合成生物学 ›› 2023, Vol. 4 ›› Issue (3): 611-627.DOI: 10.12211/2096-8280.2022-075
• 特约评述 • 上一篇
微生物组生物合成基因簇发掘方法及应用前景
赖奇龙, 姚帅, 查毓国, 白虹, 宁康
- 华中科技大学生命科学与技术学院,分子生物物理教育部重点实验室,生物信息与分子成像湖北省重点实验室,人工智能生物学研究中心,生物信息与系统生物学系,湖北 武汉 430074
-
收稿日期:2022-12-26修回日期:2022-03-10出版日期:2023-06-30发布日期:2023-07-05 -
通讯作者:白虹,宁康 -
作者简介:赖奇龙 (2000—),男,学士 。研究方向为生物信息学,人工智能生物学。 E-mail:laiqilong@hust.edu.cn白虹 (1978—),女,正高级工程师。研究方向为天然产物化学,微生物学。 E-mail:baihong@hust.edu.cn宁康 (1979—),男,教授,博士生导师。研究方向为生物信息学,微生物组学,人工智能生物学。 E-mail:ningkang@hust.edu.cn -
基金资助:国家自然科学基金(32071465);国家重点研发计划(2021YFA0910500)
Microbiome-based biosynthetic gene cluster data mining techniques and application potentials
LAI Qilong, YAO Shuai, ZHA Yuguo, BAI Hong, NING Kang
- Key Laboratory of Molecular Biophysics of the Ministry of Education,Hubei Key Laboratory of Bioinformatics and Molecular-imaging,Center of Artificial Intelligence Biology,Department of Bioinformatics and Systems Biology,College of Life Science and Technology,Huazhong University of Science and Technology,Wuhan 430074,Hubei,China
-
Received:2022-12-26Revised:2022-03-10Online:2023-06-30Published:2023-07-05 -
Contact:BAI Hong, NING Kang
摘要:
生物合成基因簇(biosynthetic gene cluster, BGC)是一类非常重要的基因集合(gene set)类型。BGC普遍存在于各类生物基因组中,并且发挥着重要的代谢和调控作用。从线性结构上来说,一个BGC中的基因通常在基因组中处于相邻的位置;从基因功能上来说,一个BGC中的基因通常共同负责一类通路,生成特定的化合物小分子。因此,BGC作为极具潜力的元件来源,在合成生物学研究中极为重要。然而从序列模式上来说,一个BGC中的基因数量众多且序列差异度大,很难通过序列同源性发掘新类型的BGC。因此,建立生物合成基因簇的智能发掘策略,系统性地发掘BGC并进行验证和转化研究,不论在理论方面还是实际应用方面,都具有非常重要的价值。本文主要基于微生物组大数据,较全面地介绍了BGC挖掘的意义和瓶颈问题,系统性地总结了当前BGC发掘中的数据资源和挖掘方法,尤其是人工智能方法,指出了干湿结合方法对于验证新发掘BGC的重要价值,同时展示了新发掘BGC的多样性和广泛应用领域。最后,展望了结合现有BGC挖掘方法和合成生物学转化,将如何在广度和宽度方面扩展目前的合成生物学研究。
中图分类号:
引用本文
赖奇龙, 姚帅, 查毓国, 白虹, 宁康. 微生物组生物合成基因簇发掘方法及应用前景[J]. 合成生物学, 2023, 4(3): 611-627.
LAI Qilong, YAO Shuai, ZHA Yuguo, BAI Hong, NING Kang. Microbiome-based biosynthetic gene cluster data mining techniques and application potentials[J]. Synthetic Biology Journal, 2023, 4(3): 611-627.

图2 BGC挖掘的整体过程(该过程包括:宏基因组数据的整合,基因和潜在BGC的预测,内源表达或异源表达、天然产物的鉴定等。本图中选用的案例是诺糖环肽A2,是从地衣Nostoc属ATCC53789中提取分离的天然产物,可作为20S蛋白酶体的抑制剂,具有抗癌活性[45])
Fig. 2 Overall process for BGC mining(This process includes the integration of metagenomic data, prediction of genes and potential BGC, endogenous or heterologous expression, identification of natural products, etc. The case chosen in this figure is Nostocyclopeptide A2, which is extracted from Nostoc sp. ATCC53789 isolated from lichen. It can be used as an inhibitor of 20S proteasome and exhibits anticancer activity[45].)
| 数据库名称 | 特色 | 网址 | 参考文献 |
|---|---|---|---|
| antiSMASH | 有关次生代谢物BGC的综合资源,集成各种分析工具 | https://antismash.secondarymetabolites.org/ | [ |
| Bactibase | 主要包括细菌及其产生的抗菌肽、细菌素等 | http://bactibase.pfba-lab-tun.org/ | [ |
| BiG-FAM | 将同源BGCs分组到基因簇家族 | https://bigfam.bioinformatics.nl/ | [ |
| ClusterMine360 | 第一个已知产物的BGC数据库 | http://www.clustermine360.ca/ | [ |
| CSDB(ClustScan Database) | 主要内容为PKS、NRPS的BGC | http://csdb.bioserv.pbf.hr/csdb/ClustScanWeb.html | [ |
| DoBISCUIT | 提供由文献给出的PKS和NRPS的BGC | http://www.bio.nite.go.jp/pks/ | [ |
| IMG-ABC | 最大的公开预测的BGC数据库 | https://img.jgi.doe.gov/abc-public | [ |
| MiBiG | 存储BGC的最小信息 | https://mibig.secondarymetabolites.org/ | [ |
| OrphanPKS | 由软件自动提取的多模块PKS序列目录 | http://sequence.stanford.edu/OrphanPKS/ | [ |
表1 代表性BGC数据库介绍
Table 1 Summary for representative BGC databases
| 数据库名称 | 特色 | 网址 | 参考文献 |
|---|---|---|---|
| antiSMASH | 有关次生代谢物BGC的综合资源,集成各种分析工具 | https://antismash.secondarymetabolites.org/ | [ |
| Bactibase | 主要包括细菌及其产生的抗菌肽、细菌素等 | http://bactibase.pfba-lab-tun.org/ | [ |
| BiG-FAM | 将同源BGCs分组到基因簇家族 | https://bigfam.bioinformatics.nl/ | [ |
| ClusterMine360 | 第一个已知产物的BGC数据库 | http://www.clustermine360.ca/ | [ |
| CSDB(ClustScan Database) | 主要内容为PKS、NRPS的BGC | http://csdb.bioserv.pbf.hr/csdb/ClustScanWeb.html | [ |
| DoBISCUIT | 提供由文献给出的PKS和NRPS的BGC | http://www.bio.nite.go.jp/pks/ | [ |
| IMG-ABC | 最大的公开预测的BGC数据库 | https://img.jgi.doe.gov/abc-public | [ |
| MiBiG | 存储BGC的最小信息 | https://mibig.secondarymetabolites.org/ | [ |
| OrphanPKS | 由软件自动提取的多模块PKS序列目录 | http://sequence.stanford.edu/OrphanPKS/ | [ |
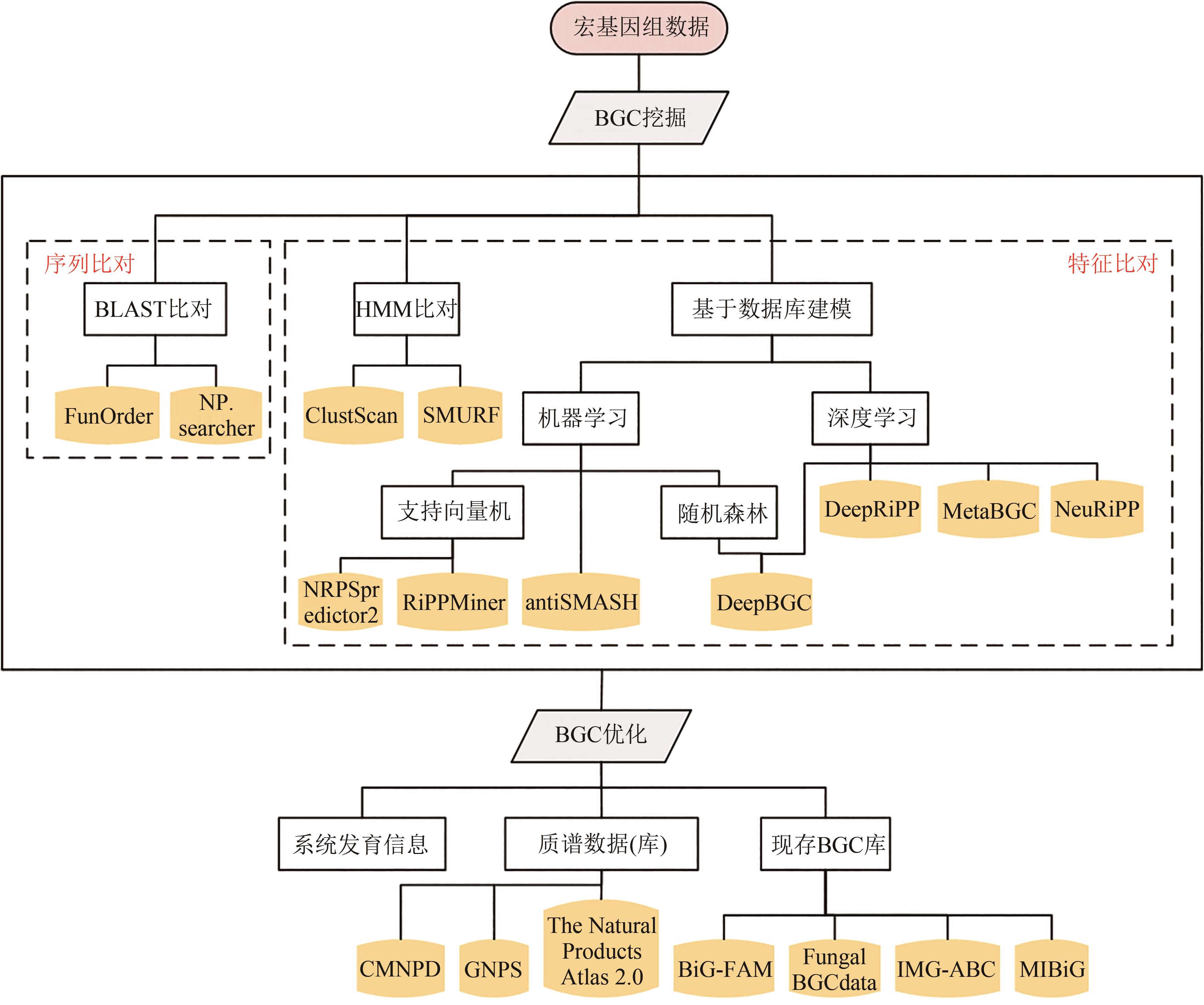
图3 BGC挖掘的一般分析流程及相关方法[从宏基因组数据中挖掘BGC,主要包括:BGC的挖掘方法(序列比对、特征比对等)和BGC的优化方法(数据库搜索、进化分析等)。其中BGC的挖掘方法主要有序列比对和特征比对两大类:序列比对主要是BLAST等方法,特征比对既包括隐马尔科夫模型(HMM)比对等传统方法,也包括基于数据模型的深度学习等方法。其中BGC的优化方法主要有数据库搜索、进化分析等:数据库搜索包括BGC序列数据库的搜索,以及BGC相关小分子质谱数据库的搜索,而进化分析的主要目标是分析BGC的演化和变异模式[54]]
Fig. 3 Overall flow for BGC analysis and mining[It mainly includes: BGC mining methods (sequence alignment, feature characterization, etc.) and BGC optimization methods (database searching, evolutionary analysis, etc.). Among them, the mining methods of BGC mainly include sequence alignment and feature characterization. Sequence alignment mainly uses BLAST and other methods, while feature characterization employs both traditional methods such as hidden Markov model (HMM) alignment and deep learning based on data model. The optimization methods of BGC mainly include database searching, evolutionary analysis, etc. Database searching includes the searching of BGC sequence database and BGC related small molecule mass spectrometry database, and the main purpose of evolutionary analysis is to analyze the evolution and variation patterns of BGC[54].]
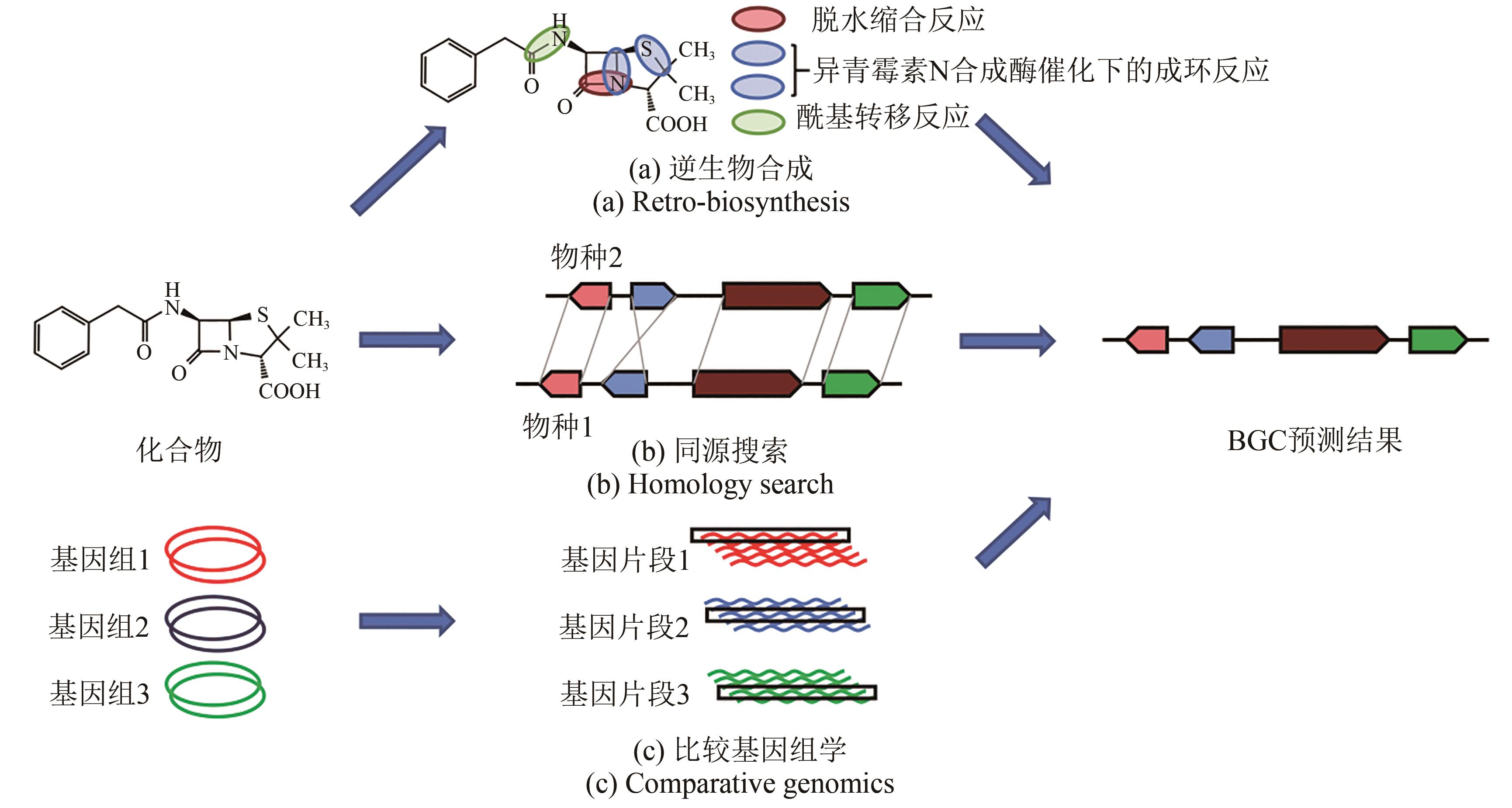
图4 建立BGC和次生代谢产物关联性的分析方法[58](a)逆生物合成:从已知化合物开始,预测生产该化合物所需的活性酶(主干酶和裁剪酶),并从这些预测中找到与基因组中需求匹配的假定簇。本图中选用的案例为青霉素G[59]。(b)同源搜索:从物种1产生的已知化合物和物种2产生的相同或相似的化合物开始,使用来自物种2的已知基因集群在物种1的基因组中搜索相似的基因集群,从而确定感兴趣的基因集群。(c)比较基因组学:从一组生物开始,其中一些生物产生目标化合物,而另一些生物则不产生,有可能在生产中识别同源基因簇,并在非生产中没有同源基因的基础上进行筛选,从而识别候选基因簇
Fig. 4 Analytical methods for establishing correlation between BGC and the production of secondary metabolites[58](a) Retro-biosynthesis: starting with a known compound but no related gene clusters identified, it is possible for predicting enzyme(s) to catalyze the synthesis of such a compound (backbone and tailoring enzymes), and with these predictions putative gene clusters matching the requirements can be found in the genome. The selected case in this figure is penicillin G[59]. (b) Homology searching: starting with a known compound produced by organism 1 and the same or similar compound produced by organism 2 with gene cluster identified, it is possible to use the known gene cluster from organism 2 to search for a similar gene cluster in the genome of organism 1, and thereby identify the gene cluster of interest. (c) Comparative genomics: starting with a group of organisms, some of which produce compounds of interest and some of which do not, it is possible to identify homologous gene clusters in the species that produce them and to screen on the basis of the absence of homologous genes in the species that does not produce them, thereby identifying candidate gene clusters.
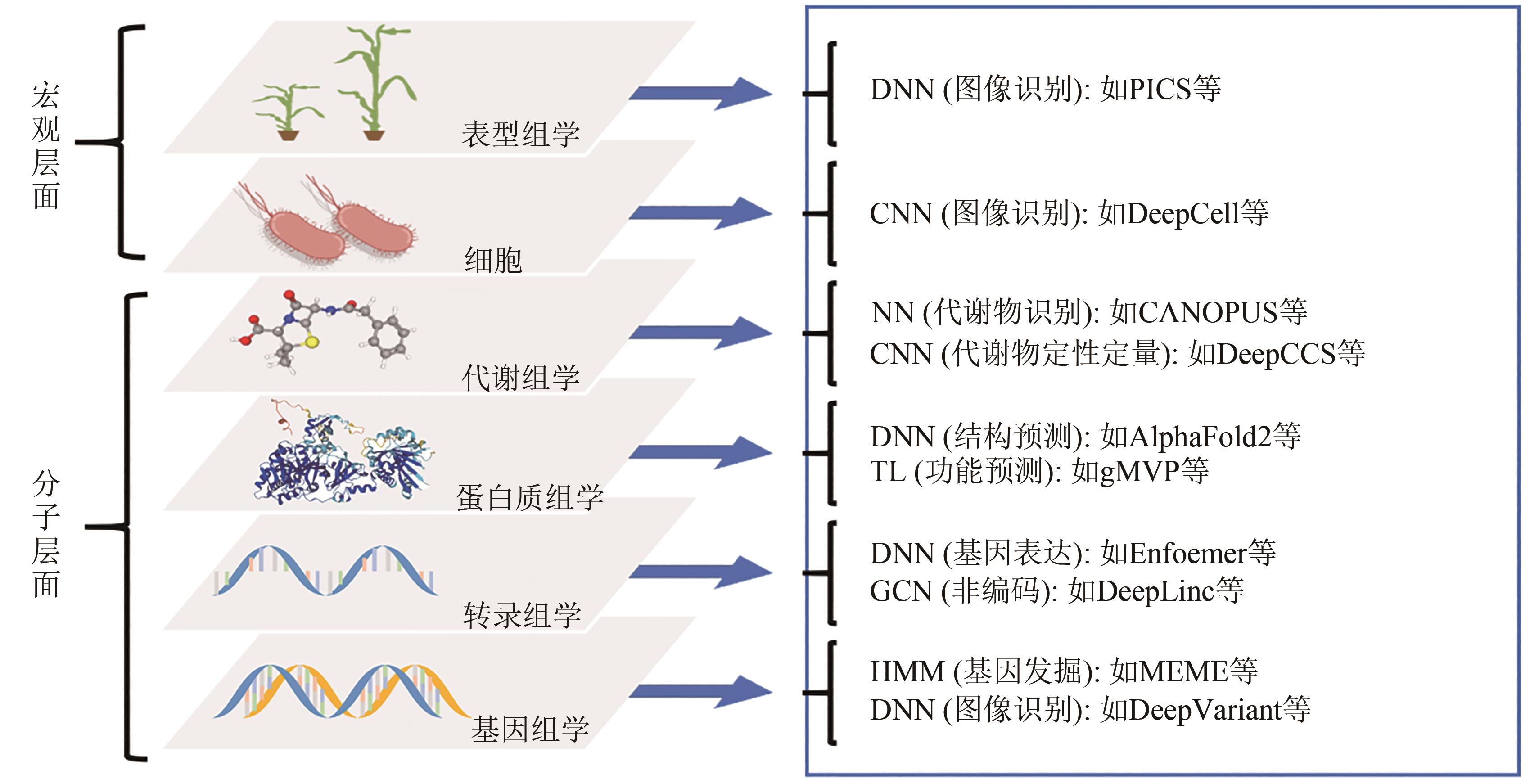
图5 序列数据的类型,以及相应的人工智能分析方法DNN—深度神经网络;CNN—卷积神经网络;NN—神经网络;TL—迁移学习;GCN—图卷及网络;HMM—隐马尔科夫模型
Fig. 5 Types of sequence data and corresponding AI analysis methodsDNN— deep neural network; CNN— convolutional neural network; NN— neural network; TL— transfer learning; GCN— graph convolutional network; HMM— hidden markov model

图6 利用人工智能进行BGC挖掘的现状和趋势(从数据出发,通过人工智能方法进行数据挖掘和模型构建,进而服务于合成生物学的转化研究,产生更多的多模态数据,形成良性循环)
Fig. 6 Status quo and trend of BGC mining using artificial intelligence(Starting from the data, data mining and model construction are carried out with artificial intelligence methods, thus serving the transformation research of synthetic biology, generating more multimodal data and forming a virtuous cycle.)
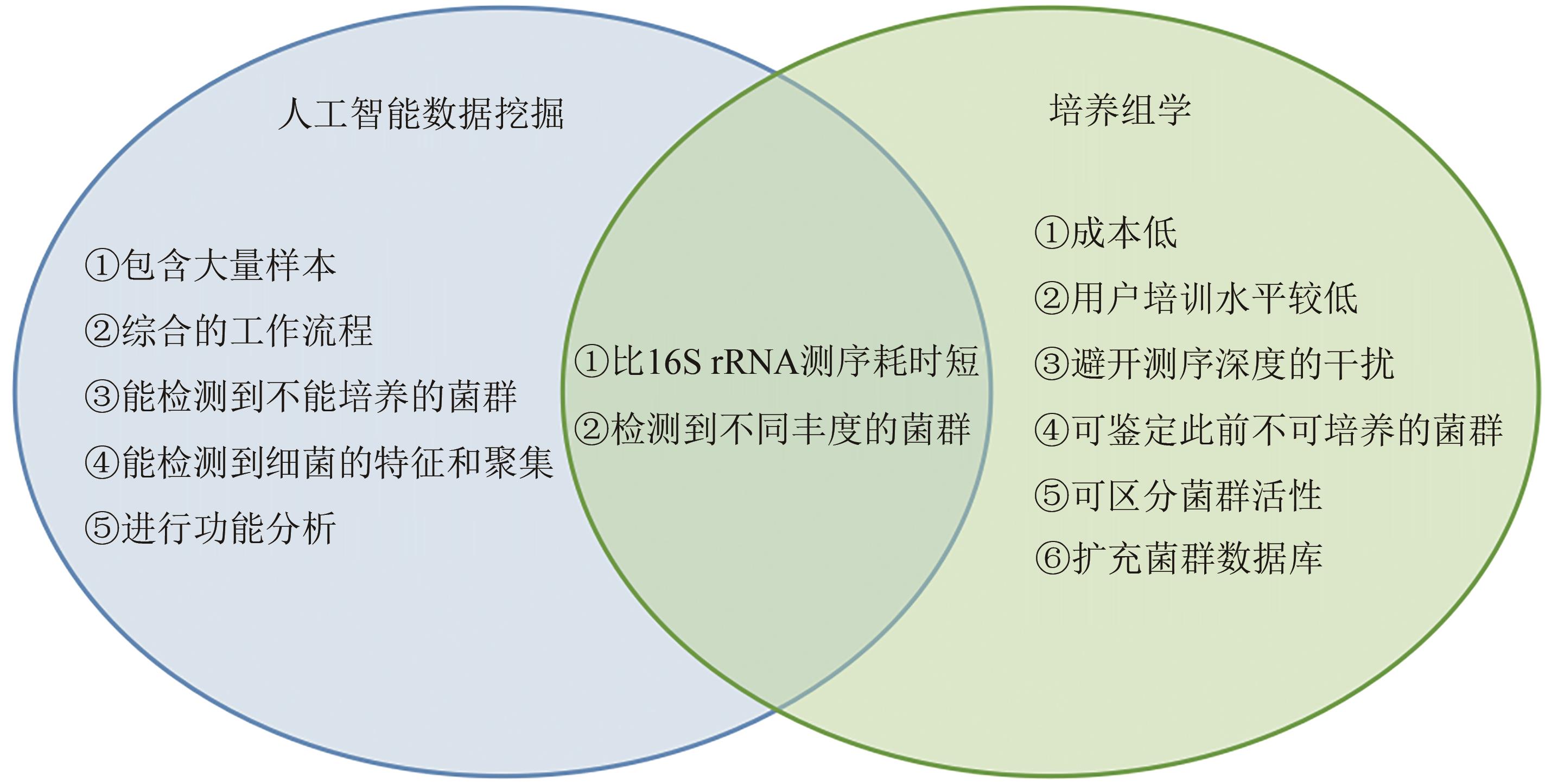
图7 人工智能数据挖掘和培养组学的各自优缺点和互补性(相关方法优缺点的罗列,是基于互相比较和与传统分子生物学方法比较的结果)
Fig. 7 Advantages, disadvantages and complementarities of artificial intelligence data mining and culturomics(The list of advantages and disadvantages of the relevant methods is based on the results of comparison with each other and with traditional molecular biological methods as well.)
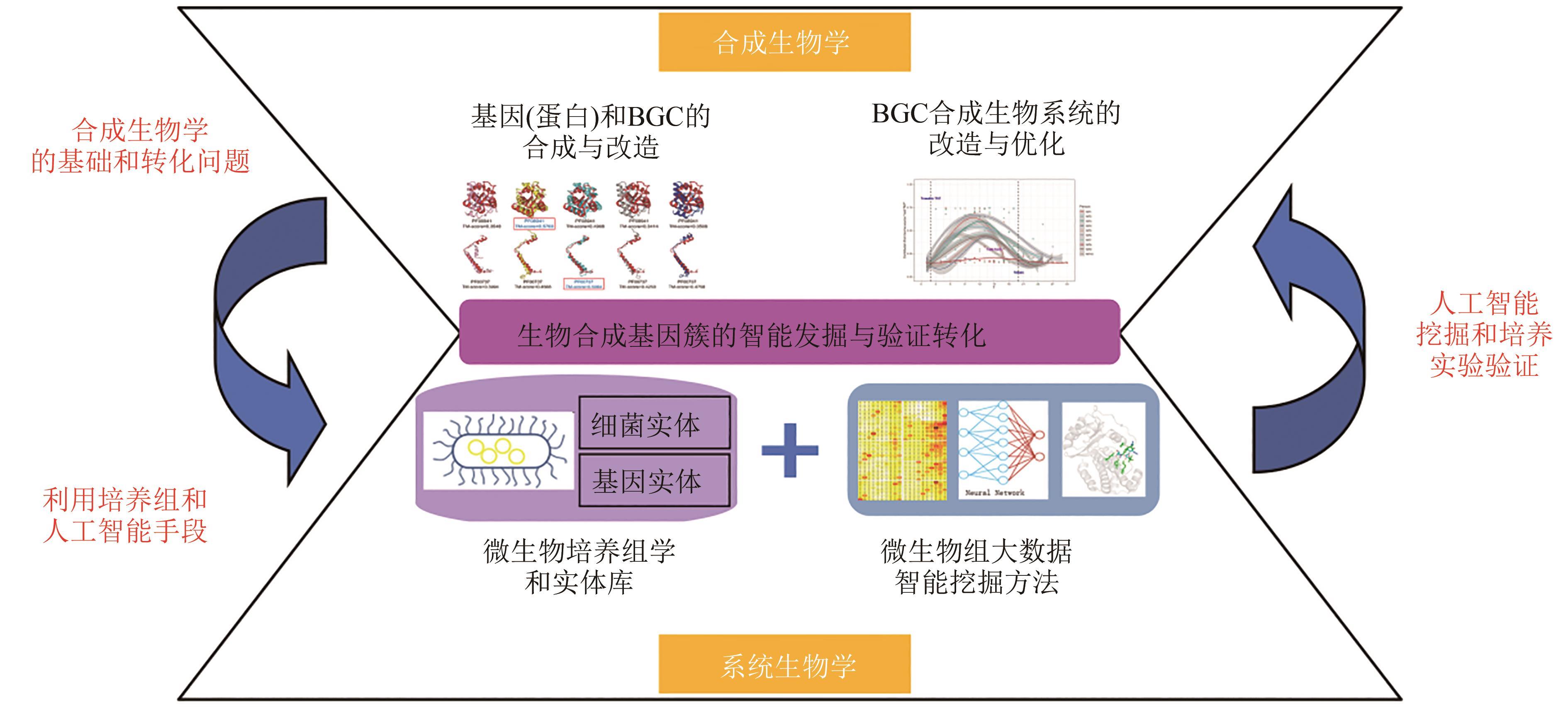
图8 BGC在系统生物学与合成生物学中的核心地位(生物合成基因簇的智能发掘与验证转化的研究,不但在数据上打通了数据库和实体库,而且在技术上打通了人工智能挖掘和培养实验验证。生物合成基因簇的智能发掘与验证转化的研究,能够紧密连接系统生物学与合成生物学,实现从数据到模型、从验证到应用的无缝转化)
Fig. 8 BGC's central role in systems biology and synthetic biology(Research on intelligent mining and verification transformation of biosynthetic gene clusters not only connects BGC database with entity database, but also connects artificial intelligence mining and culture experiment verification. Research on intelligent discovery and transformation verification for biosynthetic gene clusters can closely link systems biology and synthetic biology, and realize seamless transformation from data to model and from verification to application.)
| 1 | ZHANG L X, DEMAIN A L. Natural Products: Drug Discovery, and Therapeutic Medicines[M]. Clifton, New Jersey: Humana Totowa Press, 2005: 382. |
| 2 | SANCHEZ S, GUZMÁN-TRAMPE S, ÁVALOS M, et al. Microbial natural products[M/OL]//Natural Products in Chemical Biology. Hoboken, New Jersey, USA: John Wiley & Sons, Inc., 2012: 65-108 [2022-12-01]. . |
| 3 | LLOYD K G, STEEN A D, LADAU J, et al. Phylogenetically novel uncultured microbial cells dominate earth microbiomes[J]. mSystems, 2018, 3(5): e00055-18. |
| 4 | WOODRUFF H B. Natural products from microorganisms[J]. Science, 1980, 208(4449): 1225-1229. |
| 5 | MEDEMA M H, FISCHBACH M A. Computational approaches to natural product discovery[J]. Nature Chemical Biology, 2015, 11(9): 639-648. |
| 6 | WASCHULIN V, BORSETTO C, JAMES R, et al. Biosynthetic potential of uncultured Antarctic soil bacteria revealed through long-read metagenomic sequencing[J]. The ISME Journal, 2022, 16(1): 101-111. |
| 7 | MARTINET L, NAÔMÉ A, DEFLANDRE B, et al. A single biosynthetic gene cluster is responsible for the production of bagremycin antibiotics and ferroverdin iron chelators[J]. mBio, 2019, 10(4): e01230-19. |
| 8 | SAWANT A M, VAMKUDOTH K R. Biosynthetic process and strain improvement approaches for industrial penicillin production[J]. Biotechnology Letters, 2022, 44(2): 179-192. |
| 9 | KWON M J, STEINIGER C, CAIRNS T C, et al. Beyond the biosynthetic gene cluster paradigm: genome-wide coexpression networks connect clustered and unclustered transcription factors to secondary metabolic pathways[J]. Microbiology Spectrum, 2021, 9(2): e00898-21. |
| 10 | MARTÍN J F. Molecular control of expression of penicillin biosynthesis genes in fungi: regulatory proteins interact with a bidirectional promoter region[J]. Journal of Bacteriology, 2000, 182(9): 2355-2362. |
| 11 | MILLER B L, MILLER K Y, ROBERTI K A, et al. Position-dependent and-independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans [J]. Molecular and Cellular Biology, 1987, 7(1): 427-434. |
| 12 | DEMAIN A L, FANG A. The natural functions of secondary metabolites[M]//Advances in Biochemical Engineering/Biotechnology: History of modern biotechnologyⅠ, Berlin: Springer, 2000, 69: 1-39. |
| 13 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83(3): 770-803. |
| 14 | ARNISON P G, BIBB M J, BIERBAUM G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature[J]. Natural Product Reports, 2013, 30(1): 108-160. |
| 15 | ZHONG Z, HE B B, LI J, et al. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs)[J]. Synthetic and Systems Biotechnology, 2020, 5(3): 155-172. |
| 16 | MEDEMA M H, KOTTMANN R, YILMAZ P, et al. Minimum information about a biosynthetic gene cluster[J]. Nature Chemical Biology, 2015, 11(9): 625-631. |
| 17 | EPSTEIN S C, CHARKOUDIAN L K, MEDEMA M H. A standardized workflow for submitting data to the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository: prospects for research-based educational experiences[J]. Standards in Genomic Sciences, 2018, 13: 16. |
| 18 | KAUTSAR S A, BLIN K, SHAW S, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function[J]. Nucleic Acids Research, 2020, 48(D1): D454-D458. |
| 19 | TERLOUW B R, BLIN K, NAVARRO-MUÑOZ J C, et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters[J]. Nucleic Acids Research, 2023, 51(D1): D603-D610. |
| 20 | ALTSCHUL S F, GISH W, MILLER W, et al. Basic local alignment search tool[J]. Journal of Molecular Biology, 1990, 215(3): 403-410. |
| 21 | RABINER L, JUANG B. An introduction to hidden Markov models[J]. IEEE ASSP Magazine, 1986, 3(1): 4-16. |
| 22 | BLIN K, SHAW S, KLOOSTERMAN A M, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities[J]. Nucleic Acids Research, 2021, 49(W1): W29-W35. |
| 23 | MOHIMANI H, KERSTEN R D, LIU W T, et al. Automated genome mining of ribosomal peptide natural products[J]. ACS Chemical Biology, 2014, 9(7): 1545-1551. |
| 24 | SUGIMOTO Y, CAMACHO F R, WANG S, et al. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome[J]. Science, 2019, 366(6471): eaax9176. |
| 25 | HANNIGAN G D, PRIHODA D, PALICKA A, et al. A deep learning genome-mining strategy for biosynthetic gene cluster prediction[J]. Nucleic Acids Research, 2019, 47(18): e110. |
| 26 | RUIZ B, CHÁVEZ A, FORERO A, et al. Production of microbial secondary metabolites: regulation by the carbon source[J]. Critical Reviews in Microbiology, 2010, 36(2): 146-167. |
| 27 | O'BRIEN J, WRIGHT G D. An ecological perspective of microbial secondary metabolism[J]. Current Opinion in Biotechnology, 2011, 22(4): 552-558. |
| 28 | SEYEDSAYAMDOST M R. Toward a global picture of bacterial secondary metabolism[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(3/4): 301-311. |
| 29 | KALKREUTER E, PAN G H, CEPEDA A J, et al. Targeting bacterial genomes for natural product discovery[J]. Trends in Pharmacological Sciences, 2020, 41(1): 13-26. |
| 30 | YUAN Y J, CHENG S, BIAN G K, et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi[J]. Nature Catalysis, 2022, 5(4): 277-287. |
| 31 | BURIAN J, LIBIS V K, HERNANDEZ Y A, et al. High-throughput retrieval of target sequences from complex clone libraries using CRISPRi[J]. Nature Biotechnology, 2022: 1-5. |
| 32 | PAOLI L, RUSCHEWEYH H J, FORNERIS C C, et al. Biosynthetic potential of the global ocean microbiome[J]. Nature, 2022, 607(7917): 111-118. |
| 33 | PATEL J R, OH J S, WANG S Q, et al. Cross-kingdom expression of synthetic genetic elements promotes discovery of metabolites in the human microbiome[J]. Cell, 2022, 185(9): 1487-1505.e14. |
| 34 | DIRENÇ MUNGAN M, BLIN K, ZIEMERT N. ARTS-DB: a database for antibiotic resistant targets[J]. Nucleic Acids Research, 2022, 50(D1): D736-D740. |
| 35 | NAYFACH S, ROUX S, SESHADRI R, et al. A genomic catalog of Earth's microbiomes[J]. Nature Biotechnology, 2021, 39(4): 499-509. |
| 36 | VAN BERGEIJK D A, TERLOUW B R, MEDEMA M H, et al. Ecology and genomics of Actinobacteria: new concepts for natural product discovery[J]. Nature Reviews Microbiology, 2020, 18(10): 546-558. |
| 37 | BARBOUR A, WESCOMBE P, SMITH L. Evolution of lantibiotic salivaricins: new weapons to fight infectious diseases[J]. Trends in Microbiology, 2020, 28(7): 578-593. |
| 38 | CARRIÓN V J, PEREZ-JARAMILLO J, CORDOVEZ V, et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome[J]. Science, 2019, 366(6465): 606-612. |
| 39 | ZHAO H, FU S L, YU Y F, et al. MetaMed: Linking microbiota functions with medicine therapeutics[J]. mSystems, 2019, 4(5): e00413-19. |
| 40 | CHU J, VILA-FARRES X, BRADY S F. Bioactive synthetic-bioinformatic natural product cyclic peptides inspired by nonribosomal peptide synthetase gene clusters from the human microbiome[J]. Journal of the American Chemical Society, 2019, 141(40): 15737-15741. |
| 41 | WANG L L, RAVICHANDRAN V, YIN Y L, et al. Natural products from mammalian gut microbiota[J]. Trends in Biotechnology, 2019, 37(5): 492-504. |
| 42 | SKELLAM E. Strategies for engineering natural product biosynthesis in fungi[J]. Trends in Biotechnology, 2019, 37(4): 416-427. |
| 43 | RUTLEDGE P J, CHALLIS G L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters[J]. Nature Reviews Microbiology, 2015, 13(8): 509-523. |
| 44 | DONIA M S, CIMERMANCIC P, SCHULZE C J, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics[J]. Cell, 2014, 158(6): 1402-1414. |
| 45 | GOLAKOTI T, YOSHIDA W Y, CHAGANTY S, et al. Isolation and structure determination of nostocyclopeptides A1 and A2 from the terrestrial Cyanobacterium Nostoc sp. ATCC53789[J]. Journal of Natural Products, 2001, 64(1): 54-59. |
| 46 | HAMMAMI R, ZOUHIR A, LE LAY C, et al. BACTIBASE second release: a database and tool platform for bacteriocin characterization[J]. BMC Microbiology, 2010, 10: 22. |
| 47 | KAUTSAR S A, BLIN K, SHAW S, et al. BiG-FAM: the biosynthetic gene cluster families database[J]. Nucleic Acids Research, 2021, 49(D1): D490-D497. |
| 48 | CONWAY K R, BODDY C N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis[J]. Nucleic Acids Research, 2013, 41(D1): D402-D407. |
| 49 | DIMINIC J, ZUCKO J, RUZIC I T, et al. Databases of the thiotemplate modular systems (CSDB) and their in silico recombinants (r-CSDB)[J]. Journal of Industrial Microbiology & Biotechnology, 2013, 40(6): 653-659. |
| 50 | ICHIKAWA N, SASAGAWA M, YAMAMOTO M, et al. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters[J]. Nucleic Acids Research, 2013, 41(D1): D408-D414. |
| 51 | PALANIAPPAN K, CHEN I M A, CHU K, et al. IMG-ABC v.5.0: an update to the IMG/Atlas of biosynthetic gene clusters knowledgebase[J]. Nucleic Acids Research, 2020, 48(D1): D422-D430. |
| 52 | O'BRIEN R V, DAVIS R W, KHOSLA C, et al. Computational identification and analysis of orphan assembly-line polyketide synthases[J]. The Journal of Antibiotics, 2014, 67(1): 89-97. |
| 53 | KAUTSAR S A, VAN DER HOOFT J J J, DE RIDDER D, et al. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters[J]. GigaScience, 2021, 10(1): giaa154. |
| 54 | TRAN P N, YEN M R, CHIANG C Y, et al. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi[J]. Applied Microbiology and Biotechnology, 2019, 103(8): 3277-3287. |
| 55 | MEDEMA M H, BLIN K, CIMERMANCIC P, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences[J]. Nucleic Acids Research, 2011, 39(): W339-W346. |
| 56 | MISTRY J, FINN R D, EDDY S R, et al. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions[J]. Nucleic Acids Research, 2013, 41(12): e121. |
| 57 | EDDY S R. Profile hidden Markov models[J]. Bioinformatics, 1998, 14(9): 755-763. |
| 58 | KJÆRBØLLING I, MORTENSEN U H, VESTH T, et al. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites[J]. Fungal Genetics and Biology, 2019, 130: 107-121. |
| 59 | RABE P, KAMPS J J A G, SUTHERLIN K D, et al. X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis[J]. Science Advances, 2021, 7(34): eabh0250. |
| 60 | CHING T, HIMMELSTEIN D S, BEAULIEU-JONES B K, et al. Opportunities and obstacles for deep learning in biology and medicine[J]. Journal of the Royal Society, Interface, 2018, 15(141): 20170387. |
| 61 | JING Y K, BIAN Y M, HU Z H, et al. Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era[J]. The AAPS Journal, 2018, 20(3): 58. |
| 62 | SHEN D G, WU G R, SUK H I. Deep learning in medical image analysis[J]. Annual Review of Biomedical Engineering, 2017, 19: 221-248. |
| 63 | HAMET P, TREMBLAY J. Artificial intelligence in medicine[J]. Metabolism 2017, 69S: S36-S40. |
| 64 | JURTZ V I, JOHANSEN A R, NIELSEN M, et al. An introduction to deep learning on biological sequence data: examples and solutions[J]. Bioinformatics, 2017, 33(22): 3685-3690. |
| 65 | KANDEL M E, HE Y R, LEE Y J, et al. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments[J]. Nature Communications, 2020, 11: 6256. |
| 66 | BANNON D, MOEN E, SCHWARTZ M, et al. DeepCell Kiosk: scaling deep learning—enabled cellular image analysis with Kubernetes[J]. Nature Methods, 2021, 18(1): 43-45. |
| 67 | AVSEC Ž, AGARWAL V, VISENTIN D, et al. Effective gene expression prediction from sequence by integrating long-range interactions[J]. Nature Methods, 2021, 18(10): 1196-1203. |
| 68 | LI R Z, YANG X R. De novo reconstruction of cell interaction landscapes from single-cell spatial transcriptome data with DeepLinc[J]. Genome Biology, 2022, 23(1): 124. |
| 69 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 70 | CUI M, ZHANG D Y. Artificial intelligence and computational pathology[J]. Laboratory Investigation, 2021, 101(4): 412-422. |
| 71 | MESKO B. The role of artificial intelligence in precision medicine[J]. Expert Review of Precision Medicine and Drug Development, 2017, 2(5): 239-241. |
| 72 | CIMERMANCIC P, MEDEMA M H, CLAESEN J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters[J]. Cell, 2014, 158(2): 412-421. |
| 73 | MINOWA Y, ARAKI M, KANEHISA M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes[J]. Journal of Molecular Biology, 2007, 368(5): 1500-1517. |
| 74 | CAMACHO C, COULOURIS G, AVAGYAN V, et al. BLAST+: architecture and applications[J]. BMC Bioinformatics, 2009, 10: 421. |
| 75 | MEDEMA M H, PAALVAST Y, NGUYEN D D, et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products[J]. PLoS Computational Biology, 2014, 10(9): e1003822. |
| 76 | ALANJARY M, KRONMILLER B, ADAMEK M, et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery[J]. Nucleic Acids Research, 2017, 45(W1): W42-W48. |
| 77 | BLIN K, PEDERSEN L E, WEBER T, et al. CRISPy-web: an online resource to design sgRNAs for CRISPR applications[J]. Synthetic and Systems Biotechnology, 2016, 1(2): 118-121. |
| 78 | HERTWECK C, LUZHETSKYY A, REBETS Y, et al. Type Ⅱ polyketide synthases: gaining a deeper insight into enzymatic teamwork [J]. Natural product reports, 2007, 24(1): 162-190. |
| 79 | FENG Z Y, KALLIFIDAS D, BRADY S F. Functional analysis of environmental DNA-derived type Ⅱ polyketide synthases reveals structurally diverse secondary metabolites[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(31): 12629-12634. |
| 80 | GRAVES A, SCHMIDHUBER J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures[J]. Neural Networks, 2005, 18(5/6): 602-610. |
| 81 | MIKOLOV T, CHEN K, CORRADO G, et al. Efficient estimation of word representations in vector space[EB/OL]. arXiv, 2013: 1301.3781[2022-12-01]. . |
| 82 | BREIMAN L. Random forests[J].Machine Learning, 2001, 45(1): 5-32. |
| 83 | SCHERLACH K, HERTWECK C. Mining and unearthing hidden biosynthetic potential[J]. Nature Communications, 2021, 12(1): 3864. |
| 84 | ALBARANO L, ESPOSITO R, RUOCCO N, et al. Genome mining as new challenge in natural products discovery[J]. Marine Drugs, 2020, 18(4): 199. |
| 85 | NAVARRO-MUÑOZ J C, SELEM-MOJICA N, MULLOWNEY M W, et al. A computational framework to explore large-scale biosynthetic diversity[J]. Nature Chemical Biology, 2020, 16(1): 60-68. |
| 86 | MILLER D, STERN A, BURSTEIN D. Deciphering microbial gene function using natural language processing[J]. Nature Communications, 2022, 13: 5731. |
| 87 | HA C W Y, DEVKOTA S. The new microbiology: cultivating the future of microbiome-directed medicine[J]. American Journal of Physiology Gastrointestinal and Liver Physiology, 2020, 319(6): G639-G645. |
| 88 | LAGIER J C, DUBOURG G, MILLION M, et al. Culturing the human microbiota and culturomics[J]. Nature Reviews Microbiology, 2018, 16(9): 540-550. |
| 89 | DEMAIN A L, SANCHEZ S. Microbial drug discovery: 80 years of progress[J]. The Journal of Antibiotics, 2009, 62(1): 5-16. |
| 90 | ALMEIDA A, MITCHELL A L, BOLAND M, et al. A new genomic blueprint of the human gut microbiota[J]. Nature, 2019, 568(7753): 499-504. |
| 91 | PASOLLI E, ASNICAR F, MANARA S, et al. Extensive unexplored human microbiome diversity revealed by over 150, 000 genomes from metagenomes spanning age, geography, and lifestyle[J]. Cell, 2019, 176(3): 649-662.e20. |
| 92 | CRITS-CHRISTOPH A, DIAMOND S, BUTTERFIELD C N, et al. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis[J]. Nature, 2018, 558(7710): 440-444. |
| 93 | FIERER N. Embracing the unknown: disentangling the complexities of the soil microbiome[J]. Nature Reviews Microbiology, 2017, 15(10): 579-590. |
| 94 | BERGMANN G T, BATES S T, EILERS K G, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities[J]. Soil Biology and Biochemistry, 2011, 43(7): 1450-1455. |
| 95 | KIELAK A M, BARRETO C C, KOWALCHUK G A, et al. The ecology of acidobacteria: moving beyond genes and genomes[J]. Frontiers in Microbiology, 2016, 7: 744. |
| 96 | MORAN M A, KUJAWINSKI E B, SCHROER W F, et al. Microbial metabolites in the marine carbon cycle[J]. Nature Microbiology, 2022, 7(4): 508-523. |
| 97 | REICH H J, HONDAL R J. Why nature chose selenium[J]. ACS Chemical Biology, 2016, 11(4): 821-841. |
| 98 | KAYROUZ C M, HUANG J, HAUSER N, et al. Biosynthesis of selenium-containing small molecules in diverse microorganisms[J]. Nature, 2022, 610(7930): 199-204. |
| 99 | GONCHARENKO K V, VIT A, BLANKENFELDT W, et al. Structure of the sulfoxide synthase EgtB from the ergothioneine biosynthetic pathway[J]. Angewandte Chemie International Edition, 2015, 54(9): 2821-2824. |
| 100 | BIAN G K, DENG Z X, LIU T G. Strategies for terpenoid overproduction and new terpenoid discovery[J]. Current Opinion in Biotechnology, 2017, 48: 234-241. |
| 101 | RENNER M K, JENSEN P R, FENICAL W. Mangicols: structures and biosynthesis of a new class of sesterterpene polyols from a marine fungus of the genus Fusarium[J]. The Journal of Organic Chemistry, 2000, 65(16): 4843-4852. |
| 102 | HEINEMANN M, PANKE S. Synthetic biology—putting engineering into biology[J]. Bioinformatics, 2006, 22(22): 2790-2799. |
| 103 | LI L, LIU X C, JIANG W H, et al. Recent advances in synthetic biology approaches to optimize production of bioactive natural products in Actinobacteria[J]. Frontiers in Microbiology, 2019, 10: 2467. |
| 104 | MALICO A A, NICHOLS L, WILLIAMS G J. Synthetic biology enabling access to designer polyketides[J]. Current Opinion in Chemical Biology, 2020, 58: 45-53. |
| 105 | PYE C R, BERTIN M J, LOKEY R S, et al. Retrospective analysis of natural products provides insights for future discovery trends[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(22): 5601-5606. |
| 106 | LEE N M, HWANG S K, KIM J H, et al. Mini review: genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces [J]. Computational and Structural Biotechnology Journal, 2020, 18: 1548-1556. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

