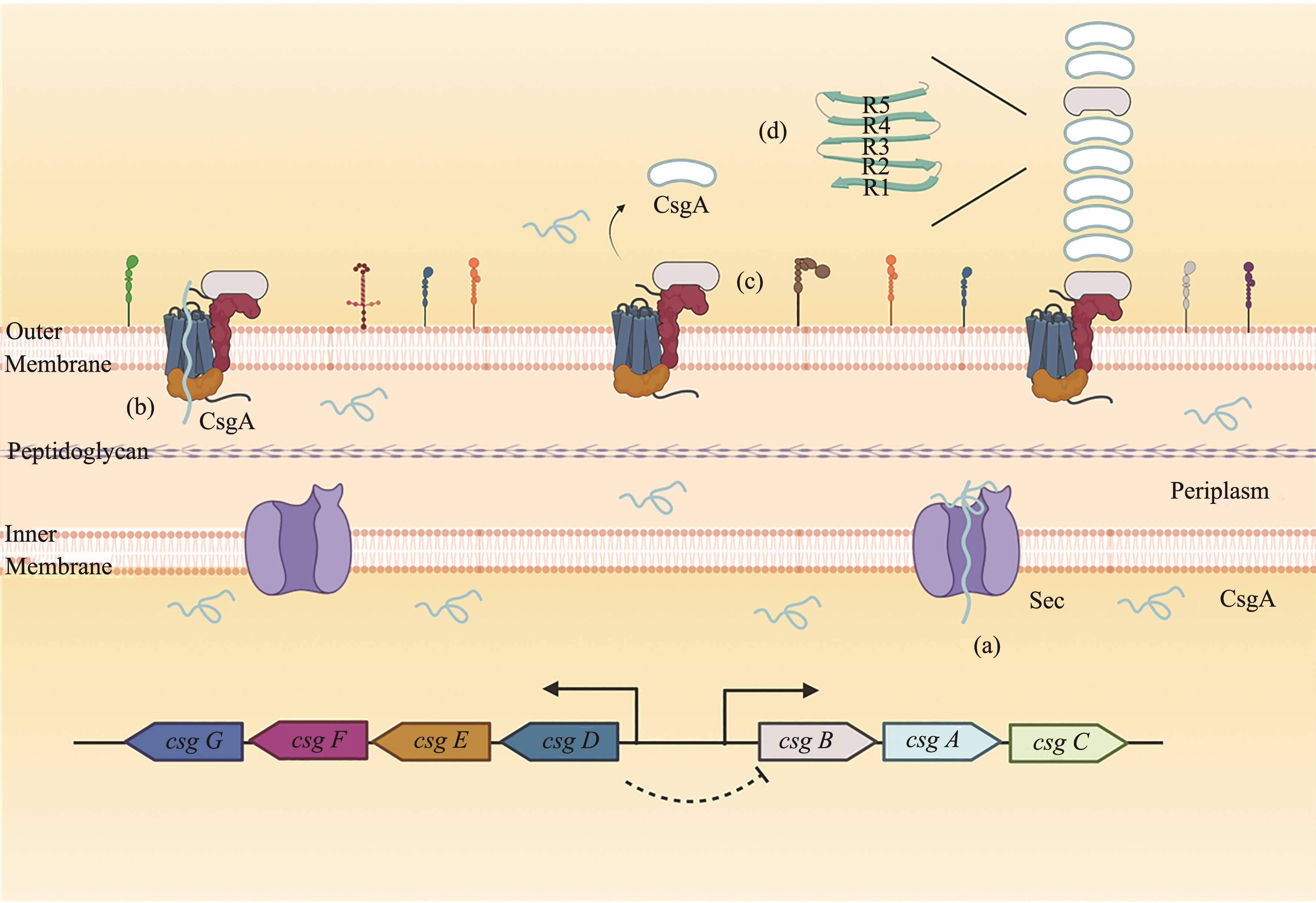
图1 curli形成的分子机制[Sec通路可将尚未折叠的CsgA转运到细胞周质,同样也可转运CsgB-C和CsgE-F跨越细菌内膜(a)[31]。当CsgA被封闭在CsgG-CsgE腔中时,由于墒增效应使得CsgA由密闭笼子扩散至外膜(b)[32]。CsgB可引发外膜表面CsgA单体成核和聚合形成curli系统(c)[33]。组成curli系统的成熟CsgA单体是典型的β折叠结构(d)[34]]
Fig. 1 The molecular mechanism for curli formation[An unfolded CsgA monomer enters the periplasm via the Sec translocon, and CsgB-C and CsgE-F are transported cross the inner membrane(a); A subunit CsgA encapsulated by a chamber of the CsgG: CsgE complex is secreted over outer membrane, which is driven by entropy increase(b); CsgB nucleated polymerization of a soluble subunit CsgA can assemble into a curli system(c); As the major subunit of the curli fiber, the mature CsgA protein is with a β-sheet-turn-β-sheet conformation (d)]
| 功能单位 | 类型 | 参考文献 |
|---|---|---|
| His Tag | 标签 | [ |
| 贻贝足蛋白 | 防水黏合剂 | [ |
| HA | 标签 | [ |
| Flag | 标签 | [ |
| 镧系元素结合标签(LBTs) | 金属结合多肽 | [ |
| A3 | 金属结合多肽 | [ |
| 流感病毒结合肽 | 结合病毒衣壳 | [ |
| 羟基磷灰石结合肽 | 矿化 | [ |
| DNA结合结构域 | 结合DNA | [ |
| 脂酶结合肽 | 结合脂酶 | [ |
| SpyTag | 结合SpyCatcher | [ |
| 金属结合域 | 结合不锈钢 | [ |
| 材料结合多肽 | 合成纳米材料 | [ |
| 几丁质结合域 | 结合几丁质 | [ |
| Mms | 结合磁颗粒 | [ |
| 4-叠氮基-L-苯丙氨酸 | 非天然氨基酸 | [ |
| 人肠三叶因子 | 治疗结肠炎 | [ |
表2 CsgA融合结构域单元实现curli功能修饰
Tab. 2 Domains-fused CsgA functionalizes curli
| 功能单位 | 类型 | 参考文献 |
|---|---|---|
| His Tag | 标签 | [ |
| 贻贝足蛋白 | 防水黏合剂 | [ |
| HA | 标签 | [ |
| Flag | 标签 | [ |
| 镧系元素结合标签(LBTs) | 金属结合多肽 | [ |
| A3 | 金属结合多肽 | [ |
| 流感病毒结合肽 | 结合病毒衣壳 | [ |
| 羟基磷灰石结合肽 | 矿化 | [ |
| DNA结合结构域 | 结合DNA | [ |
| 脂酶结合肽 | 结合脂酶 | [ |
| SpyTag | 结合SpyCatcher | [ |
| 金属结合域 | 结合不锈钢 | [ |
| 材料结合多肽 | 合成纳米材料 | [ |
| 几丁质结合域 | 结合几丁质 | [ |
| Mms | 结合磁颗粒 | [ |
| 4-叠氮基-L-苯丙氨酸 | 非天然氨基酸 | [ |
| 人肠三叶因子 | 治疗结肠炎 | [ |
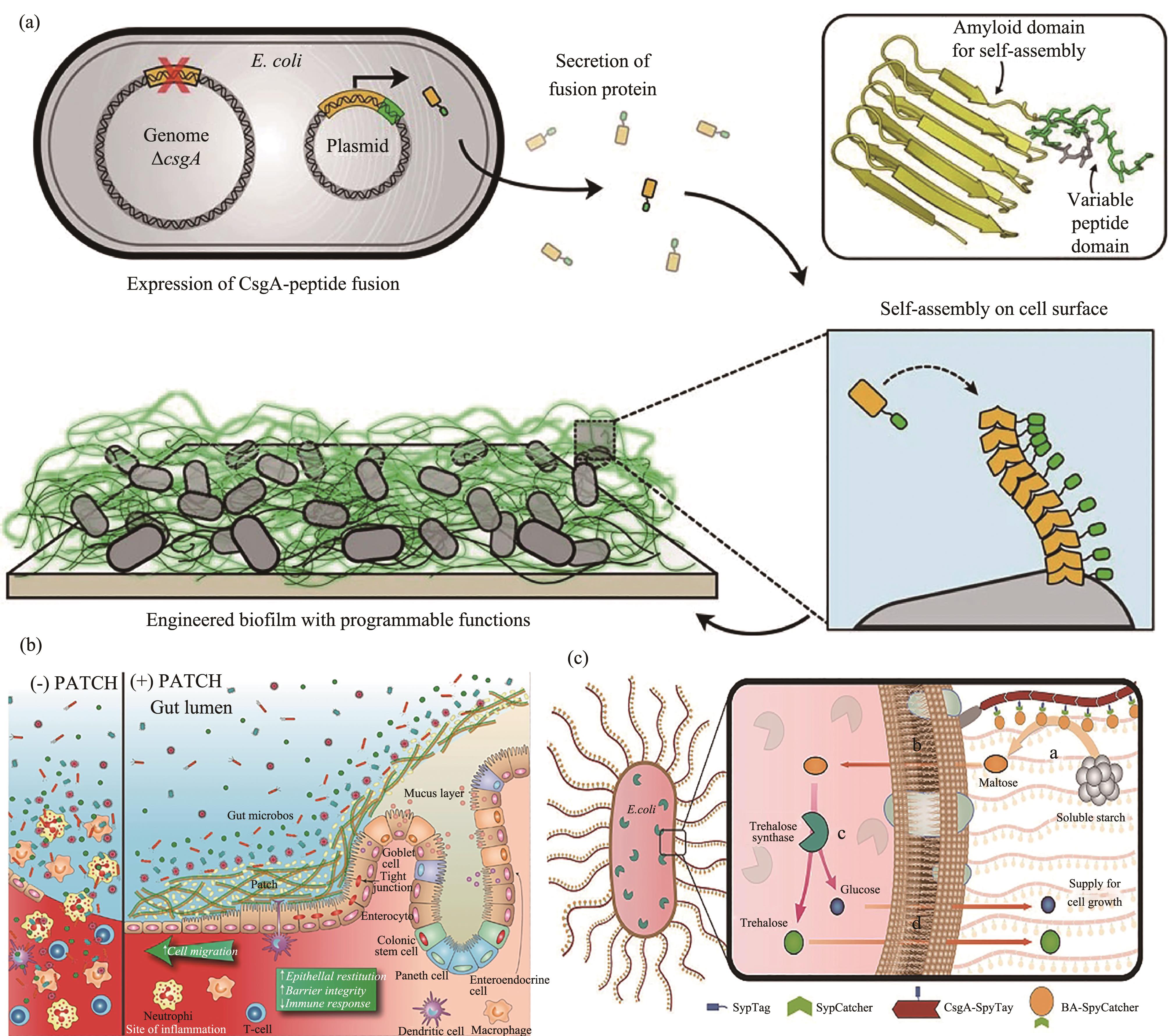
图2 与CsgA融合表达实现功能化curli系统(a)将可诱导CsgA-功能肽的蛋白基因线路导入csgA基因缺失型的宿主,可在细菌外膜自组装形成展示多种功能肽段的工程化curli[68];(b)通过将trefoil factors(TFFs)与CsgA进行融合,可以组装表面展示TFFs的curli,这种改造后的益生菌可直接用来治疗肠炎[77];(c)表面展示SpyTag的curli系统与标记SpyCatcher的淀粉水解酶结合,将胞外淀粉降解成麦芽糖,运输到胞内后由胞内的海藻糖合酶催化,实现淀粉的高效生物转化及海藻糖的合成[80]
Fig. 2 Functionalization of curli via fusion with CsgA(a) Gene circuit containing inducible expression of CsgA (with subunits engineered to display various peptide tags) was transformed into a host strain with the endogenous csgA gene deleted;(b) Fusing CsgA with trefoil factors (TFFs) led to the formation of curli nanofibers displaying TTFs. The resultant material was proven to promote intestinal barrier function and epithelial restitution;(c) SpyTag displaying curli was fused with SpyCatcher decorated β-amylase. β-amylase converted the starch into maltose. The maltose was then transported intracellularly and further catalyzed into trehalose through the intracellularly expressed trehalase
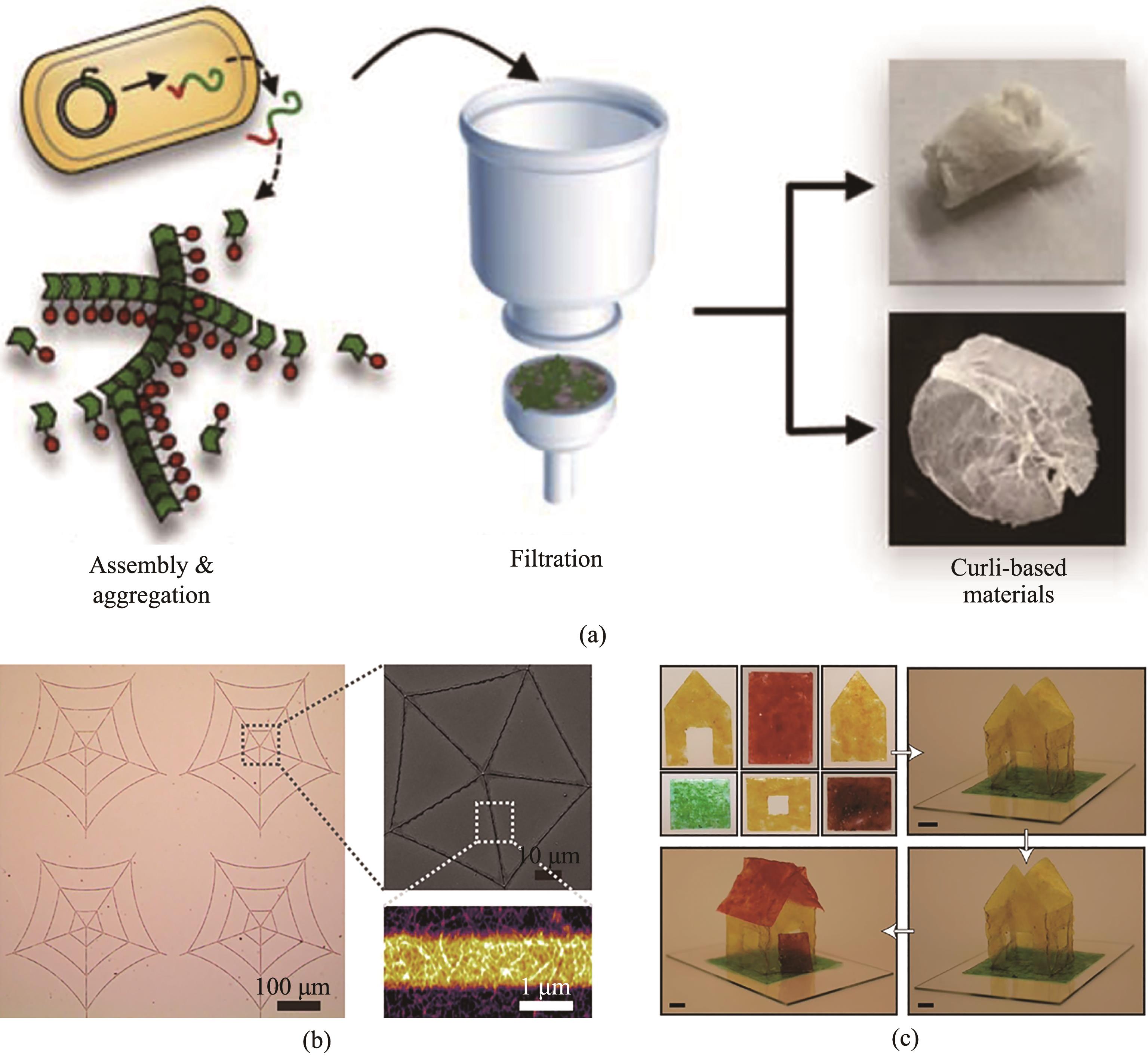
图3 利用大肠杆菌生产curli并剥离curli作为生物材料(a)利用大肠杆菌生产curli,随后通过过滤的方法实现体外剥离,再通过溶解以及重复的形式可以进行材料的进一步加工处理[81];(b)将大肠杆菌体表面的curli以及体内表达的CsgA蛋白单体混合物,通过溶解与固化可将其塑型为多种图案[83];(c)通过剥离curli并进一步加工成为水塑材料,其不但可以耐受多种有机溶剂及强酸强碱,还可以接合成多种三维形状[82]
Fig. 3 Purified curli as the materials precursors(a) Curli fiber produced by E. coli were purified using a fast and easily accessible vacuum filtration procedure. The fibers were then disassembled, reassembled into thin films, and recycled for further materials processing[81];(b) Generation of diverse patterns with a generic amyloid monomer inks (consisting of genetically engineered biofilm proteins dissolved in hexafluoroisopropanol), along with methanol-assisted curing[83];(c) Aqua plastic was produced by casting and drying purified curli under ambient conditions[82]. The resultant aqua plastic could withstand strong acid/base and organic solvents. In addition, aqua plastic could be healed and welded to form three-dimensional architectures using water
| 1 | DONLAN R M. Biofilms: microbial life on surfaces[J]. Emerging Infectious Diseases, 2002, 8(9): 881-890. |
| 2 | WATNICK P, KOLTER R. Biofilm, city of microbes[J]. Journal of Bacteriology, 2000, 182(10): 2675-2679. |
| 3 | HALL-STOODLEY L, COSTERTON J W, STOODLEY P. Bacterial biofilms: from the natural environment to infectious diseases[J]. Nature Reviews Microbiology, 2004, 2(2): 95-108. |
| 4 | WILKING J N, ANGELINI T E, SEMINARA A, et al. Biofilms as complex fluids[J]. MRS Bulletin, 2011, 36(5): 385-391. |
| 5 | KLAPPER I, RUPP C J, CARGO R, et al. Viscoelastic fluid description of bacterial biofilm material properties[J]. Biotechnology and Bioengineering, 2002, 80(3): 289-296. |
| 6 | FRANCIUS G, DOMENECH O, MINGEOT-LECLERCQ M P, et al. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin[J]. Journal of Bacteriology, 2008, 190(24): 7904-7909. |
| 7 | ARNOLDI M, FRITZ M, BÄUERLEIN E, et al. Bacterial turgor pressure can be measured by atomic force microscopy[J]. Physical Review E, Statistical Physics, Plasmas, Fluids, and Related Interdisciplinary Topics, 2000, 62(1 Pt B): 1034-1044. |
| 8 | PETERSON B W, HE Y, REN Y J, et al. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges[J]. FEMS Microbiology Reviews, 2015, 39(2): 234-245. |
| 9 | BOYER R, WELSCH G, COLLINGS E. Materials properties handbook: Titanium alloys[M]. ASM International, 1994. |
| 10 | COMMITTEE A H. Properties and selection: nonferrous alloys and special-purpose materials[M]. ASM International, 1990. |
| 11 | BORZACCHIELLO A, MAYOL L, RAMIRES P A, et al. Structural and rheological characterization of hyaluronic acid-based scaffolds for adipose tissue engineering[J]. Biomaterials, 2007, 28(30): 4399-4408. |
| 12 | EDWARDS C, MARKS R. Evaluation of biomechanical properties of human skin[J]. Clinics in Dermatology, 1995, 13(4): 375-380. |
| 13 | RATNER B D, HOFFMAN A S, SCHOEN F J, et al. Biomaterials science: an evolving, multidisciplinary endeavor[M]// Biomaterials Science. 3rd . Amsterdam: Elsevier, 2013: xxv-xxxix. |
| 14 | MURCKO A C. Patient-centered interviewing: An evidence-based method. 2nd ed[J]. Clinical Nurse Specialist, 2002, 16(6): 326. |
| 15 | LEE J, KWON H J. Measurement of stress-strain behaviour of human hair fibres using optical techniques[J]. International Journal of Cosmetic Science, 2013, 35(3): 238-243. |
| 16 | RANTONEN P J, MEURMAN J H. Viscosity of whole saliva[J]. Acta Odontologica Scandinavica, 1998, 56(4): 210-214. |
| 17 | ELERT G. Viscosity[EB/OL]// The Physics Hypertextbook, 2014. . |
| 18 | INMAN B A, ETIENNE W, RUBIN R, et al. The impact of temperature and urinary constituents on urine viscosity and its relevance to bladder hyperthermia treatment[J]. International Journal of Hyperthermia, 2013, 29(3): 206-210. |
| 19 | WANG H W, DENG H H, MA L M, et al. Influence of operating conditions on extracellular polymeric substances and surface properties of sludge flocs[J]. Carbohydrate Polymers, 2013, 92(1): 510-515. |
| 20 | KÖRSTGENS V, FLEMMING H C, WINGENDER J, et al. Uniaxial compression measurement device for investigation of the mechanical stability of biofilms[J]. Journal of Microbiological Methods, 2001, 46(1): 9-17. |
| 21 | SHAW T, WINSTON M, RUPP C J, et al. Commonality of elastic relaxation times in biofilms[J]. Physical Review Letters, 2004, 93(9): 098102. |
| 22 | STOODLEY P, LEWANDOWSKI Z, BOYLE J D, et al. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology[J]. Biotechnology and Bioengineering, 1999, 65(1): 83-92. |
| 23 | PARAMONOVA E, KALMYKOWA O J, VAN DER MEI H C, et al. Impact of hydrodynamics on oral biofilm strength[J]. Journal of Dental Research, 2009, 88(10): 922-926. |
| 24 | CHEN D T N, WEN Q, JANMEY P A, et al. Rheology of soft materials[J]. Annual Review of Condensed Matter Physics, 2010, 1: 301-322. |
| 25 | TERAOKA I. Polymer solution : an introduction to physical properties[M]. New York: John Wiley & Sons, Inc, 2002. |
| 26 | HUNG C, ZHOU Y Z, PINKNER J S, et al. Escherichia coli biofilms have an organized and complex extracellular matrix structure[J]. mBio, 2013, 4(5): e00645-e00613. |
| 27 | GREENBERG E P. Bacterial communication and group behavior[J]. The Journal of Clinical Investigation, 2003, 112(9): 1288-1290. |
| 28 | ZOGAJ X, BOKRANZ W, NIMTZ M, et al. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract[J]. Infection and Immunity, 2003, 71(7): 4151-4158. |
| 29 | TURSI S A, TÜKEL Ç. Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications[J]. Microbiology and Molecular Biology Reviews: MMBR, 2018, 82(4): e00028-e00018. |
| 30 | EVANS M L, CHORELL E, TAYLOR J D, et al. The bacterial curli system possesses a potent and selective inhibitor of amyloid formation[J]. Molecular Cell, 2015, 57(3): 445-455. |
| 31 | CHAPMAN M R, ROBINSON L S, PINKNER J S, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation[J]. Science, 2002, 295(5556): 851-855. |
| 32 | NENNINGER A A, ROBINSON L S, HAMMER N D, et al. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation[J]. Molecular Microbiology, 2011, 81(2): 486-499. |
| 33 | HAMMER N D, MCGUFFIE B A, ZHOU Y Z, et al. The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation[J]. Journal of Molecular Biology, 2012, 422(3): 376-389. |
| 34 | SUNDE M, SERPELL L C, BARTLAM M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction[J]. Journal of Molecular Biology, 1997, 273(3): 729-739. |
| 35 | ZHONG C, GURRY T, CHENG A A, et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres[J]. Nature Nanotechnology, 2014, 9: 858-866. |
| 36 | CHAPMAN M R, ROBINSON L S, PINKNER J S, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation[J]. Science, 2002, 295(5556): 851-855. |
| 37 | HAMMAR M, BIAN Z, NORMARK S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(13): 6562-6566. |
| 38 | HAMMER N D, SCHMIDT J C, CHAPMAN M R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(30): 12494-12499. |
| 39 | EVANS M L, CHAPMAN M R. Curli biogenesis: order out of disorder[J]. Biochimica et Biophysica Acta, 2014, 1843(8): 1551-1558. |
| 40 | ISHIHAMA A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks[J]. FEMS Microbiology Reviews, 2010, 34(5): 628-645. |
| 41 | HAMMAR M, ARNQVIST A, BIAN Z, et al. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12[J]. Molecular Microbiology, 1995, 18(4): 661-670. |
| 42 | ZAKIKHANY K, HARRINGTON C R, NIMTZ M, et al. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium[J]. Molecular Microbiology, 2010, 77(3): 771-786. |
| 43 | NENNINGER A A, ROBINSON L S, HULTGREN S J. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(3): 900-905. |
| 44 | GOYAL P, KRASTEVA P V, VAN GERVEN N, et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG[J]. Nature, 2014, 516(7530): 250-253. |
| 45 | ROBINSON L S, ASHMAN E M, HULTGREN S J, et al. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein[J]. Molecular Microbiology, 2006, 59(3): 870-881. |
| 46 | LOFERER H, HAMMAR M, NORMARK S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG[J]. Molecular Microbiology, 1997, 26(1): 11-23. |
| 47 | JIANG J S, PENTELUTE B L, COLLIER R J, et al. Atomic structure of anthrax protective antigen pore elucidates toxin translocation[J]. Nature, 2015, 521(7553): 545-549. |
| 48 | YAN Z F, YIN M, CHEN J N, et al. Assembly and substrate recognition of curli biogenesis system[J]. Nature Communications, 2020, 11: 241. |
| 49 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 50 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 51 | WU F L, BETHKE J H, WANG M D, et al. Quantitative and synthetic biology approaches to combat bacterial pathogens[J]. Current Opinion in Biomedical Engineering, 2017, 4: 116-126. |
| 52 | AUSLÄNDER S, WIELAND M, FUSSENEGGER M. Smart medication through combination of synthetic biology and cell microencapsulation[J]. Metabolic Engineering, 2012, 14(3): 252-260. |
| 53 | YE H F, DAOUD-EL BABA M, PENG R W, et al. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice[J]. Science, 2011, 332(6037): 1565-1568. |
| 54 | SEGALL-SHAPIRO T H, SONTAG E D, VOIGT C A. Engineered promoters enable constant gene expression at any copy number in bacteria[J]. Nature Biotechnology, 2018, 36(4): 352-358. |
| 55 | KYLILIS N, TUZA Z A, STAN G B, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia[J]. Nature Communications, 2018, 9(1): 2677. |
| 56 | VILABOA N, FENNA M, MUNSON J, et al. Novel gene switches for targeted and timed expression of proteins of interest[J]. Molecular Therapy, 2005, 12(2): 290-298. |
| 57 | GAO C, HOU J S, XU P, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli [J]. Nature Communications, 2019, 10(1): 3751. |
| 58 | NOVÁK B, TYSON J J. Design principles of biochemical oscillators[J]. Nature Reviews Molecular Cell Biology, 2008, 9(12): 981-991. |
| 59 | POTVIN-TROTTIER L, LORD N D, VINNICOMBE G, et al. Synchronous long-term oscillations in a synthetic gene circuit[J]. Nature, 2016, 538(7626): 514-517. |
| 60 | CHEN A Y, DENG Z T, BILLINGS A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature Materials, 2014, 13(5): 515-523. |
| 61 | MOSER F, THAM E, GONZÁLEZ L M, et al. Light-controlled, high-resolution patterning of living engineered bacteria onto textiles, ceramics, and plastic[J]. Advanced Functional Materials, 2019, 29(30): 1901788. |
| 62 | WANG X Y, PU J H, LIU Y, et al. Immobilization of functional nano-objects in living engineered bacterial biofilms for catalytic applications[J]. National Science Review, 2019, 6(5): 929-943. |
| 63 | CAO Y, FENG Y Y, RYSER M D, et al. Programmable assembly of pressure sensors using pattern-forming bacteria[J]. Nature Biotechnology, 2017, 35(11): 1087-1093. |
| 64 | LI Y F, LI K, WANG X Y, et al. Conformable self-assembling amyloid protein coatings with genetically programmable functionality[J]. Science Advances, 2020, 6(21): eaba1425. |
| 65 | CUI M K, QI Q, GURRY T, et al. Modular genetic design of multi-domain functional amyloids: insights into self-assembly and functional properties[J]. Chemical Science, 2019, 10(14): 4004-4014. |
| 66 | WANG Y Y, AN B L, XUE B, et al. Living materials fabricated via gradient mineralization of light-inducible biofilms[J]. Nature Chemical Biology, 2021, 17(3): 351-359. |
| 67 | AN B L, WANG Y Y, JIANG X Y, et al. Programming living glue systems to perform autonomous mechanical repairs[J]. Matter, 2020, 3(6): 2080-2092. |
| 68 | NGUYEN P Q, BOTYANSZKI Z, TAY P K R, et al. Programmable biofilm-based materials from engineered curli nanofibres[J]. Nature Communications, 2014, 5: 4945. |
| 69 | TAY P K R, MANJULA-BASAVANNA A, JOSHI N S. Repurposing bacterial extracellular matrix for selective and differential abstraction of rare earth elements[J]. Green Chemistry, 2018, 20(15): 3512-3520. |
| 70 | PU J H, LIU Y, ZHANG J C, et al. Virus disinfection: virus disinfection from environmental water sources using living engineered biofilm materials[J]. Advanced Science, 2020, 7(14): 1903558. |
| 71 | ABDALI Z, AMINZARE M, ZHU X D, et al. Curli-mediated self-assembly of a fibrous protein scaffold for hydroxyapatite mineralization[J]. ACS Synthetic Biology, 2020, 9(12): 3334-3343. |
| 72 | DONG H, ZHANG W X, XUAN Q Z, et al. Binding peptide-guided immobilization of lipases with significantly improved catalytic performance using Escherichia coli BL21(DE3) biofilms as a platform[J]. ACS Applied Materials & Interfaces, 2021, 13(5): 6168-6179. |
| 73 | BOTYANSZKI Z, TAY P K R, NGUYEN P Q, et al. Engineered catalytic biofilms: site-specific enzyme immobilization onto E. coli curli nanofibers[J]. Biotechnology and Bioengineering, 2015, 112(10): 2016-2024. |
| 74 | BAO J J, LIU N, ZHU L Y, et al. Programming a biofilm-mediated multienzyme-assembly-cascade system for the biocatalytic production of glucosamine from chitin[J]. Journal of Agricultural and Food Chemistry, 2018, 66(30): 8061-8068. |
| 75 | OLMEZ T T, SAHIN KEHRIBAR E, ISILAK M E, et al. Synthetic genetic circuits for self-actuated cellular nanomaterial fabrication devices[J]. ACS Synthetic Biology, 2019, 8(9): 2152-2162. |
| 76 | PRAVESCHOTINUNT P, DORVAL COURCHESNE N M, DEN HARTOG I, et al. Tracking of engineered bacteria in vivo using nonstandard amino acid incorporation[J]. ACS Synthetic Biology, 2018, 7(6): 1640-1650. |
| 77 | PRAVESCHOTINUNT P, DURAJ-THATTE A M, GELFAT I, et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut[J]. Nature Communications, 2019, 10: 5580. |
| 78 | DURAJ-THATTE A M, PRAVESCHOTINUNT P, NASH T R, et al. Modulating bacterial and gut mucosal interactions with engineered biofilm matrix proteins[J]. Scientific Reports, 2018, 8: 3475. |
| 79 | AXPE E, DURAJ-THATTE A, CHANG Y, et al. Fabrication of amyloid curli fibers-alginate nanocomposite hydrogels with enhanced stiffness[J]. ACS Biomaterials Science & Engineering, 2018, 4(6): 2100-2105. |
| 80 | JIANG L, SONG X G, LI Y F, et al. Programming integrative extracellular and intracellular biocatalysis for rapid, robust, and recyclable synthesis of trehalose[J]. ACS Catalysis, 2018, 8(3): 1837-1842. |
| 81 | DORVAL COURCHESNE N M, DURAJ-THATTE A, TAY P K R, et al. Scalable production of genetically engineered nanofibrous macroscopic materials via filtration[J]. ACS Biomaterials Science & Engineering, 2017, 3(5): 733-741. |
| 82 | DURAJ-THATTE A M, MANJULA-BASAVANNA A, COURCHESNE N M D, et al. Water-processable, biodegradable and coatable aquaplastic from engineered biofilms[J]. Nature Chemical Biology, 2021, 17(6): 732-738. |
| 83 | LI Y F, LI K, WANG X Y, et al. Patterned amyloid materials integrating robustness and genetically programmable functionality[J]. Nano Letters, 2019, 19(12): 8399-8408. |
| 84 | GALATSIS K, WANG K L, OZKAN M, et al. Patterning and templating for nanoelectronics[J]. Advanced Materials, 2010, 22(6): 769-778. |
| 85 | CAO Y, RYSER M D, PAYNE S, et al. Collective space-sensing coordinates pattern scaling in engineered bacteria[J]. Cell, 2016, 165(3): 620-630. |
| 86 | FERNANDEZ-RODRIGUEZ J, MOSER F, SONG M, et al. Engineering RGB color vision into Escherichia coli [J]. Nature Chemical Biology, 2017, 13(7): 706-708. |
| 87 | NGUYEN P Q, COURCHESNE N M D, DURAJ-THATTE A, et al. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials[J]. Advanced Materials, 2018, 30(19): e1704847. |
| 88 | GILBERT C, TANG T C, OTT W, et al. Living materials with programmable functionalities grown from engineered microbial co-cultures[J]. Nature Materials, 2021, 20(5): 691-700. |
| 89 | LIU X Y, YUK H, LIN S T, et al. 3D printing of living responsive materials and devices[J]. Advanced Materials, 2018, 30(4): 1704821. |
| 90 | GILBERT C, ELLIS T. Biological engineered living materials: Growing functional materials with genetically programmable properties[J]. ACS Synthetic Biology, 2019, 8(1): 1-15. |
| 91 | CHEN A Y, ZHONG C, LU T K. Engineering living functional materials[J]. ACS Synthetic Biology, 2015, 4(1): 8-11. |
| 92 | YANG P D. Liquid sunlight: the evolution of photosynthetic biohybrids[J]. Nano Letters, 2021, 21(13): 5453-5456. |
| 93 | LE FEUVRE R A, SCRUTTON N S. A living foundry for synthetic biological materials: a synthetic biology roadmap to new advanced materials[J]. Synthetic and Systems Biotechnology, 2018, 3(2): 105-112. |
| 94 | TANG T C, AN B L, HUANG Y Y, et al. Materials design by synthetic biology[J]. Nature Reviews Materials, 2021, 6(4): 332-350. |
| 95 | DAI Z J, LEE A J, ROBERTS S, et al. Versatile biomanufacturing through stimulus-responsive cell-material feedback[J]. Nature Chemical Biology, 2019, 15(10): 1017-1024. |
| 96 | DAI Z J, YANG X Y, WU F L, et al. Living fabrication of functional semi-interpenetrating polymeric materials[J]. Nature Communications, 2021, 12: 3422. |
| 97 | MOLINARI S, TESORIERO R F JR, AJO-FRANKLIN C M. Bottom-up approaches to engineered living materials: Challenges and future directions[J]. Matter, 2021, 4(10): 3095-3120. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 李石开, 曾东鳌, 杜方舟, 张京钟, 余爽. 血管化类器官的构建方法及生物材料[J]. 合成生物学, 2024, 5(4): 851-866. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
