合成生物学 ›› 2023, Vol. 4 ›› Issue (2): 333-346.DOI: 10.12211/2096-8280.2022-064
合成生物学与病毒疫苗研发
申赵铃, 吴艳玲, 应天雷
- 复旦大学基础医学院,上海 200032
-
收稿日期:2022-11-21修回日期:2022-12-29出版日期:2023-04-30发布日期:2023-04-27 -
通讯作者:应天雷 -
作者简介:申赵铃 (1999—),女,博士研究生。研究方向为病原微生物的防治新策略与跨脑药物的研发。E-mail:22111010074@m.fudan.edu.cn应天雷 (1984—),男,博士,教授。研究方向为合成免疫学。E-mail:tlying@fudan.edu.cn -
基金资助:国家重点研发计划(2019YFA0904400)
Synthetic biology and viral vaccine development
SHEN Zhaoling, WU Yanling, YING Tianlei
- School of Basic Medical Sciences,Fudan University,Shanghai 200032,China
-
Received:2022-11-21Revised:2022-12-29Online:2023-04-30Published:2023-04-27 -
Contact:YING Tianlei
摘要:
中图分类号:
引用本文
申赵铃, 吴艳玲, 应天雷. 合成生物学与病毒疫苗研发[J]. 合成生物学, 2023, 4(2): 333-346.
SHEN Zhaoling, WU Yanling, YING Tianlei. Synthetic biology and viral vaccine development[J]. Synthetic Biology Journal, 2023, 4(2): 333-346.
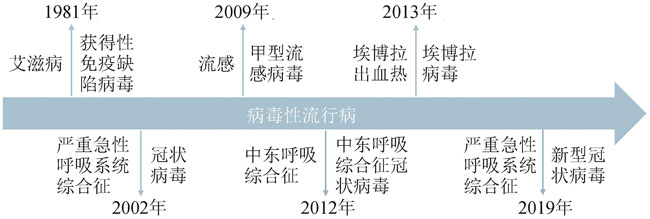
图1 病毒性流行病及相关病原体(近40年)
Fig. 1 Outbreaks of deadly viral epidemics caused by human immunodeficiency virus, SARS coronavirus, influenza A virus H1N1, MERS-CoV, Ebola virus and SARS-CoV-2, respectively, within the past four decades.
| 类型 | 优势 | 局限性 | 应用 |
|---|---|---|---|
灭活病毒疫苗 inactivated vaccines | 相对安全 热稳定性较好 免疫功能低下者和孕妇可考虑 | 免疫原性有限 需要佐剂 持续性短 诱导疾病进展 | 狂犬病 日本脑炎 甲型肝炎 |
减毒活病毒疫苗 live attenuated virus vaccines | 模拟自然感染 同时诱导先天性和适应性免疫反应 1~2次剂量后可获得终身免疫 | 热稳定性较差 免疫功能低下者禁用 有逆转为野生型病毒的可能性 | 麻疹 腮腺炎 流感 脊髓灰质炎 |
亚单位疫苗 subunit vaccines | 无传染性 高稳定性 高安全性 | 免疫原性有限 需要佐剂 | 乙型肝炎 新冠肺炎 |
DNA疫苗 DNA vaccines | 热稳定性好 刺激先天免疫反应 诱导T细胞和B细胞免疫反应 | 潜在基因组整合风险 免疫原性较弱 需要电穿孔等方式递送 | 埃博拉出血热 |
RNA疫苗 RNA vaccines | 无传染性 不存在潜在基因组整合风险 | 稳定性差 易降解性 过度产生炎性反应 | 新冠肺炎 |
病毒载体疫苗 viral vector vaccines | 应用广泛 同时诱导体液和细胞免疫反应 研发周期短 | 可能致病性 | 埃博拉出血热 |
表1 各类病毒疫苗的优缺点及其应用
Table 1 Advantages,disadvantages and the applications of different kinds of vaccines
| 类型 | 优势 | 局限性 | 应用 |
|---|---|---|---|
灭活病毒疫苗 inactivated vaccines | 相对安全 热稳定性较好 免疫功能低下者和孕妇可考虑 | 免疫原性有限 需要佐剂 持续性短 诱导疾病进展 | 狂犬病 日本脑炎 甲型肝炎 |
减毒活病毒疫苗 live attenuated virus vaccines | 模拟自然感染 同时诱导先天性和适应性免疫反应 1~2次剂量后可获得终身免疫 | 热稳定性较差 免疫功能低下者禁用 有逆转为野生型病毒的可能性 | 麻疹 腮腺炎 流感 脊髓灰质炎 |
亚单位疫苗 subunit vaccines | 无传染性 高稳定性 高安全性 | 免疫原性有限 需要佐剂 | 乙型肝炎 新冠肺炎 |
DNA疫苗 DNA vaccines | 热稳定性好 刺激先天免疫反应 诱导T细胞和B细胞免疫反应 | 潜在基因组整合风险 免疫原性较弱 需要电穿孔等方式递送 | 埃博拉出血热 |
RNA疫苗 RNA vaccines | 无传染性 不存在潜在基因组整合风险 | 稳定性差 易降解性 过度产生炎性反应 | 新冠肺炎 |
病毒载体疫苗 viral vector vaccines | 应用广泛 同时诱导体液和细胞免疫反应 研发周期短 | 可能致病性 | 埃博拉出血热 |
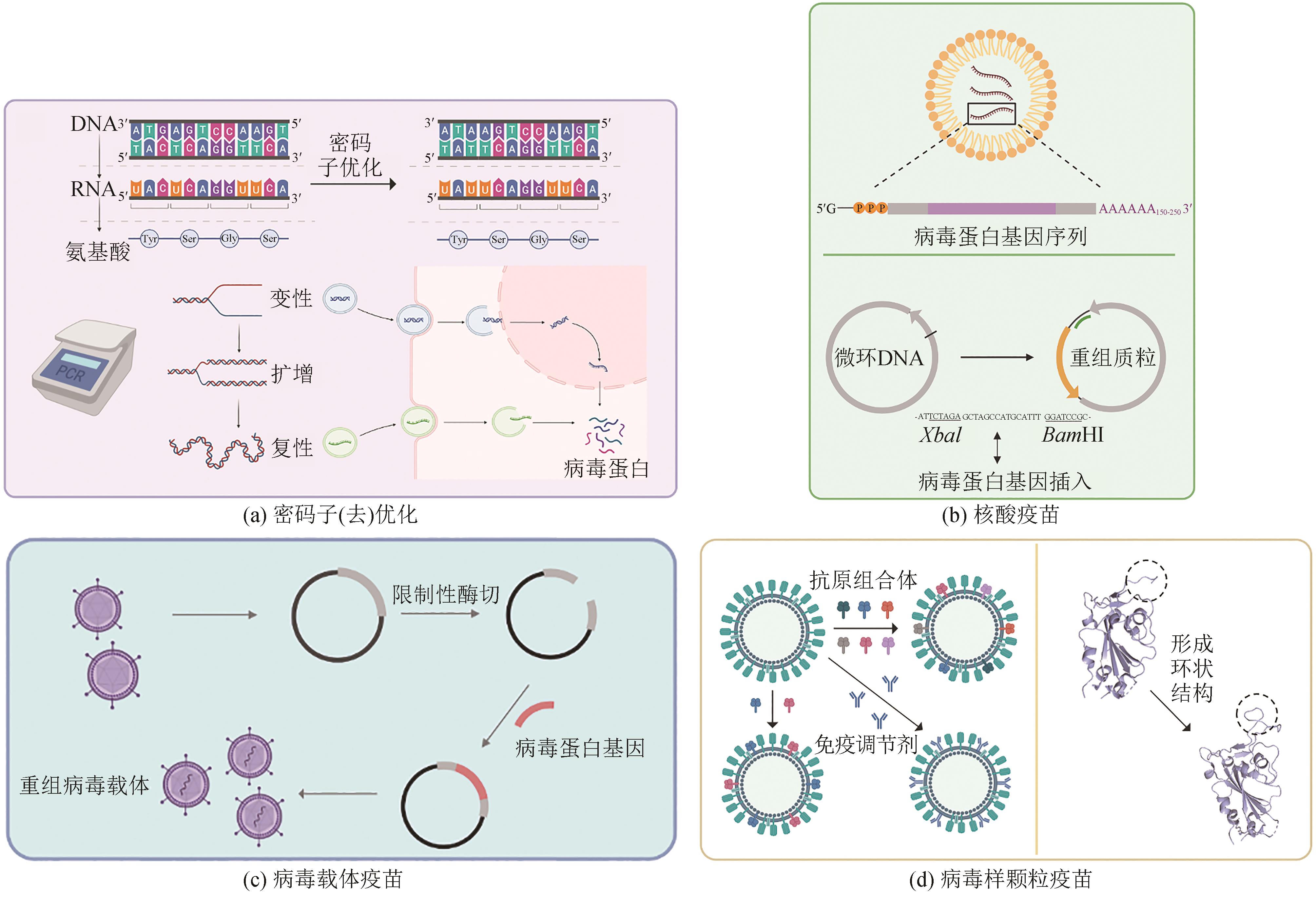
图2 合成生物技术在病毒疫苗中的应用(a) Codon optimization/deoptimization: The expression level of viral protein can be increased through codon optimization, but codon deoptimization can result in the production of live attenuated vaccines. (b) Nucleic acid vaccines: mRNA is modified with intact 5′ cap structure and 3′ polytail structure for stable expression, and microcyclic DNA is inserted into viral protein coding sequences and exogenous promoter sequences through restriction digestion sites to construct recombinant plasmids. (c) Viral vectors vaccines:viral protein coding sequences are inserted into viral vector sequences to generate recombinant viral vectors. (d) Virus-like particle (VLP) vaccines: multi-antigen VLP vaccines are developed based on GECC generation, and VLP vaccines include immunomodulators, and ring structures to accommodate antigen peptides for recognition
Fig. 2 Synthetic biology technologies for developing viral vaccines

图3 合成生物技术在病毒疫苗中的应用与优势总结(Synthetic vaccines include genomic codon-optimized vaccines, synthetic genome-based nucleic acid vaccines, viral vector-based vaccines, and virus-like particle vaccines, which can be produced on large-scale production in recombinant yeasts to shorten time for vaccine development with advantages in enhancing immunogenicity, and broadening spectrum, and improving safety and efficacy)
Fig. 3 Applications and advantges of synthetic biotechnologies in the development of synthetic vaccines
| 1 | ROYCHOUDHURY S, DAS A, SENGUPTA P, et al. Viral pandemics of the last four decades: pathophysiology, health impacts and perspectives[J]. International Journal of Environmental Research and Public Health, 2020, 17(24): 9411. |
| 2 | AKIN L, GÖZEL M G. Understanding dynamics of pandemics[J]. Turkish Journal of Medical Sciences, 2020, 50(SI-1): 515-519. |
| 3 | MEYER H, EHMANN R, SMITH G L. Smallpox in the post-eradication era[J]. Viruses, 2020, 12(2): 138. |
| 4 | AHMED R, BURTON D R. Viral vaccines: past successes and future challenges[J]. Current Opinion in Virology, 2013, 3(3): 307-308. |
| 5 | BRISSE M, VRBA S M, KIRK N, et al. Emerging concepts and technologies in vaccine development[J]. Frontiers in Immunology, 2020, 11: 583077. |
| 6 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11: 5174. |
| 7 | HO C, MORSUT L. Novel synthetic biology approaches for developmental systems[J]. Stem Cell Reports, 2021, 16(5): 1051-1064. |
| 8 | TAN X, LETENDRE J H, COLLINS J J, et al. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics[J]. Cell, 2021, 184(4): 881-898. |
| 9 | WANG N X, YUAN Z, NIU W, et al. Synthetic biology approach for the development of conditionally replicating HIV-1 vaccine[J]. Journal of Chemical Technology and Biotechnology, 2017, 92(3): 455-462. |
| 10 | GHATTAS M, DWIVEDI G, LAVERTU M, et al. Vaccine technologies and platforms for infectious diseases: current progress, challenges, and opportunities[J]. Vaccines, 2021, 9(12): 1490. |
| 11 | AWADASSEID A, WU Y L, TANAKA Y, et al. Current advances in the development of SARS-CoV-2 vaccines[J]. International Journal of Biological Sciences, 2021, 17(1): 8-19. |
| 12 | ARORA M, LAKSHMI R. Vaccines-safety in pregnancy[J]. Best Practice & Research Clinical Obstetrics & Gynaecology, 2021, 76: 23-40. |
| 13 | CURRAN M P, LEROUX-ROELS I. Inactivated split-virion seasonal influenza vaccine (Fluarix): a review of its use in the prevention of seasonal influenza in adults and the elderly[J]. Drugs, 2010, 70(12): 1519-1543. |
| 14 | CHEN H P, HUANG Z Y, CHANG S Y, et al. Immunogenicity and safety of an inactivated SARS-CoV-2 vaccine (Sinopharm BBIBP-CorV) coadministered with quadrivalent split-virion inactivated influenza vaccine and 23-valent pneumococcal polysaccharide vaccine in China: a multicentre, non-inferiority, open-label, randomised, controlled, phase 4 trial[J]. Vaccine, 2022, 40(36): 5322-5332. |
| 15 | STANFIELD B A, KOUSOULAS K G, FERNANDEZ A, et al. Rational design of live-attenuated vaccines against herpes simplex viruses[J]. Viruses, 2021, 13(8): 1637. |
| 16 | MOK D Z L, CHAN K R. The effects of pre-existing antibodies on live-attenuated viral vaccines[J]. Viruses, 2020, 12(5): 520. |
| 17 | VETTER V, DENIZER G, FRIEDLAND L R, et al. Understanding modern-day vaccines: what you need to know[J]. Annals of Medicine, 2018, 50(2): 110-120. |
| 18 | MOYLE P M, TOTH I. Modern subunit vaccines: development, components, and research opportunities[J]. ChemMedChem, 2013, 8(3): 360-376. |
| 19 | HANSSON M, NYGREN P A, STÅHL S. Design and production of recombinant subunit vaccines[J]. Biotechnology and Applied Biochemistry, 2000, 32(2): 95-107. |
| 20 | ZHANG N R, ZHENG B J, LU L, et al. Advancements in the development of subunit influenza vaccines[J]. Microbes and Infection, 2015, 17(2): 123-134. |
| 21 | AZMI F, HADI AHMAD FUAAD A AL, SKWARCZYNSKI M, et al. Recent progress in adjuvant discovery for peptide-based subunit vaccines[J]. Human Vaccines & Immunotherapeutics, 2014, 10(3): 778-796. |
| 22 | MCVOY M A. Cytomegalovirus vaccines[J]. Clinical Infectious Diseases, 2013, 57(): S196-S199. |
| 23 | PASS R F, ZHANG C P, EVANS A, et al. Vaccine prevention of maternal cytomegalovirus infection[J]. The New England Journal of Medicine, 2009, 360(12): 1191-1199. |
| 24 | TOMBÁCZ I, WEISSMAN D, PARDI N. Vaccination with messenger RNA: a promising alternative to DNA vaccination[J]. Methods in Molecular Biology, 2021, 2197: 13-31. |
| 25 | GRANT-KLEIN R J, ALTAMURA L A, BADGER C V, et al. Codon-optimized filovirus DNA vaccines delivered by intramuscular electroporation protect cynomolgus macaques from lethal Ebola and Marburg virus challenges[J]. Human Vaccines & Immunotherapeutics, 2015, 11(8): 1991-2004. |
| 26 | LEDWITH B J, MANAM S, TROILO P J, et al. Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice[J]. Intervirology, 2000, 43(4/5/6): 258-272. |
| 27 | WANG Z, TROILO P J, WANG X, et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation[J]. Gene Therapy, 2004, 11(8): 711-721. |
| 28 | SZABÓ G T, MAHINY A J, VLATKOVIC I. COVID-19 mRNA vaccines: platforms and current developments[J]. Molecular Therapy, 2022, 30(5): 1850-1868. |
| 29 | KARIKÓ K. Developing mRNA for therapy[J]. The Keio Journal of Medicine, 2022, 71(1): 31. |
| 30 | LI M Y, WANG Z N, XIE C Y, et al. Advances in mRNA vaccines[J]. International Review of Cell and Molecular Biology, 2022, 372: 295-316. |
| 31 | WANG Y, ZHANG Z Q, LUO J W, et al. mRNA vaccine: a potential therapeutic strategy[J]. Molecular Cancer, 2021, 20(1): 33. |
| 32 | FANG E Y, LIU X H, LI M, et al. Advances in COVID-19 mRNA vaccine development[J]. Signal Transduction and Targeted Therapy, 2022, 7: 94. |
| 33 | MCCANN N, O'CONNOR D, LAMBE T, et al. Viral vector vaccines[J]. Current Opinion in Immunology, 2022, 77: 102210. |
| 34 | EWER K J, LAMBE T, ROLLIER C S, et al. Viral vectors as vaccine platforms: from immunogenicity to impact[J]. Current Opinion in Immunology, 2016, 41: 47-54. |
| 35 | CHEN J D, WANG J H, ZHANG J P, et al. Advances in development and application of influenza vaccines[J]. Frontiers in Immunology, 2021, 12: 711997. |
| 36 | DE VRIES R D, RIMMELZWAAN G F. Viral vector-based influenza vaccines[J]. Human Vaccines & Immunotherapeutics, 2016, 12(11): 2881-2901. |
| 37 | KHAN M S, KIM E, MCPHERSON A, et al. Adenovirus-vectored SARS-CoV-2 vaccine expressing S1-N fusion protein[J]. Antibody Therapeutics, 2022, 5(3): 177-191. |
| 38 | TOURNIER J N, KONONCHIK J. Virus eradication and synthetic biology: changes with SARS-CoV-2?[J]. Viruses, 2021, 13(4): 569. |
| 39 | PARVATHY S T, UDAYASURIYAN V, BHADANA V. Codon usage bias[J]. Molecular Biology Reports, 2022, 49(1): 539-565. |
| 40 | XU Y C, LIU K S, HAN Y, et al. Codon usage bias regulates gene expression and protein conformation in yeast expression system P. pastoris [J]. Microbial Cell Factories, 2021, 20(1): 91. |
| 41 | NOVOA E M, PAVON-ETERNOD M, PAN T, et al. A role for tRNA modifications in genome structure and codon usage[J]. Cell, 2012, 149(1): 202-213. |
| 42 | NOVOA E M, RIBAS DE POUPLANA L. Speeding with control: codon usage, tRNAs, and ribosomes[J]. Trends in Genetics, 2012, 28(11): 574-581. |
| 43 | GUIMARAES J C, MITTAL N, GNANN A, et al. A rare codon-based translational program of cell proliferation[J]. Genome Biology, 2020, 21(1): 44. |
| 44 | FU H G, LIANG Y B, ZHONG X Q, et al. Codon optimization with deep learning to enhance protein expression[J]. Scientific Reports, 2020, 10(1): 17617. |
| 45 | FENG L Q, WANG Q, SHAN C, et al. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-CoV-2 challenge in rhesus macaques[J]. Nature Communications, 2020, 11: 4207. |
| 46 | XU S Q, YANG K P, LI R, et al. mRNA vaccine era-mechanisms, drug platform and clinical prospection[J]. International Journal of Molecular Sciences, 2020, 21(18): 6582. |
| 47 | COLEMAN J R, PAPAMICHAIL D, SKIENA S, et al. Virus attenuation by genome-scale changes in codon pair bias[J]. Science, 2008, 320(5884): 1784-1787. |
| 48 | BROADBENT A J, SANTOS C P, ANAFU A, et al. Evaluation of the attenuation, immunogenicity, and efficacy of a live virus vaccine generated by codon-pair bias de-optimization of the 2009 pandemic H1N1 influenza virus, in ferrets[J]. Vaccine, 2016, 34(4): 563-570. |
| 49 | KAPLAN B S, SOUZA C K, GAUGER P C, et al. Vaccination of pigs with a codon-pair bias de-optimized live attenuated influenza vaccine protects from homologous challenge[J]. Vaccine, 2018, 36(8): 1101-1107. |
| 50 | MARUGGI G, ZHANG C L, LI J W, et al. mRNA as a transformative technology for vaccine development to control infectious diseases[J]. Molecular Therapy, 2019, 27(4): 757-772. |
| 51 | BURNS C C, SHAW J, CAMPAGNOLI R, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region[J]. Journal of Virology, 2006, 80(7): 3259-3272. |
| 52 | SI L L, XU H, ZHOU X Y, et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines[J]. Science, 2016, 354(6316): 1170-1173. |
| 53 | JOHNSON B A, XIE X P, BAILEY A L, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis[J]. Nature, 2021, 591(7849): 293-299. |
| 54 | GOŁAWSKI M, LEWANDOWSKI P, JABŁOŃSKA I, et al. The reassessed potential of SARS-CoV-2 attenuation for COVID-19 vaccine development - a systematic review[J]. Viruses, 2022, 14(5): 991. |
| 55 | WANG Y, YANG C, SONG Y T, et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(29): e2102775118. |
| 56 | FAN R L Y, VALKENBURG S A, WONG C K S, et al. Generation of live attenuated influenza virus by using codon usage bias[J]. Journal of Virology, 2015, 89(21): 10762-10773. |
| 57 | ASRANI K H, FARELLI J D, STAHLEY M R, et al. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA[J]. RNA Biology, 2018, 15(6): 756-762. |
| 58 | TREPOTEC Z, ANEJA M K, GEIGER J, et al. Maximizing the translational yield of mRNA therapeutics by minimizing 5′-UTRs[J]. Tissue Engineering Part A, 2019, 25(1/2): 69-79. |
| 59 | VON NIESSEN A G O, POLEGANOV M A, RECHNER C, et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening[J]. Molecular Therapy, 2019, 27(4): 824-836. |
| 60 | PARDI N, HOGAN M J, WEISSMAN D. Recent advances in mRNA vaccine technology[J]. Current Opinion in Immunology, 2020, 65: 14-20. |
| 61 | LEE S, RYU J H. Influenza viruses: innate immunity and mRNA vaccines[J]. Frontiers in Immunology, 2021, 12: 710647. |
| 62 | NOORI M, NEJADGHADERI S A, ARSHI S, et al. Potency of BNT162b2 and mRNA-1273 vaccine-induced neutralizing antibodies against severe acute respiratory syndrome-CoV-2 variants of concern: a systematic review of in vitro studies[J]. Reviews in Medical Virology, 2022, 32(2): e2277. |
| 63 | FEIKIN D R, HIGDON M M, ABU-RADDAD L J, et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression[J]. The Lancet, 2022, 399(10328): 924-944. |
| 64 | XI J X, LEI L R, ZOUZAS W, et al. Nasally inhaled therapeutics and vaccination for COVID-19: developments and challenges[J]. MedComm, 2021, 2(4): 569-586. |
| 65 | ANDRADE V M, CHRISTENSEN-QUICK A, AGNES J, et al. INO-4800 DNA vaccine induces neutralizing antibodies and T cell activity against global SARS-CoV-2 variants[J]. Npj Vaccines, 2021, 6: 121. |
| 66 | SMITH T R F, PATEL A, RAMOS S, et al. Immunogenicity of a DNA vaccine candidate for COVID-19[J]. Nature Communications, 2020, 11: 2601. |
| 67 | WALTERS J N, SCHOUEST B, PATEL A, et al. Prime-boost vaccination regimens with INO-4800 and INO-4802 augment and broaden immune responses against SARS-CoV-2 in nonhuman primates[J]. Vaccine, 2022, 40(21): 2960-2969. |
| 68 | DEY A, CHOZHAVEL RAJANATHAN T M, CHANDRA H, et al. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models[J]. Vaccine, 2021, 39(30): 4108-4116. |
| 69 | KHOBRAGADE A, BHATE S, RAMAIAH V, et al. Efficacy, safety, and immunogenicity of the DNA SARS-CoV-2 vaccine (ZyCoV-D): the interim efficacy results of a phase 3, randomised, double-blind, placebo-controlled study in India[J]. The Lancet, 2022, 399(10332): 1313-1321. |
| 70 | ALMEIDA A M, EUSÉBIO D, QUEIROZ J A, et al. Minicircle DNA vaccine purification and E7 antigen expression assessment[J]. Methods in Molecular Biology, 2021, 2197: 207-222. |
| 71 | JIANG Y L, GAO X, XU K, et al. A novel cre recombinase-mediated in vivo minicircle DNA (CRIM) vaccine provides partial protection against Newcastle disease virus[J]. Applied and Environmental Microbiology, 2019, 85(14): e00407-e00419. |
| 72 | ZHOU X W, JIANG X, QU M Y, et al. Engineering antiviral vaccines[J]. ACS Nano, 2020, 14(10): 12370-12389. |
| 73 | TATSIS N, ERTL H C J. Adenoviruses as vaccine vectors[J]. Molecular Therapy, 2004, 10(4): 616-629. |
| 74 | SAKURAI F, TACHIBANA M, MIZUGUCHI H. Adenovirus vector-based vaccine for infectious diseases[J]. Drug Metabolism and Pharmacokinetics, 2022, 42: 100432. |
| 75 | SHIRLEY J L, DE JONG Y P, TERHORST C, et al. Immune responses to viral gene therapy vectors[J]. Molecular Therapy, 2020, 28(3): 709-722. |
| 76 | TOMORI O, KOLAWOLE M O. Ebola virus disease: current vaccine solutions[J]. Current Opinion in Immunology, 2021, 71: 27-33. |
| 77 | YAMAZAKI K I, DE MORA K, SAITOH K. BioBrick-based 'quick gene assembly' in vitro [J]. Synthetic Biology, 2017, 2(1): ysx003. |
| 78 | SHETTY R P, ENDY D, KNIGHT T F. Engineering BioBrick vectors from BioBrick parts[J]. Journal of Biological Engineering, 2008, 2: 5. |
| 79 | NOORAEI S, BAHRULOLUM H, HOSEINI Z S, et al. Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers[J]. Journal of Nanobiotechnology, 2021, 19(1): 59. |
| 80 | PATEL J M, KIM M C, VARTABEDIAN V F, et al. Protein transfer-mediated surface engineering to adjuvantate virus-like nanoparticles for enhanced anti-viral immune responses[J]. Nanomedicine: Nanotechnology, Biology, and Medicine, 2015, 11(5): 1097-1107. |
| 81 | MOHSEN M O, ZHA L S, CABRAL-MIRANDA G, et al. Major findings and recent advances in virus-like particle (VLP)-based vaccines[J]. Seminars in Immunology, 2017, 34: 123-132. |
| 82 | LAMPINEN V, HEINIMÄKI S, LAITINEN O H, et al. Modular vaccine platform based on the norovirus-like particle[J]. Journal of Nanobiotechnology, 2021, 19(1): 25. |
| 83 | COHEN A A, GNANAPRAGASAM P N P, LEE Y E, et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice[J]. Science, 2021, 371(6530): 735-741. |
| 84 | CHARLTON HUME H K, VIDIGAL J, CARRONDO M J T, et al. Synthetic biology for bioengineering virus-like particle vaccines[J]. Biotechnology and Bioengineering, 2019, 116(4): 919-935. |
| 85 | TEYMENNET-RAMÍREZ K V, MARTÍNEZ-MORALES F, TREJO-HERNÁNDEZ M R. Yeast surface display system: strategies for improvement and biotechnological applications[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 794742. |
| 86 | KUMAR R, KUMAR P. Yeast-based vaccines: new perspective in vaccine development and application[J]. FEMS Yeast Research, 2019, 19(2): foz007. |
| 87 | POLLET J, CHEN W H, VERSTEEG L, et al. SARS‑CoV-2 RBD219-N1C1: a yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice[J]. Human Vaccines & Immunotherapeutics, 2021, 17(8): 2356-2366. |
| 88 | LEE J, LIU Z Y, CHEN W H, et al. Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain-based vaccine candidate, RBD219-N1C1[J]. Applied Microbiology and Biotechnology, 2021, 105(10): 4153-4165. |
| 89 | ZHAO Q J, TOWNE V, BROWN M, et al. In-depth process understanding of RECOMBIVAX HB® maturation and potential epitope improvements with redox treatment: multifaceted biochemical and immunochemical characterization[J]. Vaccine, 2011, 29(45): 7936-7941. |
| 90 | WANG J W, RODEN R B. Virus-like particles for the prevention of human papillomavirus-associated malignancies[J]. Expert Review of Vaccines, 2013, 12(2): 129-141. |
| 91 | VENTER J C, GLASS J I, C A Ⅲ HUTCHISON, et al. Synthetic chromosomes, genomes, viruses, and cells[J]. Cell, 2022, 185(15): 2708-2724. |
| 92 | WANG W H, ERAZO E M, ISHCOL M R C, et al. Virus-induced pathogenesis, vaccine development, and diagnosis of novel H7N9 avian influenza A virus in humans: a systemic literature review[J]. The Journal of International Medical Research, 2020, 48(1): 300060519845488. |
| 93 | TRIPATHI N K, SHRIVASTAVA A. Recent developments in recombinant protein-based dengue vaccines[J]. Frontiers in Immunology, 2018, 9: 1919. |
| 94 | PENG X L, CHENG J S Y, GONG H L, et al. Advances in the design and development of SARS-CoV-2 vaccines[J]. Military Medical Research, 2021, 8(1): 67. |
| 95 | CALZAS C, CHEVALIER C. Innovative mucosal vaccine formulations against influenza A virus infections[J]. Frontiers in Immunology, 2019, 10: 1605. |
| 96 | NAGY G, EMODY L, PÁL T. Strategies for the development of vaccines conferring broad-spectrum protection[J]. International Journal of Medical Microbiology: IJMM, 2008, 298(5/6): 379-395. |
| 97 | SÁNCHEZ-SAMPEDRO L, PERDIGUERO B, MEJÍAS-PÉREZ E, et al. The evolution of poxvirus vaccines[J]. Viruses, 2015, 7(4): 1726-1803. |
| 98 | CUNNINGHAM A L, GARÇON N, LEO O, et al. Vaccine development: from concept to early clinical testing[J]. Vaccine, 2016, 34(52): 6655-6664. |
| 99 | LI Z H, SONG S, HE M Z, et al. Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity[J]. Nature Communications, 2018, 9: 5360. |
| 100 | DE GROOT A S, MOISE L, TERRY F, et al. Better epitope discovery, precision immune engineering, and accelerated vaccine design using immunoinformatics tools[J]. Frontiers in Immunology, 2020, 11: 442. |
| 101 | ANTIA R, AHMED H, BULL J J. Directed attenuation to enhance vaccine immunity[J]. PLoS Computational Biology, 2021, 17(2): e1008602. |
| 102 | HAMMARLUND E, LEWIS M W, HANSEN S G, et al. Duration of antiviral immunity after smallpox vaccination[J]. Nature Medicine, 2003, 9(9): 1131-1137. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
