合成生物学 ›› 2024, Vol. 5 ›› Issue (6): 1419-1436.DOI: 10.12211/2096-8280.2024-016
基于极端微生物的生物制造
邵明威1, 孙思勉1, 杨时茂1, 陈国强1,2,3
- 1.清华大学生命科学学院,北京 100084
2.清华大学合成与系统生物学中心,北京 100084
3.清华大学化学工程系,教育部工业生物催化重点实验室,北京 100084
-
收稿日期:2024-02-04修回日期:2024-04-27出版日期:2024-12-31发布日期:2025-01-10 -
通讯作者:陈国强 -
作者简介:邵明威 (2001—),男,博士研究生。研究方向为盐单胞菌进化系统构建与应用。E-mail:shaomingwei@phalab.org陈国强 (1963—),男,博士,教授。研究方向为微生物聚羟基脂肪酸酯(PHA)的合成、代谢和应用。E-mail:chengq@mail.tsinghua.edu.cn -
基金资助:国家自然科学基金(32130001)
Bioproduction based on extremophiles
SHAO Mingwei1, SUN Simian1, YANG Shimao1, CHEN Guoqiang1,2,3
- 1.School of Life Sciences,Tsinghua University,Beijing 100084,China
2.Center for Synthetic and Systems Biology,Tsinghua University,Beijing 100084,China
3.MOE Key Lab of Industrial Biocatalysis,Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
-
Received:2024-02-04Revised:2024-04-27Online:2024-12-31Published:2025-01-10 -
Contact:CHEN Guoqiang
摘要:
以微生物或酶为基础的生物制造,正以其绿色、环保、可持续等优势逐渐替代以化石燃料为原料的传统化工生产模式。然而,传统工业生物技术存在易染菌、设备复杂、难以连续发酵等劣势。相较而言,“下一代工业生物技术”(NGIB)利用以嗜盐菌、嗜热菌和嗜酸碱菌等极端微生物作为底盘细胞,使用廉价底物生产多种高附加值产品,具有开放、无需灭菌、连续发酵等优点。本文介绍了嗜盐菌、嗜热菌和嗜酸碱菌极端微生物的定义以及在高盐、高温、极度酸碱等极端环境下快速生长的特性。随后总结了目前极端微生物的基因工程手段例如启动子工程、以CRISPR为代表的基因编辑技术、命运共同体策略、稳定质粒载体等,代谢工程手段例如增加碳源前体、敲除旁路代谢、减少副产物、提高转运等,以及极端微生物生产的多种产品例如PHA、蛋白质、氨基酸及小分子衍生物等。同时概括了目前在极端微生物底盘细胞改造过程中仍存在的问题,如缺乏多种优秀的质粒载体、质粒转化效率低、缺乏高效基因编辑技术以及其他非嗜盐菌生长发酵周期较长等,并提出了相应的解决策略。最后展望了如何充分利用不同类型极端微生物的特性生产优势产品,推动下一代工业生物技术的发展与完善,实现绿色、环保、可持续的生物制造。
中图分类号:
引用本文
邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436.
SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles[J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436.
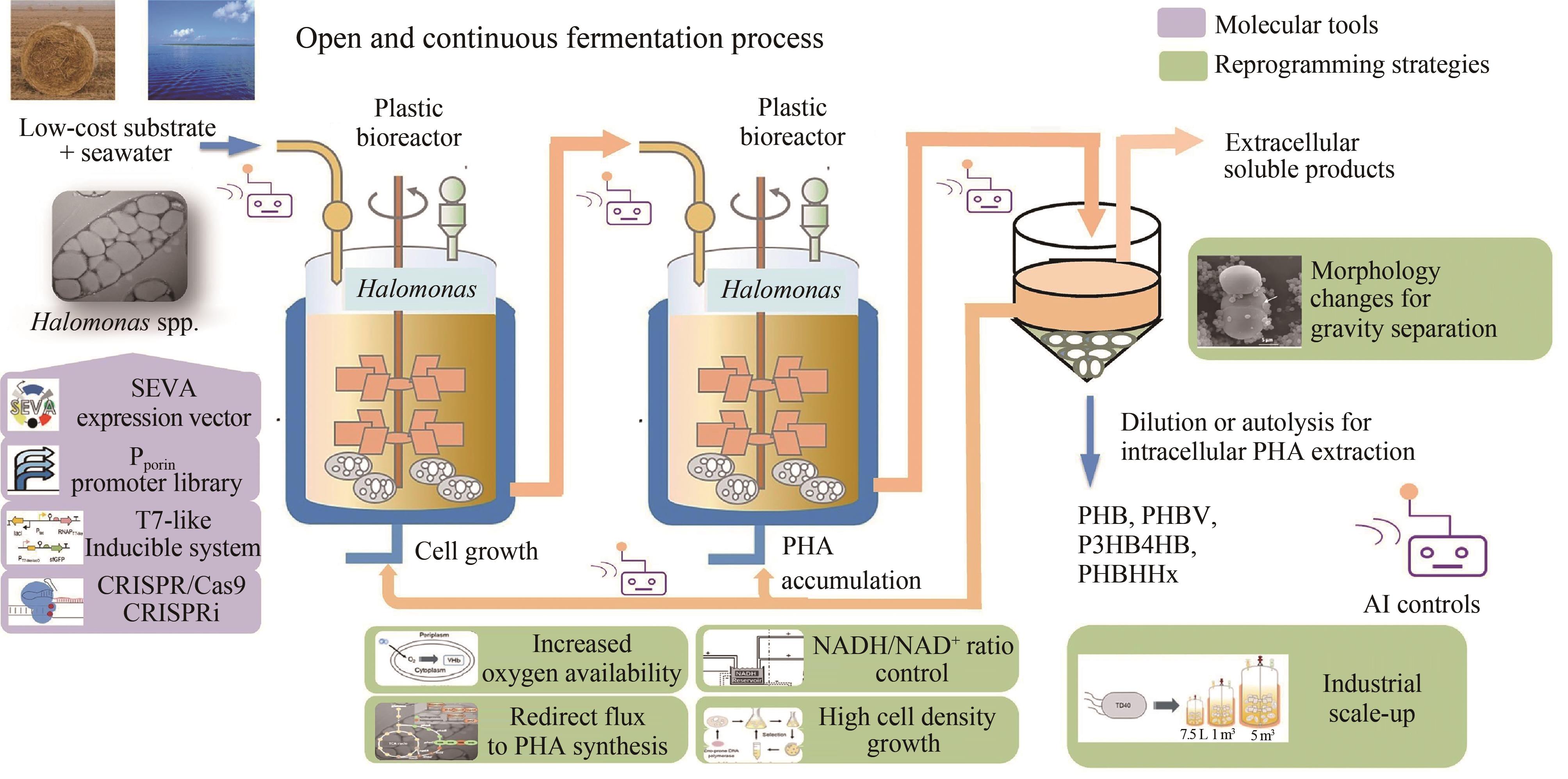
图1 基于以盐单胞菌为代表的极端微生物下一代工业生物技术示意图[12](使用重组盐单胞菌作为底盘细胞,在塑料或其他低成本生物反应器中利用海水和廉价的底物进行开放、无需灭菌和连续发酵,生产各种类型的PHA、蛋白质和胞外小分子产物)
Fig. 1 Next generation industrial biotechnology based on extremophiles represented by Halomonas spp. [12](Production of intracellular PHA products or proteins and extracelullar small molecules by recombinant Halomonas grown on low-cost substrates in seawater under open unsterile and continuous processes conducted in plastic or other low cost bioreactors)
| 类型 | 生长环境 | 代表微生物 | 生产应用 | 参考文献 |
|---|---|---|---|---|
| 嗜盐菌 | 高盐高碱(NaCl>30 g/L, pH 8~10) | 盐单胞菌:H. bluephagenesis TD01,H.campaniensis LS21,H. smyrnensis AAD6 | 生产PHA(以H. bluephagenesis为例:细胞干重大于80 g/L,PHB质量分数大于90%),四氢嘧啶[以H. bluephagenesis为例,生产滴度达85 g/L,生产速率达1 g/(L·h)]等 | [ |
| 嗜热菌 | 高温(>45 ℃) | 热细菌:Fervidobacterium thermophilum,Thermoanaerobacterium saccharolyticum | 生产生物燃料[以Thermoanaerobacter sp.X514为例,正丁醇生产滴度达357 mg/L,生产速率达2.975 g/(L·h)],分离热稳定酶(纤维素酶,角质溶解酶)等 | [ |
| 嗜酸/碱菌 | 极酸或极碱(pH<3,pH>10) | 嗜酸氧化亚铁硫杆菌Acidithiobacillus ferrooxidans,嗜碱细菌Clostridium alkalicellulosi | 生产有机酸(以Issatchenkia orientalis SD108为例,琥珀酸生产滴度达到11.63 g/L) | [ |
表1 各种类型极端微生物的特性与生产应用
Table 1 Characteristics and production applications of various types of extremophiles
| 类型 | 生长环境 | 代表微生物 | 生产应用 | 参考文献 |
|---|---|---|---|---|
| 嗜盐菌 | 高盐高碱(NaCl>30 g/L, pH 8~10) | 盐单胞菌:H. bluephagenesis TD01,H.campaniensis LS21,H. smyrnensis AAD6 | 生产PHA(以H. bluephagenesis为例:细胞干重大于80 g/L,PHB质量分数大于90%),四氢嘧啶[以H. bluephagenesis为例,生产滴度达85 g/L,生产速率达1 g/(L·h)]等 | [ |
| 嗜热菌 | 高温(>45 ℃) | 热细菌:Fervidobacterium thermophilum,Thermoanaerobacterium saccharolyticum | 生产生物燃料[以Thermoanaerobacter sp.X514为例,正丁醇生产滴度达357 mg/L,生产速率达2.975 g/(L·h)],分离热稳定酶(纤维素酶,角质溶解酶)等 | [ |
| 嗜酸/碱菌 | 极酸或极碱(pH<3,pH>10) | 嗜酸氧化亚铁硫杆菌Acidithiobacillus ferrooxidans,嗜碱细菌Clostridium alkalicellulosi | 生产有机酸(以Issatchenkia orientalis SD108为例,琥珀酸生产滴度达到11.63 g/L) | [ |
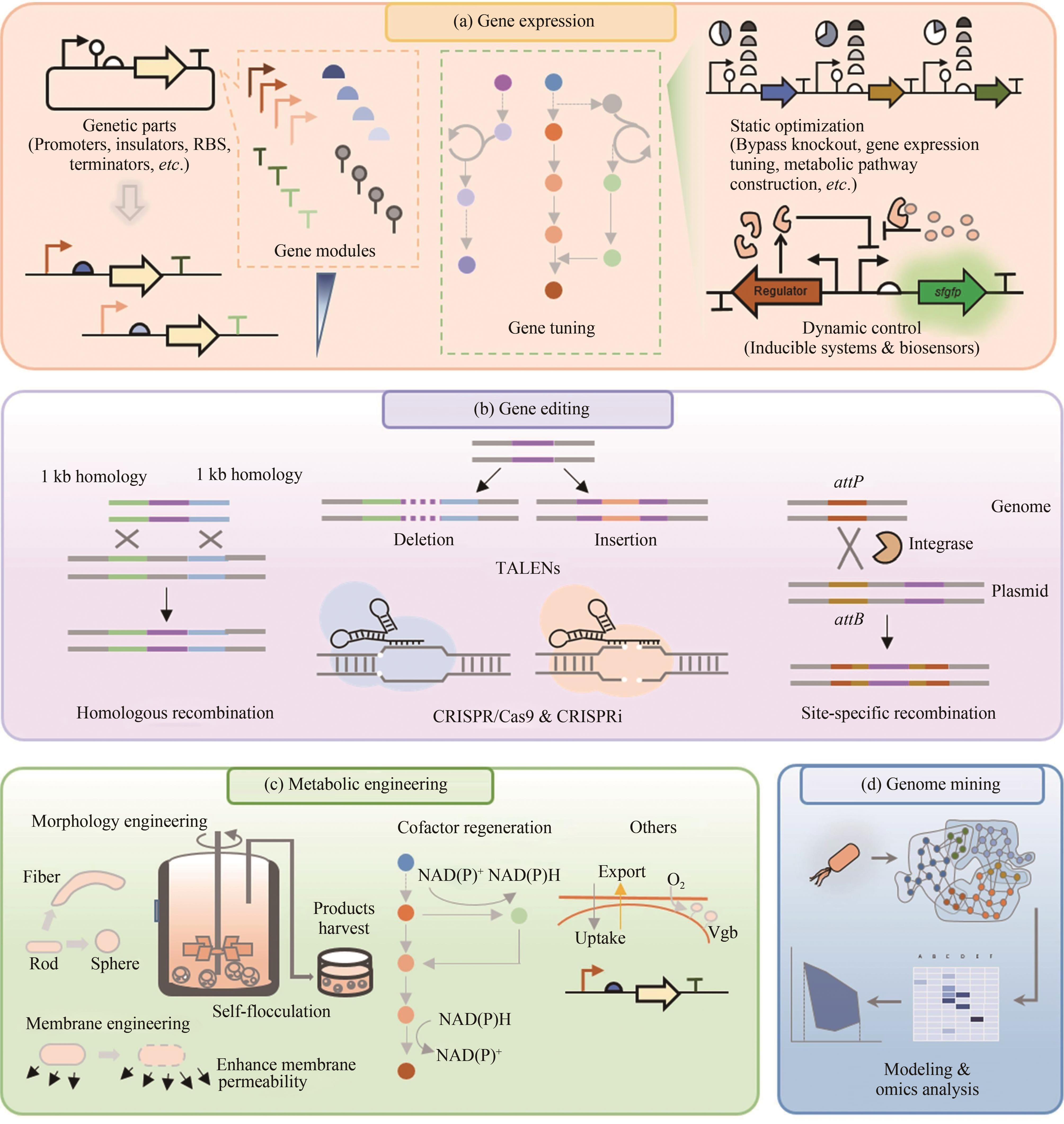
图2 极端微生物的基因工程与代谢工程[51](a)基因表达调控元件和工具。基因元件包括启动子、绝缘子、核糖体结合位点和终止子;基因调控包括静态调控(例如旁路敲除),通路过表达以及动态调控(例如化学诱导系统,生物传感器)。(b)基因编辑技术包括同源重组、TALENs、CRISPR/Cas9和特异位点重组。(c)代谢工程包括形态学工程、膜工程、调控NAD(P)+/NAD(P)H比例、表达血红蛋白。(d)基因组挖掘。利用组学数据构建代谢模型预测新的元件、酶和通路
Fig. 2 Genetic and metabolic engineering of extremophiles[51](a) Gene expression regulatory elements and tools. Genetic elements include promoters, insulators, ribosome binding sites, and terminators. Gene regulation includes static regulation such as bypass knockout, pathway overexpression, and dynamic regulation such as chemical-induction system, biosensors; (b) Gene editing tools include homologous recombination, TALENs, CRISPR/Cas9 and specific-site recombination; (c) Metabolic engineering includes morphological engineering, membrane engineering, regulation of NAD(P)+/NAD(P)H ratio, and expression of hemoglobin; (D) Genome mining. The applications of omics data to build metabolic models for prediction of new elements, enzymes, and pathways
| 工程化改造思路 | 具体策略 | 参考文献 |
|---|---|---|
| 基因表达元件工程 | porin启动子,类T7表达系统,响应十种小分子诱导物的多重诱导系统 | [ |
| 基因编辑技术 | CRISPR/Cas9系统,CRISPRi系统,CRISPR/AID系统,sRNA调控系统 | [ |
| 生物传感器与动态调控 | 感知油酸、群体感应信号分子AHL调控系统,荧光定量PHA含量(qPHA) | [ |
| 命运共同体策略 | 将PHA合成基因插入必需基因ompW启动子后共表达 | [ |
| 形态学工程 | 抑制细胞骨架基因mreB,ftsZ的表达 | [ |
| 稳定质粒表达载体 | 基于盐单胞菌内源性质粒中的hbpB/hbpC毒素-抗毒素系统 | [ |
表2 盐单胞菌的基因工程改造技术
Table 2 Genetic engineering of Halomonas spp.
| 工程化改造思路 | 具体策略 | 参考文献 |
|---|---|---|
| 基因表达元件工程 | porin启动子,类T7表达系统,响应十种小分子诱导物的多重诱导系统 | [ |
| 基因编辑技术 | CRISPR/Cas9系统,CRISPRi系统,CRISPR/AID系统,sRNA调控系统 | [ |
| 生物传感器与动态调控 | 感知油酸、群体感应信号分子AHL调控系统,荧光定量PHA含量(qPHA) | [ |
| 命运共同体策略 | 将PHA合成基因插入必需基因ompW启动子后共表达 | [ |
| 形态学工程 | 抑制细胞骨架基因mreB,ftsZ的表达 | [ |
| 稳定质粒表达载体 | 基于盐单胞菌内源性质粒中的hbpB/hbpC毒素-抗毒素系统 | [ |
| 产物 | 碳源/底物 | 改造策略 | 产量 | 参考文献 |
|---|---|---|---|---|
| PHB | 葡萄糖,淀粉水解物,厨余垃圾混合碳源 | 限氧,限氮;平衡NADH+/NAD比例,补充乙酸 | 细胞干重大于80 g/L,PHB质量分数大于90% | [ |
| 3-羟基丁酸与4-羟基丁酸共聚物P(3HB-co-4HB) | 葡萄糖,葡萄糖酸盐废弃物,废弃玉米浆,γ-丁内酯 | 构建两条相互关联的4-羟基丁酸(4HB)生物合成途径,表达4-羟基丁酸转移酶基因orfZ;敲除琥珀酸半醛脱氢酶基因gadD;构建数学模型与理性计算辅助设计;敲除外膜相关基因lpxL和lpxM | 7 L发酵罐产生26.3 g/L细胞干重,包含质量分数60.5%的P(3HB-co-4HB),其中4HB的摩尔分数为17.04%;5 L发酵罐中产生100 g/L细胞干重包含质量分数60.4%的 P(3HB-co-4HB),其中4HB的摩尔分数为13.5%;敲除外膜菌H. bluephagenesis WZY254在7 L发酵罐产生84 g/L细胞干重包含质量分数81%的P(3HB-co-4HB),其中4HB的摩尔分数为26% | [ |
| 3-羟基丁酸与3-羟基戊酸共聚物(PHBV) | 葡萄糖,葡萄糖酸钠 | 敲低或敲除2-甲基柠檬酸合成酶基因prpC;敲除TCA循环相关基因sdhE和icl;在染色体上表达编码磷酸烯醇式丙酮酸羧化酶基因ppc | 摇瓶中6.3 g/L细胞干重,包含质量分数65%的PHBV,其中3HV摩尔分数达到35% | [ |
| 3-羟基丁酸与3-羟基己酸共聚物(PHBHHx) | 葡萄糖,己酸钠 | 表达来自Aeromonas caviae FA440的PHA合成酶基因phaCac和烯酰辅酶-A水合酶基因phaJac | 7 L发酵罐产生33.1 g/L细胞干重,包含质量分数50.32%的P(3HB-co-37.23% 3HHx) ,3HHx摩尔比例可以在0%~37%范围调控 | [ |
| PHA颗粒相蛋白(PhaP) | 葡萄糖 | 敲除phaC基因,在基因组上过表达phaP基因 | PhaP累积量占比19%,产量为1.86 g/L | [ |
| 淀粉酶,葡萄糖苷酶,PHA,小分子氨基酸(L-苏氨酸,L-赖氨酸) | 淀粉 | 筛选合适的信号肽和连接子将过表达的α-淀粉酶和葡萄糖苷酶分泌到胞外,异源表达5个L-苏氨酸合成基因和外排转运蛋白,敲除内运转运蛋白和L-苏氨酸脱氢酶;过表达L-赖氨酸合成相关基因,解除底物抑制效应,增强L-赖氨酸外排能力 | 以淀粉作为唯一碳源生产PHA、四氢嘧啶、苏氨酸等多种产品 | [ |
| 葡萄糖 | 7 L发酵罐生产苏氨酸,产量33 g/L;7 L发酵罐生产赖氨酸,产量22.59 g/L | [ | ||
| 戊二胺 | 赖氨酸 | 异源表达赖氨酸脱羧酶基因CadA, LdcC | 7 L发酵罐生产戊二胺,产量118 g/L | [ |
| 四氢嘧啶 | 葡萄糖 | 理性调控和四氢嘧啶合成相关的ectABC、lysC和asd三个基因簇,提高前体供应,增强产物转运系统,优化培养条件 | 7 L发酵罐生产四氢嘧啶,产量85 g/L | [ |
| 5-氨基戊酸 | 赖氨酸 | 敲除gabT基因,在基因组上表达三个拷贝的dvaBA基因 | 7 L发酵罐生产5-氨基戊酸,产量67.4 g/L | [ |
| 3-羟基丙酸 | 葡萄糖,1,3-丙二醇 | 理性调控3-羟基丙酸合成相关的AldDHb和AdhP基因,敲除降解基因DddA | 7 L发酵罐生产3-羟基丙酸,产量154 g/L | [ |
| 乙偶姻 | 丙酮酸 | 异源表达枯草芽孢杆菌α-乙酰乳酸合酶基因alsS和α-乙酰乳酸脱羧酶基因alsD | 全细胞催化生产乙偶姻,产量85.84 g/L | [ |
| 衣康酸 | 柠檬酸 | 表达顺乌头酸脱羧酶编码基因cadA和顺乌头酸酶编码基因acn;表达分子伴侣GroESL;增加编码限速酶基因acn的拷贝数以及弱化竞争途径 | 摇瓶中生产衣康酸,产量63.60 g/L | [ |
| 甲羟戊酸 | 葡萄糖 | 敲除phaB和phaC基因;异源表达甲羟戊酸合成基因HMG-CoA合成酶和HMG-CoA-还原酶;CIRSPRi技术敲低50个候选基因;引入非氧糖酵解通路(NOG通路)减少碳损失 | 5 L发酵罐中生产甲羟戊酸,产量121 g/L | [ |
表3 基于盐单胞菌生产的各种产物
Table 3 Various products based on Halomonas spp.
| 产物 | 碳源/底物 | 改造策略 | 产量 | 参考文献 |
|---|---|---|---|---|
| PHB | 葡萄糖,淀粉水解物,厨余垃圾混合碳源 | 限氧,限氮;平衡NADH+/NAD比例,补充乙酸 | 细胞干重大于80 g/L,PHB质量分数大于90% | [ |
| 3-羟基丁酸与4-羟基丁酸共聚物P(3HB-co-4HB) | 葡萄糖,葡萄糖酸盐废弃物,废弃玉米浆,γ-丁内酯 | 构建两条相互关联的4-羟基丁酸(4HB)生物合成途径,表达4-羟基丁酸转移酶基因orfZ;敲除琥珀酸半醛脱氢酶基因gadD;构建数学模型与理性计算辅助设计;敲除外膜相关基因lpxL和lpxM | 7 L发酵罐产生26.3 g/L细胞干重,包含质量分数60.5%的P(3HB-co-4HB),其中4HB的摩尔分数为17.04%;5 L发酵罐中产生100 g/L细胞干重包含质量分数60.4%的 P(3HB-co-4HB),其中4HB的摩尔分数为13.5%;敲除外膜菌H. bluephagenesis WZY254在7 L发酵罐产生84 g/L细胞干重包含质量分数81%的P(3HB-co-4HB),其中4HB的摩尔分数为26% | [ |
| 3-羟基丁酸与3-羟基戊酸共聚物(PHBV) | 葡萄糖,葡萄糖酸钠 | 敲低或敲除2-甲基柠檬酸合成酶基因prpC;敲除TCA循环相关基因sdhE和icl;在染色体上表达编码磷酸烯醇式丙酮酸羧化酶基因ppc | 摇瓶中6.3 g/L细胞干重,包含质量分数65%的PHBV,其中3HV摩尔分数达到35% | [ |
| 3-羟基丁酸与3-羟基己酸共聚物(PHBHHx) | 葡萄糖,己酸钠 | 表达来自Aeromonas caviae FA440的PHA合成酶基因phaCac和烯酰辅酶-A水合酶基因phaJac | 7 L发酵罐产生33.1 g/L细胞干重,包含质量分数50.32%的P(3HB-co-37.23% 3HHx) ,3HHx摩尔比例可以在0%~37%范围调控 | [ |
| PHA颗粒相蛋白(PhaP) | 葡萄糖 | 敲除phaC基因,在基因组上过表达phaP基因 | PhaP累积量占比19%,产量为1.86 g/L | [ |
| 淀粉酶,葡萄糖苷酶,PHA,小分子氨基酸(L-苏氨酸,L-赖氨酸) | 淀粉 | 筛选合适的信号肽和连接子将过表达的α-淀粉酶和葡萄糖苷酶分泌到胞外,异源表达5个L-苏氨酸合成基因和外排转运蛋白,敲除内运转运蛋白和L-苏氨酸脱氢酶;过表达L-赖氨酸合成相关基因,解除底物抑制效应,增强L-赖氨酸外排能力 | 以淀粉作为唯一碳源生产PHA、四氢嘧啶、苏氨酸等多种产品 | [ |
| 葡萄糖 | 7 L发酵罐生产苏氨酸,产量33 g/L;7 L发酵罐生产赖氨酸,产量22.59 g/L | [ | ||
| 戊二胺 | 赖氨酸 | 异源表达赖氨酸脱羧酶基因CadA, LdcC | 7 L发酵罐生产戊二胺,产量118 g/L | [ |
| 四氢嘧啶 | 葡萄糖 | 理性调控和四氢嘧啶合成相关的ectABC、lysC和asd三个基因簇,提高前体供应,增强产物转运系统,优化培养条件 | 7 L发酵罐生产四氢嘧啶,产量85 g/L | [ |
| 5-氨基戊酸 | 赖氨酸 | 敲除gabT基因,在基因组上表达三个拷贝的dvaBA基因 | 7 L发酵罐生产5-氨基戊酸,产量67.4 g/L | [ |
| 3-羟基丙酸 | 葡萄糖,1,3-丙二醇 | 理性调控3-羟基丙酸合成相关的AldDHb和AdhP基因,敲除降解基因DddA | 7 L发酵罐生产3-羟基丙酸,产量154 g/L | [ |
| 乙偶姻 | 丙酮酸 | 异源表达枯草芽孢杆菌α-乙酰乳酸合酶基因alsS和α-乙酰乳酸脱羧酶基因alsD | 全细胞催化生产乙偶姻,产量85.84 g/L | [ |
| 衣康酸 | 柠檬酸 | 表达顺乌头酸脱羧酶编码基因cadA和顺乌头酸酶编码基因acn;表达分子伴侣GroESL;增加编码限速酶基因acn的拷贝数以及弱化竞争途径 | 摇瓶中生产衣康酸,产量63.60 g/L | [ |
| 甲羟戊酸 | 葡萄糖 | 敲除phaB和phaC基因;异源表达甲羟戊酸合成基因HMG-CoA合成酶和HMG-CoA-还原酶;CIRSPRi技术敲低50个候选基因;引入非氧糖酵解通路(NOG通路)减少碳损失 | 5 L发酵罐中生产甲羟戊酸,产量121 g/L | [ |
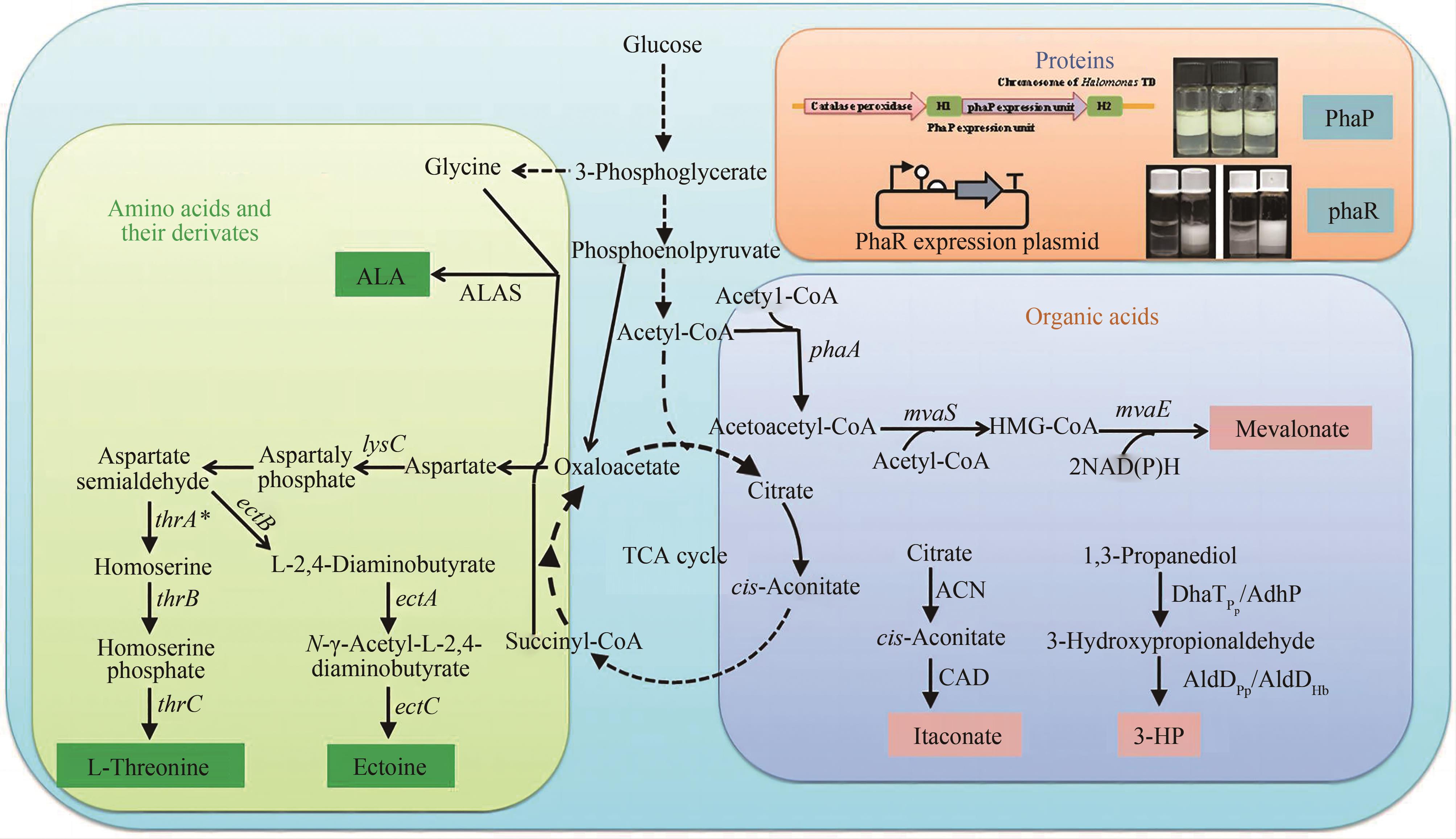
图3 利用盐单胞菌生产各类化合物[107](利用盐单胞菌目生产各种类型的化合物,例如蛋白质、氨基酸及衍生物、有机酸)ALAS—5-氨基乙酰丙酸合酶;lysC—编码天冬氨酸酶;thrA*—编码来自大肠杆菌MG1655的G433R高丝氨酸脱氢酶突变体;thrB—编码高丝氨酸激酶;thrC—编码L-苏氨酸合酶;ectA—编码L-2,4-二氨基丁酸乙酰转移酶;ectB—编码L-2,4-丁二酸转氨酶;ectC—编码四氢嘧啶合酶;ACN—乌头酸酶;CAD—顺乌头酸脱羧酶;DhaTPp和AdhP—醇脱氢酶和醛脱氢酶;AldDPp—来自恶臭假单胞菌KT2440的醛脱氢酶;AldDHb—来自嗜盐单胞菌的醛脱氢酶;phaA—编码3-酮硫醇酶;mvaS—编码HMG-CoA合酶;mvaE—编码HMG-CoA还原酶
Fig. 3 Diverse chemical compounds produced by Halomonas spp. [107](H. bluephagenesis has been engineered to produce non-PHA chemicals including proteins, amino acids, and their derivates, organic acids.) ALAS—5-aminolevulinic acid synthase; lysC—gene of aspartokinase; thrA*—gene of homoserine dehydrogenase mutant at G433R from Escherichia coli MG1655; thrB—gene of homoserine kinase; thrC—gene of L-threonine synthase; ectA—encoding L-2,4-diaminobutyrate acetyltransferase; ectB—encoding L-2,4-diaminobutyrate transaminase; ectC—encoding ectoine synthase; ACN—aconitase; CAD—cis-aconitate decarboxylase; DhaTPp and AdhP—alcohol dehydrogenase and aldehyde dehydrogenase; AldDPp—aldehyde dehydrogenase from Pseudomonas putida KT2440; AldDHb—aldehyde dehydrogenase from Halomonas bluephagenesis; phaA—encoding 3-ketothiolase; mvaS—encoding HMG-CoA synthase; mvaE—encoding HMG-CoA reductase
| 类型 | 未来产品 | 优势 |
|---|---|---|
| 嗜冷菌 | 蛋白质或酶 | 在胞内不容易形成包涵体 |
| 嗜热菌 | 挥发性小分子化合物 | 直接蒸馏提纯产品,简化处理工艺 |
| 嗜酸菌 | 酸性化合物(例如有机酸) | 耐受高浓度酸性产物 |
| 嗜碱菌 | 碱性化合物(例如赖氨酸) | 耐受高浓度碱性产物 |
| 嗜盐菌 | 饲料蛋白与酸性小分子化合物 | 开放式发酵,无需灭菌,耐渗透压 |
表4 未来基于极端微生物生产的产品
Table 4 Products based on extremophiles in the future
| 类型 | 未来产品 | 优势 |
|---|---|---|
| 嗜冷菌 | 蛋白质或酶 | 在胞内不容易形成包涵体 |
| 嗜热菌 | 挥发性小分子化合物 | 直接蒸馏提纯产品,简化处理工艺 |
| 嗜酸菌 | 酸性化合物(例如有机酸) | 耐受高浓度酸性产物 |
| 嗜碱菌 | 碱性化合物(例如赖氨酸) | 耐受高浓度碱性产物 |
| 嗜盐菌 | 饲料蛋白与酸性小分子化合物 | 开放式发酵,无需灭菌,耐渗透压 |
| 1 | GAVRILESCU M, CHISTI Y. Biotechnology - a sustainable alternative for chemical industry[J]. Biotechnology Advances, 2005, 23(7/8): 471-499. |
| 2 | KIRCHER M. Bioeconomy - present status and future needs of industrial value chains[J]. New Biotechnology, 2021, 60: 96-104. |
| 3 | RITTMANN B E. Opportunities for renewable bioenergy using microorganisms[J]. Biotechnology and Bioengineering, 2008, 100(2): 203-212. |
| 4 | LEVI P G, CULLEN J M. Mapping global flows of chemicals: from fossil fuel feedstocks to chemical products[J]. Environmental Science & Technology, 2018, 52(4): 1725-1734. |
| 5 | SHELDON R A, BRADY D. Green chemistry, biocatalysis, and the chemical industry of the future[J]. ChemSusChem, 2022, 15(9): e202102628. |
| 6 | GARTLAND K M, BRUSCHI F, DUNDAR M, et al. Progress towards the ‘golden age’ of biotechnology[J]. Current Opinion in Biotechnology, 2013, 24(): S6-S13. |
| 7 | CHEN G Q, JIANG X R. Next generation industrial biotechnology based on extremophilic bacteria[J]. Current Opinion in Biotechnology, 2018, 50: 94-100. |
| 8 | ROBINSON C J, CARBONELL P, JERVIS A J, et al. Rapid prototyping of microbial production strains for the biomanufacture of potential materials monomers[J]. Metabolic Engineering, 2020, 60: 168-182. |
| 9 | DE LORENZO V, KRASNOGOR N, SCHMIDT M. For the sake of the bioeconomy: define what a Synthetic Biology Chassis is![J]. New Biotechnology, 2021, 60: 44-51. |
| 10 | ZHANG X, LIN Y N, CHEN G Q. Halophiles as chassis for bioproduction[J]. Advanced Biosystems, 2018, 2(11): 1800088. |
| 11 | YU L P, WU F Q, CHEN G Q. Next-generation industrial biotechnology-transforming the current industrial biotechnology into competitive processes[J]. Biotechnology Journal, 2019, 14(9): e1800437. |
| 12 | TAN D, WANG Y, TONG Y, et al. Grand challenges for industrializing polyhydroxyalkanoates (PHAs)[J]. Trends in Biotechnology, 2021, 39(9): 953-963. |
| 13 | ORELLANA R, MACAYA C, BRAVO G, et al. Living at the frontiers of life: extremophiles in Chile and their potential for bioremediation[J]. Frontiers in Microbiology, 2018, 9: 2309. |
| 14 | DALMASSO C, OGER P, SELVA G, et al. Thermococcus piezophilus sp. nov., a novel hyperthermophilic and piezophilic archaeon with a broad pressure range for growth, isolated from a deepest hydrothermal vent at the Mid-Cayman Rise[J]. Systematic and Applied Microbiology, 2016, 39(7): 440-444. |
| 15 | FELLER G. Protein folding at extreme temperatures: current issues[J]. Seminars in Cell & Developmental Biology, 2018, 84: 129-137. |
| 16 | RAMPELOTTO P H. Extremophiles and extreme environments[J]. Life, 2013, 3(3): 482-485. |
| 17 | REKADWAD B, GONZALEZ J M. Multidisciplinary involvement and potential of thermophiles[J]. Folia Microbiologica, 2019, 64(3): 389-406. |
| 18 | REKADWAD B N, LI W J, GONZALEZ J M, et al. Extremophiles: the species that evolve and survive under hostile conditions[J]. 3 Biotech, 2023, 13(9): 316. |
| 19 | MESBAH N M, WIEGEL J. Life under multiple extreme conditions: diversity and physiology of the halophilic alkalithermophiles[J]. Applied and Environmental Microbiology, 2012, 78(12): 4074-4082. |
| 20 | CHEN X B, YIN J, YE J W, et al. Engineering Halomonas bluephagenesis TD01 for non-sterile production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate)[J]. Bioresource Technology, 2017, 244(Pt 1): 534-541. |
| 21 | OBULISAMY P K, MEHARIYA S. Polyhydroxyalkanoates from extremophiles: a review[J]. Bioresource Technology, 2021, 325: 124653. |
| 22 | QUILLAGUAMÁN J, GUZMÁN H, VAN-THUOC D, et al. Synthesis and production of polyhydroxyalkanoates by halophiles: current potential and future prospects[J]. Applied Microbiology and Biotechnology, 2010, 85(6): 1687-1696. |
| 23 | EDBEIB M F, WAHAB R A, HUYOP F. Halophiles: biology, adaptation, and their role in decontamination of hypersaline environments[J]. World Journal of Microbiology & Biotechnology, 2016, 32(8): 135. |
| 24 | MARGESIN R, SCHINNER F. Bioremediation (natural attenuation and biostimulation) of diesel-oil-contaminated soil in an alpine glacier skiing area[J]. Applied and Environmental Microbiology, 2001, 67(7): 3127-3133. |
| 25 | MA H, ZHAO Y Q, HUANG W Z, et al. Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine[J]. Nature Communications, 2020, 11(1): 3313. |
| 26 | DELGADO-GARCÍA M, VALDIVIA-URDIALES B, AGUILAR-GONZÁLEZ C N, et al. Halophilic hydrolases as a new tool for the biotechnological industries[J]. Journal of the Science of Food and Agriculture, 2012, 92(13): 2575-2580. |
| 27 | YIN J, CHEN J C, WU Q, et al. Halophiles, coming stars for industrial biotechnology[J]. Biotechnology Advances, 2015, 33(7): 1433-1442. |
| 28 | TAN D, WU Q, CHEN J C, et al. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates[J]. Metabolic Engineering, 2014, 26: 34-47. |
| 29 | TAO W, LV L, CHEN G Q. Engineering Halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi[J]. Microbial Cell Factories, 2017, 16(1): 48. |
| 30 | YE J W, HUANG W Z, WANG D S, et al. Pilot scale-up of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) production by Halomonas bluephagenesis via cell growth adapted optimization process[J]. Biotechnology Journal, 2018, 13(5): e1800074. |
| 31 | TAN D, XUE Y S, AIBAIDULA G, et al. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01[J]. Bioresource Technology, 2011, 102(17): 8130-8136. |
| 32 | YUE H T, LING C, YANG T, et al. A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates[J]. Biotechnology for Biofuels, 2014, 7(1): 108. |
| 33 | SARILMISER H K, ATES O, OZDEMIR G, et al. Effective stimulating factors for microbial levan production by Halomonas smyrnensis AAD6T[J]. Journal of Bioscience and Bioengineering, 2015, 119(4): 455-463. |
| 34 | SOMAYAJI A, DHANJAL C R, LINGAMSETTY R, et al. An insight into the mechanisms of homeostasis in extremophiles[J]. Microbiological Research, 2022, 263: 127115. |
| 35 | NARSING RAO M P, LUO Z H, DONG Z Y, et al. Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles[J]. Environmental Research, 2022, 209: 112888. |
| 36 | ZHU D C, ADEBISI W A, AHMAD F, et al. Recent development of extremophilic bacteria and their application in biorefinery[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 483. |
| 37 | SAXENA R, DHAKAN D B, MITTAL P, et al. Metagenomic analysis of hot springs in central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments[J]. Frontiers in Microbiology, 2017, 7: 2123. |
| 38 | DAMER B, DEAMER D. The hot spring hypothesis for an origin of life[J]. Astrobiology, 2020, 20(4): 429-452. |
| 39 | LV Z B, DING J X, WANG H, et al. Isolation of a novel thermophilic methanogen and the evolutionary history of the class Methanobacteria [J]. Biology, 2022, 11(10): 1514. |
| 40 | KIM J S, KLUSKENS L D, DE VOS W M, et al. Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin[J]. Journal of Molecular Biology, 2004, 335(3): 787-797. |
| 41 | ATALAH J, CÁCERES-MORENO P, ESPINA G, et al. Thermophiles and the applications of their enzymes as new biocatalysts[J]. Bioresource Technology, 2019, 280: 478-488. |
| 42 | KHONGTO B, LAOTENG K, TONGTA A. Fermentation process development of recombinant Hansenula polymorpha for gamma-linolenic acid production[J]. Journal of Microbiology and Biotechnology, 2010, 20(11): 1555-1562. |
| 43 | BHANDIWAD A, SHAW A J, GUSS A, et al. Metabolic engineering of Thermoanaerobacterium saccharolyticum for n-butanol production[J]. Metabolic Engineering, 2014, 21: 17-25. |
| 44 | TIAN L, CONWAY P M, CERVENKA N D, et al. Metabolic engineering of Clostridium thermocellum for n-butanol production from cellulose[J]. Biotechnology for Biofuels, 2019, 12: 186. |
| 45 | KRULWICH T A, SACHS G, PADAN E. Molecular aspects of bacterial pH sensing and homeostasis[J]. Nature Reviews Microbiology, 2011, 9(5): 330-343. |
| 46 | LEIGH M B, WU W M, CARDENAS E, et al. Microbial communities biostimulated by ethanol during uranium (Ⅵ) bioremediation in contaminated sediment as shown by stable isotope probing[J]. Frontiers of Environmental Science & Engineering, 2015, 9(3): 453-464. |
| 47 | ARULAZHAGAN P, AL-SHEKRI K, HUDA Q, et al. Biodegradation of polycyclic aromatic hydrocarbons by an acidophilic Stenotrophomonas maltophilia strain AJH1 isolated from a mineral mining site in Saudi Arabia[J]. Extremophiles, 2017, 21(1): 163-174. |
| 48 | XIAO H, SHAO Z Y, JIANG Y, et al. Exploiting Issatchenkia orientalis SD108 for succinic acid production[J]. Microbial Cell Factories, 2014, 13: 121. |
| 49 | JUNG H, INABA Y, BANTA S. Genetic engineering of the acidophilic chemolithoautotroph Acidithiobacillus ferrooxidans [J]. Trends in Biotechnology, 2022, 40(6): 677-692. |
| 50 | SOUSA J A, SOROKIN D Y, BIJMANS M F, et al. Ecology and application of haloalkaliphilic anaerobic microbial communities[J]. Applied Microbiology and Biotechnology, 2015, 99(22): 9331-9336. |
| 51 | YE J W, LIN Y N, YI X Q, et al. Synthetic biology of extremophiles: a new wave of biomanufacturing[J]. Trends in Biotechnology, 2023, 41(3): 342-357. |
| 52 | LI T T, LI T, JI W Y, et al. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp[J]. Biotechnology Journal, 2016, 11(2): 219-227. |
| 53 | SHEN R, YIN J, YE J W, et al. Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2018, 7(8): 1897-1906. |
| 54 | OLSON D G, MALONEY M, LANAHAN A A, et al. Identifying promoters for gene expression in Clostridium thermocellum [J]. Metabolic Engineering Communications, 2015, 2: 23-29. |
| 55 | ZHANG Y T, LIU H L, LIU Y J, et al. A promoter engineering-based strategy enhances polyhydroxyalkanoate production in Pseudomonas putida KT2440[J]. International Journal of Biological Macromolecules, 2021, 191: 608-617. |
| 56 | SUN W H, JIANG B, ZHAO D Y, et al. Integration of metabolic pathway manipulation and promoter engineering for the fine-tuned biosynthesis of malic acid in Bacillus coagulans [J]. Biotechnology and Bioengineering, 2021, 118(7): 2597-2608. |
| 57 | KERNAN T, MAJUMDAR S, LI X Z, et al. Engineering the iron-oxidizing chemolithoautotroph Acidithiobacillus ferrooxidans for biochemical production[J]. Biotechnology and Bioengineering, 2016, 113(1): 189-197. |
| 58 | WERNICK D G, PONTRELLI S P, POLLOCK A W, et al. Sustainable biorefining in wastewater by engineered extreme alkaliphile Bacillus marmarensis [J]. Scientific Reports, 2016, 6: 20224. |
| 59 | MA Y Y, YE J W, LIN Y N, et al. Flux optimization using multiple promoters in Halomonas bluephagenesis as a model chassis of the next generation industrial biotechnology[J]. Metabolic Engineering, 2024, 81: 249-261. |
| 60 | WANG L J, JIANG X R, HOU J, et al. Engineering Halomonas bluephagenesis via small regulatory RNAs[J]. Metabolic Engineering, 2022, 73: 58-69. |
| 61 | LIU X, LI D J, YAN X, et al. Rapid quantification of polyhydroxyalkanoates accumulated in living cells based on green fluorescence protein-labeled phasins: the qPHA method[J]. Biomacromolecules, 2022, 23(10): 4153-4166. |
| 62 | ZHAO D H, CAI L, WU J H, et al. Improving polyhydroxyalkanoate production by knocking out the genes involved in exopolysaccharide biosynthesis in Haloferax mediterranei [J]. Applied Microbiology and Biotechnology, 2013, 97(7): 3027-3036. |
| 63 | MARX C J, LIDSTROM M E. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria[J]. BioTechniques, 2002, 33(5): 1062-1067. |
| 64 | XU J Q, YANG S, YANG L R. Vibrio natriegens as a host for rapid biotechnology[J]. Trends in Biotechnology, 2022, 40(4): 381-384. |
| 65 | INABA Y, BANERJEE I, KERNAN T, et al. Transposase-mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans [J]. Applied and Environmental Microbiology, 2018, 84(21): e01381-18. |
| 66 | ZHAO C H, ZHENG T R, FENG Y H, et al. Engineered Halomonas spp. for production of L-lysine and cadaverine[J]. Bioresource Technology, 2022, 349: 126865. |
| 67 | XU T, CHEN J Y, MITRA R, et al. Deficiency of exopolysaccharides and O-antigen makes Halomonas bluephagenesis self-flocculating and amenable to electrotransformation[J]. Communications Biology, 2022, 5(1): 623. |
| 68 | WANG J T, WEI J W, LI H J, et al. High-efficiency genome editing of an extreme thermophile Thermus thermophilus using endogenous type Ⅰ and type Ⅲ CRISPR-Cas systems[J]. mLife, 2022, 1(4): 412-427. |
| 69 | BOST J, RECALDE A, WAßMER B, et al. Application of the endogenous CRISPR-Cas type Ⅰ-D system for genetic engineering in the thermoacidophilic archaeon Sulfolobus acidocaldarius [J]. Frontiers in Microbiology, 2023, 14: 1254891. |
| 70 | LIN L, CHEN J Y, MITRA R, et al. Optimising PHBV biopolymer production in haloarchaea via CRISPRi-mediated redirection of carbon flux[J]. Communications Biology, 2021, 4(1): 1007. |
| 71 | LAN L H, ZHAO H, CHEN J C, et al. Engineering Halomonas spp. as a low-cost production host for production of bio-surfactant protein PhaP[J]. Biotechnology Journal, 2016, 11(12): 1595-1604. |
| 72 | YASUI K, KANO Y, TANAKA K, et al. Improvement of bacterial transformation efficiency using plasmid artificial modification[J]. Nucleic Acids Research, 2009, 37(1): e3. |
| 73 | SUZUKI T, YASUI K. Plasmid artificial modification: a novel method for efficient DNA transfer into bacteria[J]. Methods in Molecular Biology, 2011, 765: 309-326. |
| 74 | JI M K, ZHENG T R, WANG Z Y, et al. PHB production from food waste hydrolysates by Halomonas bluephagenesis Harboring PHB operon linked with an essential gene[J]. Metabolic Engineering, 2023, 77: 12-20. |
| 75 | JIANG X R, YAO Z H, CHEN G Q. Controlling cell volume for efficient PHB production by Halomonas [J]. Metabolic Engineering, 2017, 44: 30-37. |
| 76 | WANG Z Y, QIN Q, ZHENG Y F, et al. Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties[J]. Metabolic Engineering, 2021, 67: 53-66. |
| 77 | REN K, ZHAO Y Q, CHEN G Q, et al. Construction of a stable expression system based on the endogenous hbpB/hbpC toxin-antitoxin system of Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2024, 13(1): 61-67. |
| 78 | QIN Q, LING C, ZHAO Y Q, et al. CRISPR/Cas9 editing genome of extremophile Halomonas spp[J]. Metabolic Engineering, 2018, 47: 219-229. |
| 79 | MA Y Y, ZHENG X R, LIN Y N, et al. Engineering an oleic acid-induced system for Halomonas, E. coli and Pseudomonas [J]. Metabolic Engineering, 2022, 72: 325-336. |
| 80 | DE FOUCHÉCOUR F, SÁNCHEZ-CASTAÑEDA A K, SAULOU-BÉRION C, et al. Process engineering for microbial production of 3-hydroxypropionic acid[J]. Biotechnology Advances, 2018, 36(4): 1207-1222. |
| 81 | JIANG X R, YAN X, YU L P, et al. Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis [J]. Nature Communications, 2021, 12(1): 1513. |
| 82 | LIN Y N, GUAN Y Y, DONG X, et al. Engineering Halomonas bluephagenesis as a chassis for bioproduction from starch[J]. Metabolic Engineering, 2021, 64: 134-145. |
| 83 | ZHANG L Z, LIN Y N, YI X Q, et al. Engineering low-salt growth Halomonas bluephagenesis for cost-effective bioproduction combined with adaptive evolution[J]. Metabolic Engineering, 2023, 79: 146-158. |
| 84 | CHEN J, LI W, ZHANG Z Z, et al. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source[J]. Microbial Cell Factories, 2018, 17(1): 102. |
| 85 | CHEN Y, CHEN X Y, DU H T, et al. Chromosome engineering of the TCA cycle in Halomonas bluephagenesis for production of copolymers of 3-hydroxybutyrate and 3-hydroxyvalerate (PHBV)[J]. Metabolic Engineering, 2019, 54: 69-82. |
| 86 | SHAW A J, PODKAMINER K K, DESAI S G, et al. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(37): 13769-13774. |
| 87 | DU H T, ZHAO Y Q, WU F Q, et al. Engineering Halomonas bluephagenesis for L-threonine production[J]. Metabolic Engineering, 2020, 60: 119-127. |
| 88 | BELL S C, TURNER J M. Bacterial catabolism of threonine. Threonine degradation initiated by L-threonine-NAD+ oxidoreductase[J]. Biochemical Journal, 1976, 156(2): 449-458. |
| 89 | MICHETTI D, BRANDSDAL B O, BON D, et al. A comparative study of cold- and warm-adapted endonucleases A using sequence analyses and molecular dynamics simulations[J]. PLoS One, 2017, 12(2): e0169586. |
| 90 | JOSHI S, SATYANARAYANA T. Biotechnology of cold-active proteases[J]. Biology, 2013, 2(2): 755-783. |
| 91 | MESBAH N M. Industrial biotechnology based on enzymes from extreme environments[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 870083. |
| 92 | ELLEUCHE S, SCHRÖDER C, SAHM K, et al. Extremozymes: biocatalysts with unique properties from extremophilic microorganisms[J]. Current Opinion in Biotechnology, 2014, 29: 116-123. |
| 93 | XIAN L, WANG F, LUO X, et al. Purification and characterization of a highly efficient calcium-independent α-amylase from Talaromyces pinophilus 1-95[J]. PLoS One, 2015, 10(3): e0121531. |
| 94 | SHARMA A, KAWARABAYASI Y, SATYANARAYANA T. Acidophilic bacteria and Archaea: acid stable biocatalysts and their potential applications[J]. Extremophiles, 2012, 16(1): 1-19. |
| 95 | VESTER J K, GLARING M A, STOUGAARD P. Discovery of novel enzymes with industrial potential from a cold and alkaline environment by a combination of functional metagenomics and culturing[J]. Microbial Cell Factories, 2014, 13: 72. |
| 96 | FU X Z, TAN D, AIBAIDULA G, et al. Development of Halomonas TD01 as a host for open production of chemicals[J]. Metabolic Engineering, 2014, 23: 78-91. |
| 97 | LING C, QIAO G Q, SHUAI B W, et al. Engineering NADH/NAD+ ratio in Halomonas bluephagenesis for enhanced production of polyhydroxyalkanoates (PHA)[J]. Metabolic Engineering, 2018, 49: 275-286. |
| 98 | YE J W, HU D K, CHE X M, et al. Engineering of Halomonas bluephagenesis for low cost production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) from glucose[J]. Metabolic Engineering, 2018, 47: 143-152. |
| 99 | WANG Z Y, ZHENG Y F, JI M K, et al. Hyperproduction of PHA copolymers containing high fractions of 4-hydroxybutyrate (4HB) by outer membrane-defected Halomonas bluephagenesis grown in bioreactors[J]. Microbial Biotechnology, 2022, 15(5): 1586-1597. |
| 100 | WANG H, YE J W, CHEN X Y, et al. Production of PHA copolymers consisting of 3-hydroxybutyrate and 3-hydroxyhexanoate (PHBHHx) by recombinant Halomonas bluephagenesis [J]. Chemical Engineering Journal, 2023, 466: 143261. |
| 101 | ZHANG J, ZHANG X, MAO Y F, et al. Substrate profiling and tolerance testing of Halomonas TD01 suggest its potential application in sustainable manufacturing of chemicals[J]. Journal of Biotechnology, 2020, 316: 1-5. |
| 102 | ZHANG J, JIN B, HONG K Q, et al. Cell catalysis of citrate to itaconate by engineered Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2021, 10(11): 3017-3027. |
| 103 | ZHANG J, YUAN Y, WANG Z W, et al. Metabolic engineering of Halomonas bluephagenesis for high-level mevalonate production from glucose and acetate mixture[J]. Metabolic Engineering, 2023, 79: 203-213. |
| 104 | ZHENG M Y, CUI Z Z, ZHANG J, et al. Efficient acetoin production from pyruvate by engineered Halomonas bluephagenesis whole-cell biocatalysis[J]. Frontiers of Chemical Science and Engineering, 2023, 17(4): 425-436. |
| 105 | HU Q T, SUN S M, ZHANG Z N, et al. Ectoine hyperproduction by engineered Halomonas bluephagenesis [J]. Metabolic Engineering, 2024, 82: 238-249. |
| 106 | YANG F, WANG H, ZHAO C H, et al. Metabolic engineering of Halomonas bluephagenesis for production of five carbon molecular chemicals derived from L-lysine[J]. Metabolic Engineering, 2024, 81: 227-237. |
| 107 | CHEN G Q, ZHANG X, LIU X, et al. Halomonas spp., as chassis for low-cost production of chemicals[J]. Applied Microbiology and Biotechnology, 2022, 106(21): 6977-6992. |
| 108 | MARTÍNEZ-GARCÍA E, FRAILE S, ALGAR E, et al. SEVA 4.0: an update of the Standard European Vector Architecture database for advanced analysis and programming of bacterial phenotypes[J]. Nucleic Acids Research, 2023, 51(D1): D1558-D1567. |
| 109 | GURDO N, VOLKE D C, NIKEL P I. Merging automation and fundamental discovery into the design-build-test-learn cycle of nontraditional microbes[J]. Trends in Biotechnology, 2022, 40(10): 1148-1159. |
| 110 | JOHNSTON C, MARTIN B, FICHANT G, et al. Bacterial transformation: distribution, shared mechanisms and divergent control[J]. Nature Reviews Microbiology, 2014, 12(3): 181-196. |
| 111 | VAN BELJOUW S P B, SANDERS J, RODRÍGUEZ-MOLINA A, et al. RNA-targeting CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2023, 21(1): 21-34. |
| 112 | VOLKE D C, ORSI E, NIKEL P I. Emergent CRISPR-Cas-based technologies for engineering non-model bacteria[J]. Current Opinion in Microbiology, 2023, 75: 102353. |
| 113 | ARROYO-OLARTE R D, BRAVO RODRÍGUEZ R, MORALES-RÍOS E. Genome editing in bacteria: CRISPR-Cas and beyond[J]. Microorganisms, 2021, 9(4): 844. |
| 114 | ZHANG L, ZHAO R, JIA D C, et al. Engineering Clostridium ljungdahlii as the gas-fermenting cell factory for the production of biofuels and biochemicals[J]. Current Opinion in Chemical Biology, 2020, 59: 54-61. |
| 115 | NORMAN R O J, MILLAT T, WINZER K, et al. Progress towards platform chemical production using Clostridium autoethanogenum [J]. Biochemical Society Transactions, 2018, 46(3): 523-535. |
| 116 | BRAUTASET T, JAKOBSEN Ø M, DEGNES K F, et al. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase Ⅰ and Ⅱ and their roles for L-lysine production from methanol at 50 ℃[J]. Applied Microbiology and Biotechnology, 2010, 87(3): 951-964. |
| 117 | CUI L Y, WANG S S, GUAN C G, et al. Breeding of methanol-tolerant Methylobacterium extorquens AM1 by atmospheric and room temperature plasma mutagenesis combined with adaptive laboratory evolution[J]. Biotechnology Journal, 2018, 13(6): e1700679. |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 应汉杰, 柳东, 王振宇, 沈涛, 庄伟, 朱晨杰. 工业生物制造与“碳中和”目标探讨[J]. 合成生物学, 2025, 6(1): 1-7. |
| [3] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [4] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [5] | 刘晓悦, 王盼娣, 吴刚, 刘芳. 基因工程辅助萝卜硫苷在十字花科作物中的高效生物合成[J]. 合成生物学, 2025, 6(1): 136-156. |
| [6] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [7] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [8] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [9] | 张以恒, 陈雪梅, 石婷. 生物制造的市本率(PC值):定义与应用[J]. 合成生物学, 2025, 6(1): 8-17. |
| [10] | 张以恒. 中国哲学思想“道法术器”对生物制造的启示[J]. 合成生物学, 2024, 5(6): 1231-1241. |
| [11] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [12] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [13] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [14] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [15] | 刘建明, 张炽坚, 张冰, 曾安平. 巴氏梭菌作为工业底盘细胞高效生产1,3-丙二醇——从代谢工程和菌种进化到过程工程和产品分离[J]. 合成生物学, 2024, 5(6): 1386-1403. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
