合成生物学 ›› 2020, Vol. 1 ›› Issue (1): 7-28.DOI: 10.12211/2096-8280.2020-057
合成生物学重要研究方向进展
丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进
- 教育部合成生物学前沿科学中心,系统生物工程教育部重点实验室,天津大学化工学院,天津 300072
-
收稿日期:2020-04-23修回日期:2020-05-13出版日期:2020-02-29发布日期:2020-09-24 -
通讯作者:元英进 -
作者简介:丁明珠,女,博士,副研究员,研究方向为合成生物学,人工混菌体系设计合成。E-mail:mzding@tju.edu.cn
元英进(1963—),男,教授,博士生导师,研究方向为合成生物学及人工基因组化学合成。E-mail: yjyuan@tju.edu.cn -
基金资助:国家自然科学基金(21621004)
Significant research progress in synthetic biology
DING Mingzhu, LI Bingzhi, WANG Ying, XIE Zexiong, LIU Duo, YUAN Yingjin
- Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (Ministry of Education), School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China
-
Received:2020-04-23Revised:2020-05-13Online:2020-02-29Published:2020-09-24 -
Contact:YUAN Yingjin
摘要:
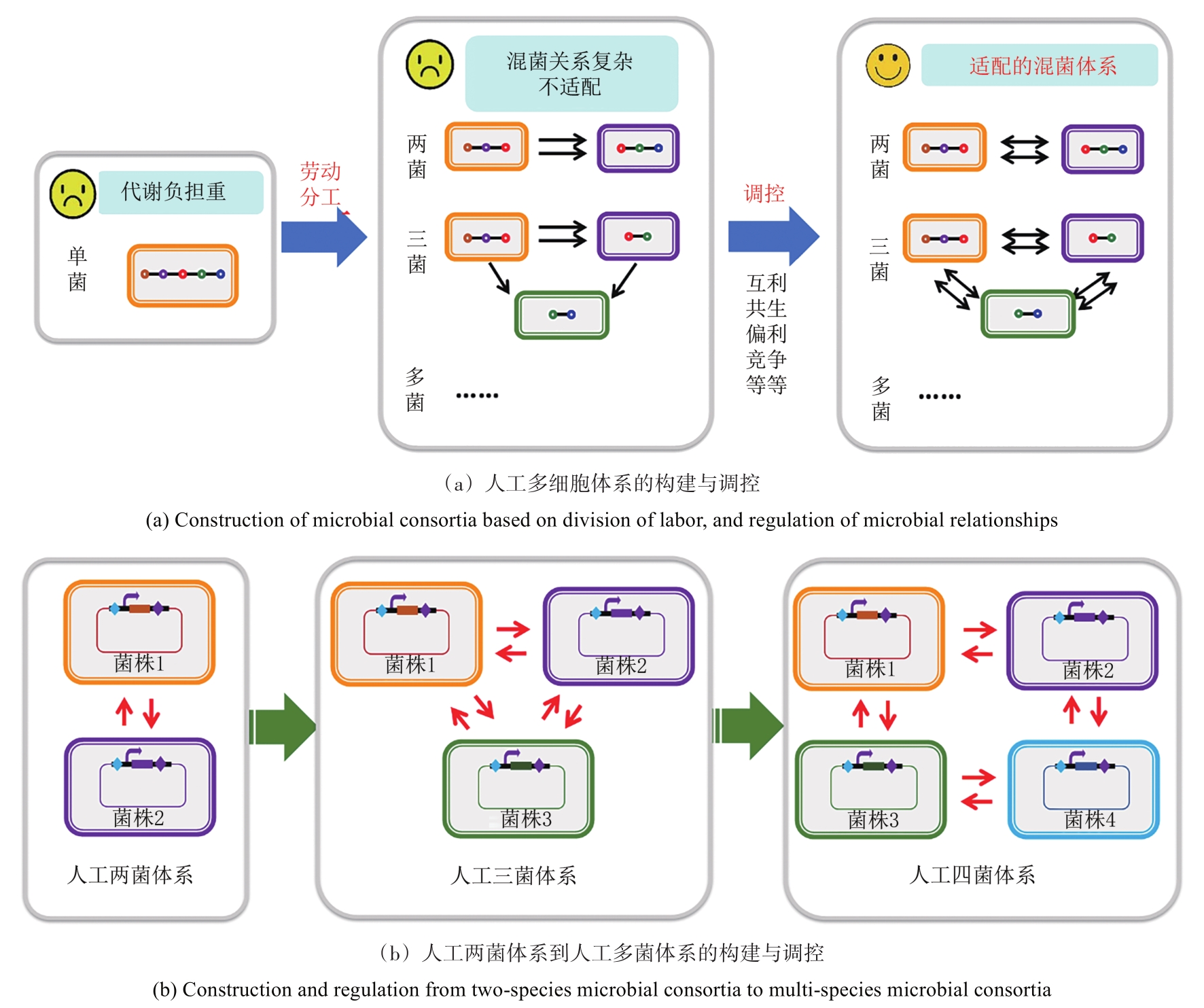
合成生物学作为一个新兴的交叉学科领域,随着DNA合成技术的进步和合成生物学理念的深入,多个研究方向取得了长足发展。本文主要对基因回路、基因组设计合成、细胞工厂和人工多细胞体系的进展进行了综述。可设计构建的人工基因线路的复杂度逐步提升,人工控制更加精细;组装技术取得快速进展的同时,人工基因组的设计深度也在不断拓展,设计合成的人工基因组由支原体拓展向大肠杆菌,甚至真核生物酿酒酵母,推动了生物进化演化的研究;细胞工厂的设计构建在逐步挑战代谢途径更长、复杂程度更高的化合物的合成,模块化和正交化策略对复杂细胞工厂构建的支撑作用日益明显,鲁棒性和适配性成为细胞工厂构建需要考虑的重要问题;人工多细胞体系的设计构建已经从设计构建两菌体系向多菌体系扩展,通过多种原则进行设计,实现更加复杂的预期功能。本文也对合成生物学与其他学科交叉融合产生的一些新研究方向进行了简介。
中图分类号:
引用本文
丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28.
DING Mingzhu, LI Bingzhi, WANG Ying, XIE Zexiong, LIU Duo, YUAN Yingjin. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28.
| 1 | LEPROUST E M, PECK B J, SPIRIN K, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process[J]. Nucleic Acids Research, 2010, 38 (8): 2522-2540. |
| 2 | PALLUK S, ARLOW D H, DE ROND T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36 (7): 645-650. |
| 3 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403 (6767): 335-338. |
| 4 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403 (6767): 339-342. |
| 5 | RANTASALO A, KUIVANEN J, PENTTILA M, et al. Synthetic toolkit for complex genetic circuit engineering in Saccharomyces cerevisiae [J]. ACS Synthtic Biology, 2018, 7 (6): 1573-1587. |
| 6 | ZONG Y, ZHANG H M, LYU C, et al. Insulated transcriptional elements enable precise design of genetic circuits[J]. Nature Communications, 2017, 8 (1): 52. |
| 7 | SHETTY R P, ENDY D, KNIGHT T F JR. Engineering BioBrick vectors from BioBrick parts[J]. J. Biol. Eng., 2008, 2: 5. |
| 8 | LEE T S, KRUPA R A, ZHANG F Z, et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression[J]. Journal of Biological Engineering, 2011, 5 (1): 12. |
| 9 | AGMON N, MITCHELL L A, CAI Y Z, et al. Yeast golden gate (yGG) for the efficient assembly of s-cerevisiae transcription Units[J]. ACS Synthetic Biology, 2015, 4 (7): 853-859. |
| 10 | WEBER E, ENGLER C, GRUETZNER R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PLoS One, 2011, 6 (2): e16765. |
| 11 | BERVOETS I, BREMPT M VAN, NEROM K VAN, et al. A sigma factor toolbox for orthogonal gene expression in Escherichia coli [J]. Nucleic Acids Research, 2018, 46 (4): 2133-2144. |
| 12 | REDDEN H, ALPER H S. The development and characterization of synthetic minimal yeast promoters[J]. Nature Communications, 2015, 6: e7810. |
| 13 | ROGERS J K, TAYLOR N D, CHURCH G M. Biosensor-based engineering of biosynthetic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 84-91. |
| 14 | TEMME K, HILL R, SEGALL-SHAPIRO T H, et al. Modular control of multiple pathways using engineered orthogonal T7 polymerases[J]. Nucleic Acids Research, 2012, 40 (17): 8773-8781. |
| 15 | MORSE N J, WAGNER J M, REED K B, et al. T7 polymerase expression of guide RNAs in vivo allows exportable CRISPR-Cas9 editing in multiple yeast hosts[J]. ACS Synthetic Biology, 2018, 7 (4): 1075-1084. |
| 16 | LIAO W, LIU B, CHANG C C, et al. Functional characterization of insulation effect for synthetic gene circuits in mammalian cells[J]. ACS Synthetic Biology, 2018, 7 (2): 412-418. |
| 17 | BATEY R T, GILBERT S D, MONTANGE R K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine[J]. Nature, 2004, 432 (7015): 411-415. |
| 18 | BADORREK C S, GHERGHE C M, WEEKS K M. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization[J]. PNAS, 2006, 103 (37): 13640-13645. |
| 19 | MOON T S, LOU C B, TAMSIR A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491 (7423): 249-253. |
| 20 | SIUTI P, YAZBEK J, LU T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature Biotechnology, 2013, 31 (5): 448-452. |
| 21 | STRICKER J, COOKSON S, BENNETT M R, et al. A fast, robust and tunable synthetic gene oscillator[J]. Nature, 2008, 456 (7221): 516-519. |
| 22 | ANDREWS L B, NIELSEN A A K, VOIGT C A. Cellular checkpoint control using programmable sequential logic[J]. Science, 2018, 361 (6408): eaap8987. |
| 23 | LIAO M J, DIN M O, TSINRING L, et al. Rock-paper-scissors: engineered population dynamics increase genetic stability[J]. Science, 2019, 365 (6457): 1045-1049. |
| 24 | POTVIN-TROTTIER L, LORD N D, VINNICOMBE G, et al. Synchronous long-term oscillations in a synthetic gene circuit[J]. Nature, 2016, 538 (7626): 514-517. |
| 25 | GAO X J, CHONG L S, KIM M S, et al. Programmable protein circuits in living cells[J]. Science, 2018, 361 (6408): 1252-1258. |
| 26 | ADAMALA K P, MARTIN-ALARCON D A, GUTHRIE-HONEA K R, et al. Engineering genetic circuit interactions within and between synthetic minimal cells[J]. Nature Chemistry, 2017, 9 (5): 431-439. |
| 27 | PURCELL O, WANG J, SIUTI P, et al. Encryption and steganography of synthetic gene circuits[J]. Nature Communications, 2018, 9 (1): 4942. |
| 28 | CHEN M T, WEISS R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana [J]. Nature Biotechnology, 2005, 23 (12): 1551-1555. |
| 29 | YOU L C, COX R S, WEISS R, et al. Programmed population control by cell-cell communication and regulated killing[J]. Nature, 2004, 428 (6985): 868-871. |
| 30 | BALAGADDE F K, SONG H, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem[J]. Molecular Systems Biology, 2008, 4: 187. |
| 31 | LIU C L, FU X F, LIU L L, et al. Sequential establishment of stripe patterns in an expanding cell population[J]. Science, 2011, 334 (6053): 238-241. |
| 32 | TAMSIR A, TABOR J J, VOIGT C A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'[J]. Nature, 2011, 469 (7329): 212-215. |
| 33 | REGOT S, MACIA J, CONDE N, et al. Distributed biological computation with multicellular engineered networks[J]. Nature, 2011, 469 (7329): 207-211. |
| 34 | TODA S, BLAUCH L R, TANG S K Y, et al. Programming self-organizing multicellular structures with synthetic cell-cell signaling[J]. Science, 2018, 361 (6398): 156-162. |
| 35 | ZHANG F Z, CAROTHERS J M, KEASLING J D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids[J]. Nature Biotechnology, 2012, 30 (4): 354-359. |
| 36 | CHOU H H, KEASLING J D. Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4: 2595. |
| 37 | TAO R K, ZHAO Y Z, CHU H Y, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism[J]. Nature Methods, 2017, 14 (9): 720-728. |
| 38 | TAY P K R, NGUYEN P Q, JOSHI N S. A Synthetic circuit for mercury bioremediation using self assembling functional amyloids[J]. ACS Synthetic Biology, 2017, 6 (10): 1841-1850. |
| 39 | WAN X Y, VOLPETTI F, PETROVA E, et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals[J]. Nature Chemical Biology, 2019, 15 (5): 540-548. |
| 40 | GALLAGHER R R, PATEL J R, INTERIANO A L, et al. Multilayered genetic safeguards limit growth of microorganisms to defined environments[J]. Nucleic Acids Research, 2015, 43 (3): 1945-1954. |
| 41 | CHAN C T Y, LEE J W, CAMERON D E, et al. 'Deadman' and 'Passcode' microbial kill switches for bacterial containment[J]. Nature Chemical Biology, 2016, 12 (2): 82-86. |
| 42 | ROYBAL K T, RUPP L J, MORSUT L, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits[J]. Cell, 2016, 164 (4): 770-779. |
| 43 | ROYBAL K T, WILLIAMS J Z, MORSUT L, et al. Engineering T cells with customized therapeutic response programs using synthetic notch receptors[J]. Cell, 2016, 167 (2): 419-432. |
| 44 | MIMEE M, NADEAU P, HAYWARD A, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health[J]. Science, 2018, 360 (6391): 915-918. |
| 45 | SHAO J W, XUE S, YU G L, et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice[J]. Science Translational Medicine, 2017, 9 (387): eaal2298. |
| 46 | TIGGES M, DENERVAUD N, GREBER D, et al. A synthetic low-frequency mammalian oscillator[J]. Nucleic Acids Research, 2010, 38 (8): 2702-2711. |
| 47 | TIGGES M, MARQUEZ-LAGO T T, STELLING J, et al. A tunable synthetic mammalian oscillator[J]. Nature, 2009, 457 (7227): 309-312. |
| 48 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329 (5987): 52-56. |
| 49 | HUTCHISON C A, CHUANG R Y, NOSKOV V N, et al. Design and synthesis of a minimal bacterial genome[J]. Science, 2016, 351 (6280): aad6253. |
| 50 | OSTROV N, LANDON M, GUELL M, et al. Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353 (6301): 819-822. |
| 51 | FREDENS J, WANG K, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569 (7757): 514-518. |
| 52 | ANNALURU N, MULLER H, MITCHELL L A, et al. Total synthesis of a functional designer eukaryotic chromosome[J]. Science, 2014, 344 (6179): 55-58. |
| 53 | XIE Z X, LI B Z, MITCHELL L A, et al. Perfect designer chromosome Ⅴ and behavior of a ring derivative[J]. Science, 2017, 355 (6329): eaaf4704. |
| 54 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G, et al. Design of a synthetic yeast genome[J]. Science, 2017, 355 (6329): 1040-1044. |
| 55 | MITCHELL L A, WANG A, STRACQUADANIO G, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: syn Ⅵ and beyond[J]. Science, 2017, 355 (6329): eaaf4831. |
| 56 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355 (6329): eaaf4791. |
| 57 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355 (6329): eaaf4706. |
| 58 | ZHANG W, ZHAO G, LUO Z, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355 (6329): eaaf3981. |
| 59 | XIE Z X, LIU D, LI B Z, et al. Design and chemical synthesis of eukaryotic chromosomes[J]. Chemical Society Reviews, 2017, 46 (23): 7191-7207. |
| 60 | ISAACS F J, CARR P A, WANG H H, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement[J]. Science, 2011, 333 (6040): 348-353. |
| 61 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533 (7603): 420-424. |
| 62 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551 (7681): 464-471. |
| 63 | BAO Z, HAMEDIRAD M, XUE P, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision[J]. Nature Biotechnology, 2018, 36 (6): 505-508. |
| 64 | JIA B, WU Y, LI B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9 (1): 1933. |
| 65 | WANG J, XIE Z X, MA Y, et al. Ring synthetic chromosome ⅤSCRaMbLE[J]. Nature Communications, 2018, 9 (1): 3783. |
| 66 | NAKAMURA C E, WHITED G M. Metabolic engineering for the microbial production of 1,3-propanediol[J]. Current Opinion in Biotechnology, 2003, 14 (5): 454-459. |
| 67 | RO D K, PARADISE E M, OUELLET M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast[J]. Nature, 2006, 440 (7086): 940-943. |
| 68 | MARTIN V J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids[J]. Nature Biotechnology, 2003, 21 (7): 796-802. |
| 69 | NAKAGAWA A, MATSUMURA E, KOYANAGI T, et al. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli [J]. Nature Communications, 2016, 7: 10390. |
| 70 | LUO X, REITER M A, D'ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567 (7746): 123-126. |
| 71 | CHEN X L, GAO C, GUO L, et al. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals[J]. Chemical Reviews, 2018, 118 (1): 4-72. |
| 72 | DU Y L, ALKHALAF L M, RYAN K S. In vitro reconstitution of indolmycin biosynthesis reveals the molecular basis of oxazolinone assembly[J]. PNAS, 2015, 112 (9): 2717-2722. |
| 73 | LUO Y, HUANG H, LIANG J, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster[J]. Nature Communications, 2013, 4: 2894. |
| 74 | TU L, SU P, ZHANG Z, et al. GenomGenome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis [J]. Nature Communications, 2020, 11 (1): 971. |
| 75 | LAU W, SATTELY E S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone[J]. Science, 2015, 349 (6253): 1224-1228. |
| 76 | HAMMER S C, KNIGHT A M, ARNOLD F H. Design and evolution of enzymes for non-natural chemistry[J]. Current Opinion in Green and Sustainable Chemistry, 2017, 7: 23-30. |
| 77 | CHEN K, ARNOLD F H. Engineering new catalytic activities in enzymes[J]. Nature Catalysis, 2020, 3 (3): 203-213. |
| 78 | LEAVER-FAY A, TYKA M, LEWIS S M, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules[J]. Methods Enzymol, 2011, 487: 545-574. |
| 79 | DOU J Y, VOROBIEVA A A, SHEFFLER W, et al. De novo design of a fluorescence-activating beta-barrel[J]. Nature, 2018, 561 (7724): 485-491. |
| 80 | BOYKEN S E, BENHAIM M A, BUSCH F, et al. De novo design of tunable, pH-driven conformational changes[J]. Science, 2019, 364 (6441): 658-664. |
| 81 | CHEN Z B, BOYKEN S E, JIA M X, et al. Programmable design of orthogonal protein heterodimers[J]. Nature, 2019, 565 (7737): 106-111. |
| 82 | HAMMER S C, KUBIK G, WATKINS E, et al. Anti-Markovnikov alkene oxidation by metal-oxo-mediated enzyme catalysis[J]. Science, 2017, 358 (6360): 215-218. |
| 83 | KAN S B J, HUANG X, GUMULYA Y, et al. Genetically programmed chiral organoborane synthesis[J]. Nature, 2017, 552 (7683): 132-136. |
| 84 | KAN S B, LEWIS R D, CHEN K, et al. Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life[J]. Science, 2016, 354 (6315): 1048-1051. |
| 85 | IGNEA C, PONTINI M, MOTAWIA M S, et al. Synthesis of 11-carbon terpenoids in yeast using protein and metabolic engineering[J]. Nature Chemical Biology, 2018, 14 (12): 1090-1098. |
| 86 | FURUBAYASHI M, IKEZUMI M, TAKAICHI S, et al. A highly selective biosynthetic pathway to non-natural C50 carotenoids assembled from moderately selective enzymes[J]. Nature Communications, 2015, 6: 7534. |
| 87 | XIONG M, SCHNEIDERMAN D K, BATES F S, et al. Scalable production of mechanically tunable block polymers from sugar[J]. PNAS, 2014, 111 (23): 8357-8362. |
| 88 | TAI Y S, XIONG M, JAMBUNATHAN P, et al. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products[J]. Nature Chemical Biology, 2016, 12 (4): 247-253. |
| 89 | YAO Y F, WANG C S, QIAO J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway[J]. Metabolic Engineering, 2013, 19: 79-87. |
| 90 | AJIKUMAR P K, XIAO W H, TYO K E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330 (6000): 70-74. |
| 91 | XU P, GU Q, WANG W, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 92 | QIN J, ZHOU Y J, KRIVORUCHKO A, et al. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine[J]. Nature Communications, 2015, 6: 8224. |
| 93 | XU P, QIAO K, AHN W S, et al. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals[J]. PNAS, 2016, 113 (39): 10848-10853. |
| 94 | FANG H, LI D, KANG J, et al. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12 [J]. Nature Communications, 2018, 9 (1): 4917. |
| 95 | SRINIVASAN P, SMOLKE C D. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids[J]. Nature Communications, 2019, 10 (1): 3634. |
| 96 | SWEETLOVE L J, FERNIE A R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation[J]. Nature Communications, 2018, 9 (1): 2136. |
| 97 | HAMMER S K, AVALOS J L. Harnessing yeast organelles for metabolic engineering[J]. Nature Chemical Biology, 2017, 13 (8): 823-832. |
| 98 | BERNER N, REUTTER K R, WOLF D H. Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path[J]. Annual Review of Biochemistry, 2018, 87: 751-782. |
| 99 | CHEN S J, WU X, WADAS B, et al. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes[J]. Science, 2017, 355 (6323): aal3655. |
| 100 | PENG B, NIELSEN L K, KAMPRANIS S C, et al. Engineered protein degradation of farnesyl pyrophosphate synthase is an effective regulatory mechanism to increase monoterpene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2018, 47: 83-93. |
| 101 | PENG B, PLAN M R, CHRYSANTHOPOULOS P, et al. A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2017, 39: 209-219. |
| 102 | MORSUT L, ROYBAL K T, XIONG X, et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors[J]. Cell, 2016, 164 (4): 780-791. |
| 103 | KENT R, DIXON N. Systematic evaluation of genetic and environmental factors affecting performance of translational riboswitches[J]. ACS Synthetic Biology, 2019, 8 (4): 884-901. |
| 104 | DAHLMAN J E, ABUDAYYEH O O, JOUNG J, et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease[J]. Nature Biotechnology, 2015, 33 (11): 1159-1161. |
| 105 | PANDIT A V, SRINIVASAN S, MAHADEVAN R. Redesigning metabolism based on orthogonality principles[J]. Nature Communications, 2017, 8: 15188. |
| 106 | DE LORENZO V. Beware of metaphors: chasses and orthogonality in synthetic biology[J]. Bioengineered Bugs, 2011, 2 (1): 3-7. |
| 107 | LIU H, LU T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli [J]. Metabolic Engineering, 2015, 29: 135-141. |
| 108 | IGNEA C, RAADAM M H, MOTAWIA M S, et al. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate[J]. Nature Communications, 2019, 10 (1): 3799. |
| 109 | JI D, WANG L, HOU S, et al. Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide[J]. Journal of the American Chemical Society, 2011, 133 (51): 20857-20862. |
| 110 | LIU Y, FENG Y, WANG L, et al. Structural insights into phosphite dehydrogenase variants favoring a non-natural redox cofactor[J]. ACS Catalysis, 2019, 9 (3): 1883-1887. |
| 111 | WANG L, JI D, LIU Y, et al. Synthetic cofactor-linked metabolic circuits for selective energy transfer[J]. ACS Catalysis, 2017, 7 (3): 1977-1983. |
| 112 | GUO X, LIU Y, WANG Q, et al. Non-natural cofactor and formate-driven reductive carboxylation of pyruvate[J]. Angewandte Chemie International Edition, 2020, 59 (8): 3143-3146. |
| 113 | LIU G S, LI T, ZHOU W, et al. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction[J]. Metabolic Engineering, 2020, 57: 151-161. |
| 114 | BALDI N, DYKSTRA J C, LUTTIK M A H, et al. Functional expression of a bacterial alpha-ketoglutarate dehydrogenase in the cytosol of Saccharomyces cerevisiae [J]. Metabolic Engineering, 2019, 56: 190-197. |
| 115 | LV X, WANG F, ZHOU P, et al. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae [J]. Nature Communications, 2016, 7: 12851. |
| 116 | SADRE R, KUO P, CHEN J, et al. Cytosolic lipid droplets as engineered organelles for production and accumulation of terpenoid biomaterials in leaves[J]. Nature Communications, 2019, 10 (1): 853. |
| 117 | ZHAO C, KIM Y, ZENG Y, et al. Co-compartmentation of terpene biosynthesis and storage via synthetic droplet[J]. ACS Synthetic Biology, 2018, 7 (3): 774-781. |
| 118 | ZHOU Y J, BUIJS N A, ZHU Z, et al. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition[J]. Journal of the American Chemical Society, 2016, 138 (47): 15368-15377. |
| 119 | KIM J E, JANG I S, SON S H, et al. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway[J]. Metabolic Engineering, 2019, 56: 50-59. |
| 120 | CHEN R, YANG S, ZHANG L, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23 (3): 100879. |
| 121 | KLEI I J VAN DER, VEENHUIS M. Yeast peroxisomes: function and biogenesis of a versatile cell organelle[J]. Trends Microbiol., 1997, 5 (12): 502-509. |
| 122 | CROSS L L, PAUDYAL R, KAMISUGI Y, et al. Towards designer organelles by subverting the peroxisomal import pathway[J]. Nature Communications, 2017, 8 (1): 454. |
| 123 | STEINKUHLER J, KNORR R L, ZHAO Z, et al. Controlled division of cell-sized vesicles by low densities of membrane-bound proteins[J]. Nature Communications, 2020, 11 (1): 905. |
| 124 | KERFELD C A, AUSSIGNARGUES C, ZARZYCKI J, et al. Bacterial microcompartments[J]. Nat. Rev. Microbiol., 2018, 16 (5): 277-290. |
| 125 | HAGEN A, SUTTER M, SLOAN N, et al. Programmed loading and rapid purification of engineered bacterial microcompartment shells[J]. Nature Communications, 2018, 9 (1): 2881. |
| 126 | LEE M J, MANTELL J, BROWN I R, et al. De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments[J]. Nature Communications, 2018, 9 (1): 3413. |
| 127 | FERLEZ B, SUTTER M, KERFELD C A. A designed bacterial microcompartment shell with tunable composition and precision cargo loading[J]. Metabolic Engineering, 2019, 54: 286-291. |
| 128 | GIESSEN T W, SILVER P A. Widespread distribution of encapsulin nanocompartments reveals functional diversity[J]. Nature Microbiology, 2017, 2 (6): 17029. |
| 129 | BUGAJ L J, CHOKSI A T, MESUDA C K, et al. Optogenetic protein clustering and signaling activation in mammalian cells[J]. Nature Methods, 2013, 10 (3): 249-252. |
| 130 | GIL A A, LAPTENOK S P, IULIANO J N, et al. Photoactivation of the BLUF protein PixD Probed by the site-specific incorporation of fluorotyrosine residues[J]. Journal of the American Chemical Society, 2017, 139 (41): 14638-14648. |
| 131 | LAU Y H, GIESSEN T W, ALTENBURG W J, et al. Prokaryotic nanocompartments form synthetic organelles in a eukaryote[J]. Nature Communications, 2018, 9 (1): 1311. |
| 132 | ZHAO E M, SUEK N, WILSON M Z, et al. Light-based control of metabolic flux through assembly of synthetic organelles[J]. Nature Chemical Biology, 2019, 15 (6): 589-597. |
| 133 | RUGBJERG P, SOMMER M O A. Overcoming genetic heterogeneity in industrial fermentations[J]. Nature Biotechnology, 2019, 37 (8): 869-876. |
| 134 | RUGBJERG P, SARUP-LYTZEN K, NAGY M, et al. Synthetic addiction extends the productive life time of engineered Escherichia coli populations[J]. PNAS, 2018, 115 (10): 2347-2352. |
| 135 | XIAO Y, BOWEN C H, LIU D, et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis[J]. Nature Chemical Biology, 2016, 12 (5): 339-344. |
| 136 | RAMAN S, ROGERS J K, TAYLOR N D, et al. Evolution-guided optimization of biosynthetic pathways[J]. PNAS, 2014, 111 (50): 17803-17808. |
| 137 | GUPTA A, REIZMAN I M, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nature Biotechnology, 2017, 35 (3): 273-279. |
| 138 | DAHL R H, ZHANG F, ALONSO-GUTIERREZ J, et al. Engineering dynamic pathway regulation using stress-response promoters[J]. Nature Biotechnology, 2013, 31 (11): 1039-1046. |
| 139 | LIANG C, ZHANG X, WU J, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit[J]. Metabolic Engineering, 2020, 57: 239-246. |
| 140 | CERONI F, BOO A, FURINI S, et al. Burden-driven feedback control of gene expression[J]. Nature Methods, 2018, 15 (5): 387-393. |
| 141 | JULLESSON D, DAVID F, PFLEGER B, et al. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals[J]. Biotechnology Advances, 2015, 33 (7): 1395-1402. |
| 142 | SANDBERG T E, SALAZAR M J, WENG L L, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology[J]. Metabolic Engineering, 2019, 56: 1-16. |
| 143 | FLETCHER E, FEIZI A, BISSCHOPS M M M, et al. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments[J]. Metabolic Engineering, 2017, 39: 19-28. |
| 144 | DEATHERAGE D E, KEPNER J L, BENNETT A F, et al. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures[J]. PNAS, 2017, 114 (10): E1904-E1912. |
| 145 | MUNDHADA H, SEOANE J M, SCHNEIDER K, et al. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution[J]. Metabolic Engineering, 2017, 39: 141-150. |
| 146 | RODRíGUEZ-VERDUGO A, CARRILLO-CISNEROS D, GONZáLEZ-GONZáLEZ A, et al. Different tradeoffs result from alternate genetic adaptations to a common environment[J]. PNAS, 2014, 111 (33): 12121. |
| 147 | CASPETA L, CHEN Y, GHIACI P, et al. Altered sterol composition renders yeast thermotolerant[J]. Science, 2014, 346 (6205): 75-78. |
| 148 | THORWALL S, SCHWARTZ C, CHARTRON J W, et al. Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis[J]. Nature Chemical Biology, 2020, 16 (2): 113-121. |
| 149 | CALERO P, NIKEL P I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms[J]. Microbial Biotechnology, 2019, 12 (1): 98-124. |
| 150 | KUMAR V, PARK S. Potential and limitations of Klebsiella pneumoniae as a microbial cell factory utilizing glycerol as the carbon source[J]. Biotechnology Advances, 2018, 36 (1): 150-167. |
| 151 | CHARUBIN K, BENNETT R K, FAST A G, et al. Engineering Clostridium organisms as microbial cell-factories: challenges & opportunities[J]. Metabolic Engineering, 2018, 50: 173-191. |
| 152 | TAN D, WU Q, CHEN J C, et al. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates[J]. Metabolic Engineering, 2014, 26: 34-47. |
| 153 | BRENNER K, YOU L, ARNOLD F H. Engineering microbial consortia: a new frontier in synthetic biology[J]. Trends in Biotechnology, 2008, 26 (9): 483-489. |
| 154 | JONES J A, WANG X. Use of bacterial co-cultures for the efficient production of chemicals[J]. Current Opinion in Biotechnology, 2018, 53: 33-38. |
| 155 | SONG H, DING M Z, JIA X Q, et al. Synthetic microbial consortia: from systematic analysis to construction and applications[J]. Chemical Society Reviews, 2014, 43 (20): 6954-6981. |
| 156 | HAYS S G, PATRICK W G, ZIESACK M, et al. Better together: engineering and application of microbial symbioses[J]. Current Opinion in Biotechnology, 2015, 36: 40-49. |
| 157 | KONG W T, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14 (8): 821-829. |
| 158 | JAROSZ D F, BROWN J C S, WALKER G A, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism[J]. Cell, 2014, 158 (5): 1083-1093. |
| 159 | ZELEZNIAK A, ANDREJEV S, PONOMAROVA O, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities[J]. PNAS, 2015, 112 (20): 6449-6454. |
| 160 | WINTERMUTE E H, SILVER P A. Dynamics in the mixed microbial concourse[J]. Genes & Development, 2010, 24 (23): 2603-2614. |
| 161 | BRENNER K, KARIG D K, WEISS R, et al. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium[J]. PNAS, 2007, 104 (44): 17300-17304. |
| 162 | FAUST K, RAES J. Microbial interactions: from networks to models[J]. Nature Reviews Microbiology, 2012, 10 (8): 538-550. |
| 163 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37 (2): 181-197. |
| 164 | SCOTT S R, DIN M O, BITTIHN P, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated[J]. Nature Microbiology, 2017, 2 (8): 17083. |
| 165 | SHOU W Y, RAM S, VILAR J M G. Synthetic cooperation in engineered yeast populations[J]. PNAS, 2007, 104 (6): 1877-1882. |
| 166 | HU B, DU J, ZOU R Y, et al. An environment-sensitive synthetic microbial ecosystem[J]. PLoS One, 2010, 5 (5): e10619. |
| 167 | WANG E X, DING M Z, MA Q, et al. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation[J]. Microbial Cell Factories, 2016, 15: 21. |
| 168 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor[J]. Microbial Cell Factories, 2019, 18 (1): 35. |
| 169 | LIU Y, DING M Z, LING W, et al. A three-species microbial consortium for power generation[J]. Energy & Environmental Science, 2017, 10 (7): 1600-1609. |
| 170 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33 (4): 377-383. |
| 171 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids[J]. Metabolic Engineering, 2016, 35: 55-63. |
| 172 | PANDE S, SHITUT S, FREUND L, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria[J]. Nature Communications, 2015, 6: 6238. |
| 173 | BENOMAR S, RANAVA D, CARDENAS M L, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species[J]. Nature Communications, 2015, 6: 6283. |
| 174 | WEBER W, BABA M, FUSSENEGGER M. Synthetic ecosystems based on airborne inter- and intrakingdom communication[J]. PNAS, 2007, 104 (25): 10435-10440. |
| 175 | CHEN Z Y, SUN X X, LI Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols[J]. Metabolic Engineering, 2017, 39: 102-109. |
| 176 | JONES J A, VERNACCHIO V R, COLLINS S M, et al. Complete biosynthesis of Anthocyanins using E. coli polycultures[J]. Mbio., 2017, 8 (3): e00621-00617. |
| 177 | QIAN X, CHEN L, SUI Y, et al. Biotechnological potential and applications of microbial consortia[J]. Biotechnology Advances, 2020, 40: 107500. |
| 178 | SGOBBA E, WENDISCH V F. Synthetic microbial consortia for small molecule production[J]. Current Opinion in Biotechnology, 2019, 62: 72-79. |
| 179 | ZHANG H, PEREIRA B, LI Z, et al. Engineering Escherichia coli coculture systems for the production of biochemical products[J]. PNAS, 2015, 112 (27): 8266-8271. |
| 180 | LIU X, LI X B, JIANG J L, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metabolic Engineering, 2018, 47: 243-253. |
| 181 | ZHAO C H, SINUMVAYO J P, ZHANG Y P, et al. Design and development of a "Y-shaped" microbial consortium capable of simultaneously utilizing biomass sugars for efficient production of butanol[J]. Metabolic Engineering, 2019, 55: 111-119. |
| 182 | AKDEMIR H, SILVA A, ZHA J, et al. Production of pyranoanthocyanins using Escherichia coli co-cultures[J]. Metabolic Engineering, 2019, 55: 290-298. |
| 183 | GUO X Y, LI Z H, WANG X N, et al. De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering[J]. Biotechnology and Bioengineering, 2019, 116 (12): 3349-3359. |
| 184 | MINTY J J, SINGER M E, SCHOLZ S A, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass[J]. PNAS, 2013, 110 (36): 14592-14597. |
| 185 | SCHOLZ S A, GRAVES I, MINTY J J, et al. Production of cellulosic organic acids via synthetic fungal consortia[J]. Biotechnology and Bioengineering, 2018, 115 (4): 1096-1100. |
| 186 | TSAI S L, GOYAL G, CHEN W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production[J]. Applied and Environmental Microbiology, 2010, 76 (22): 7514-7520. |
| 187 | WEN Z Q, MINTON N P, ZHANG Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium[J]. Metabolic Engineering, 2017, 39: 38-48. |
| 188 | GILBERT E S, WALKER A W, KEASLING J D. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion[J]. Applied Microbiology and Biotechnology, 2003, 61 (1): 77-81. |
| 189 | ZHANG H, YANG C, LI C K, et al. Functional assembly of a microbial consortium with autofluorescent and mineralizing activity for the biodegradation of organophosphates[J]. Journal of Agricultural and Food Chemistry, 2008, 56 (17): 7897-7902. |
| 190 | MARTINEZ I, MOHAMED M E S, ROZAS D, et al. Engineering synthetic bacterial consortia for enhanced desulfurization and revalorization of oil sulfur compounds[J]. Metabolic Engineering, 2016, 35: 46-54. |
| 191 | IBRAR M, ZHANG H. Construction of a hydrocarbon-degrading consortium and characterization of two new lipopeptides biosurfactants[J]. Science of the Total Environment, 2020, 714: 136400. |
| 192 | ERLICH Y, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355 (6328): 950-953. |
| 193 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36 (7): 660-660. |
| 194 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20 (8): 456-466. |
| 195 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494 (7435): 77-80. |
| 196 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337 (6102): 1628. |
| 197 | KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory[J]. Nature Biotechnology, 2020, 38 (1): 39-43. |
| 198 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie-International Edition, 2015, 54 (8): 2552-2555. |
| 199 | TAKAHASHI C N, NGUYEN B H, STRAUSS K, et al. Demonstration of end-to-end automation of DNA data storage[J]. Scientific Reports, 2019, 9 (1): 4998. |
| 200 | CHOI Y, RYU T, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9 (1): 6582. |
| 201 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37 (10): 1237. |
| 202 | 陈为刚, 黄刚, 李炳志, 等. 音视频文件的DNA信息存储[J]. 中国科学:生命科学, 2020, 50 (1): 81-85. |
| CHEN W G, HUANG G, LI B Z, et al. DNA information storage for audio and video files (in Chinese)[J]. SCIENTIA SINICA Vitae, 2020, 50(1): 81-85. | |
| 203 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440 (7082): 297-302. |
| 204 | 钱璐璐, 汪颖, 张钊, 等. DNA纳米结构仿中国地图[J]. 科学通报, 2006, 51 (24): 2860-2863. |
| QIAN L L, WANG Y, ZHANG Z, et al. Imating China map with DNA nanostructure[J]. Chinese Science Bulletin, 2006, 51 (24): 2860-2863. | |
| 205 | DOUGLAS S M, CHOU J J, SHIH W M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination[J]. PNAS, 2007, 104 (16): 6644-6648. |
| 206 | ANDERSEN E S, DONG M, NIELSEN M M, et al. Self-assembly of a nanoscale DNA box with a controllable lid[J]. Nature, 2009, 459 (7243): 73-76. |
| 207 | HAN D R, PAL S, NANGREAVE J, et al. DNA origami with complex curvatures in three-dimensional space[J]. Science, 2011, 332 (6027): 342-346. |
| 208 | DU Y, JIANG Q, BEZIERE N, et al. DNA-nanostructure-gold-nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy[J]. Advanted Materials, 2016, 28 (45): 10000-10007. |
| 209 | ZHANG Y N, WANG F, CHAO J, et al. DNA origami cryptography for secure communication[J]. Nature Communications, 2019, 10 (1): 5469. |
| 210 | BAZRAFSHAN A, MEYER T A, SU H Q, et al. Tunable DNA origami motors translocate ballistically over μm distances at nm/s speeds[J]. Angewandte Chemie-International Edition, 2020, 132: 2-10. |
| 211 | YOUNG T S, SCHULTZ P G. Beyond the canonical 20 amino acids: expanding the genetic lexicon[J]. Journal of Biological Chemistry, 2010, 285 (15): 11039-11044. |
| 212 | DEEPANKUMAR K, SHON M, NADARAJAN S P, et al. Enhancing thermostability and organic solvent tolerance of w-transaminase through global incorporation of fluorotyrosine[J]. Advanced Synthesis & Catalysis, 2014, 356 (5): 993-998. |
| 213 | DRIENOVSKA I, MAYER C, DULSON C, et al. A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue[J]. Nature Chemistry, 2018, 10 (9): 946-952. |
| 214 | ROVNER A J, HAIMOVICH A D, KATZ S R, et al. Recoded organisms engineered to depend on synthetic amino acids[J]. Nature, 2015, 518 (7537): 89-93. |
| 215 | MANDELL D J, LAJOIE M J, MEE M T, et al. Biocontainment of genetically modified organisms by synthetic protein design[J]. Nature, 2015, 518 (7537): 55-60. |
| 216 | YU Y, LV X, LI J, et al. Defining the role of tyrosine and rational tuning of oxidase activity by genetic incorporation of unnatural tyrosine analogs[J]. Journal of the American Chemical Society, 2015, 137 (14): 4594-4597. |
| 217 | YOUNG D D, SCHULTZ P G. Playing with the molecules of life[J]. ACS Chemical Biology, 2018, 13 (4): 854-870. |
| 218 | ZHANG Y, PTACIN J L, FISCHER E C, et al. A semi-synthetic organism that stores and retrieves increased genetic information[J]. Nature, 2017, 551 (7682): 644-647. |
| 219 | DUNN M R, JIMENEZ R M, CHAPUT J C. Analysis of aptamer discovery and technology[J]. Nature Reviews Chemistry, 2017, 1 (10): 0076. |
| 220 | CHEN T, HONGDILOKKUL N, LIU Z X, et al. The expanding world of DNA and RNA[J]. Current Opinion in Chemical Biology, 2016, 34: 80-87. |
| 221 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363 (6429): 884-887. |
| 222 | EREMEEVA E, HERDEWIJN P. Non canonical genetic material[J]. Current Opinion in Biotechnology, 2019, 57: 25-33. |
| 223 | KUK S K, SINGH R K, NAM D H, et al. Photoelectrochemical reduction of carbon dioxide to methanol through a highly efficient enzyme cascade[J]. Angewandte Chemie-International Edition, 2017, 56 (14): 3827-3832. |
| 224 | SOKOL K P, ROBINSON W E, WARNAN J, et al. Bias-free photoelectrochemical water splitting with photosystem II on a dye-sensitized photoanode wired to hydrogenase[J]. Nature Energy, 2018, 3 (11): 944-951. |
| 225 | SAKIMOTO K K, WONG A B, YANG P D. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production[J]. Science, 2016, 351 (6268): 74-77. |
| 226 | GUO J L, SUASTEGUI M, SAKIMOTO K K, et al. Light-driven fine chemical production in yeast biohybrids[J]. Science, 2018, 362 (6416): 813-816. |
| 227 | WANG X Y, PU J H, AN B L, et al. Programming cells for dynamic assembly of Iiorganic nano-objects with spatiotemporal control[J]. Advanced Materials, 2018, 30 (16): e1705968. |
| 228 | WANG X Y, PU J H, LIU Y, et al. Immobilization of functional nano-objects in living engineered bacterial biofilms for catalytic applications[J]. National Science Review, 2019, 6 (5): 929-943. |
| 229 | JIN X, HONG S H. Cell-free protein synthesis for producing ‘difficult-to-express’ proteins[J]. Biochemical Engineering Journal, 2018, 138: 156-164. |
| 230 | SHINODA T, SHINYA N, ITO K, et al. Cell-free methods to produce structurally intact mammalian membrane proteins[J]. Scientific Reports, 2016, 6: 30442. |
| 231 | MARSHALL R, MAXWELL C S, COLLINS S P, et al. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system[J]. Molecular Cell, 2018, 69 (1): 146-157.e3. |
| 232 | MARTIN R W, MAJEWSKA N I, CHEN C X, et al. Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies[J]. ACS Synthetic Biology, 2017, 6 (7): 1370-1379. |
| 233 | RAMOS-BENITEZ M J, LOPEZ-CRUZ L M, AGUAYO V, et al. Cell-free expression, purification and immunoreactivity assessment of recombinant Fasciola hepatica saposin-like protein-2[J]. Molecular Biology Reports, 2018, 45 (5): 1551-1556. |
| 234 | GAO W, CHO E, LIU Y, et al. Advances and challenges in cell-free incorporation of unnatural amino acids into proteins[J]. Front Pharmacol, 2019, 10: 611. |
| 235 | ZIMMERMAN E S, HEIBECK T H, GILL A, et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system[J]. Bioconjugate Chemistry, 2014, 25 (2): 351-361. |
| 236 | HOFFMANN B, LOHR F, LAGUERRE A, et al. Protein labeling strategies for liquid-state NMR spectroscopy using cell-free synthesis[J]. Progress in Nuclear Magnetic Resonance Spectroscopy, 2018, 105: 1-22. |
| 237 | PEREZ J G, STARK J C, JEWETT M C. Cell-free synthetic biology: engineering beyond the cell[J]. Cold Spring Harbor Perspectives in Biology, 2016, 8 (12): a023853. |
| 238 | ADACHI J, KATSURA K, SEKI E, et al. Cell-free protein synthesis using S30 extracts from Escherichia coli RFzero strains for efficient incorporation of non-natural amino acids into proteins[J]. International Journal of Molecular Sciences, 2019, 20 (3): 492. |
| [1] | 应汉杰, 柳东, 王振宇, 沈涛, 庄伟, 朱晨杰. 工业生物制造与“碳中和”目标探讨[J]. 合成生物学, 2025, 6(1): 1-7. |
| [2] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||