合成生物学 ›› 2020, Vol. 1 ›› Issue (1): 29-43.DOI: 10.12211/2096-8280.2020-063
生物逆向工程设计在合成生物学中的应用
陈欣懋, 欧阳颀
- 北京大学物理学院,北京 100871
-
收稿日期:2020-05-06修回日期:2020-05-16出版日期:2020-02-29发布日期:2020-07-07 -
通讯作者:欧阳颀 -
作者简介:陈欣懋(1994-),女,博士研究生,研究方向为定量系统生物学和合成生物学。E-mail: xmaochen@pku.edu.cn
欧阳颀(1955-),男,博士,教授,中国科学院院士,研究方向为定量系统生物学、生物网络动力学、生物系统中的非线性问题、生物微流体技术。E-mail: qi@pku.edu.cn -
基金资助:国家自然科学基金(11774011)
The application of biological reverse engineering in synthetic biology
CHEN Xinmao, OUYANG Qi
- School of Physics, Peking University, Beijing 100871, China
-
Received:2020-05-06Revised:2020-05-16Online:2020-02-29Published:2020-07-07 -
Contact:OUYANG Qi
摘要:
合成生物学是一门涉及生物学、生物工程学、系统生物学、数学、物理、化学与信息科学的新生的交叉学科。它的目的是在工程化思想的指导下有目的地、可预测地设计人造生命系统。经过近二十年的蓬勃发展,合成生物学取得了重大成就,但依旧面临复杂系统理性设计的困难。在系统生物学中,运用数学物理等知识根据网络功能来研究功能背后网络结构的方法被称为逆向工程。系统生物学逆向工程的研究思路与合成生物学设计过程的一致性,启发了我们利用逆向工程指导合成生物学的理性设计。逆向工程应用到合成生物学,将大大降低复杂功能回路的设计难度。本文从合成生物学的设计思路与问题出发,根据本文作者研究团队近十年来在逆向工程研究中的经验,归纳总结了目前逆向工程设计在合成生物学中的应用方法,包括网络穷举方法、子网络拼接方法、从离散模型到连续模型的方法,论证了逆向工程指导合成生物学理性设计的可行性与有效性,分析了目前逆向工程设计在合成生物学中的发展瓶颈。
中图分类号:
引用本文
陈欣懋, 欧阳颀. 生物逆向工程设计在合成生物学中的应用[J]. 合成生物学, 2020, 1(1): 29-43.
CHEN Xinmao, OUYANG Qi. The application of biological reverse engineering in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 29-43.
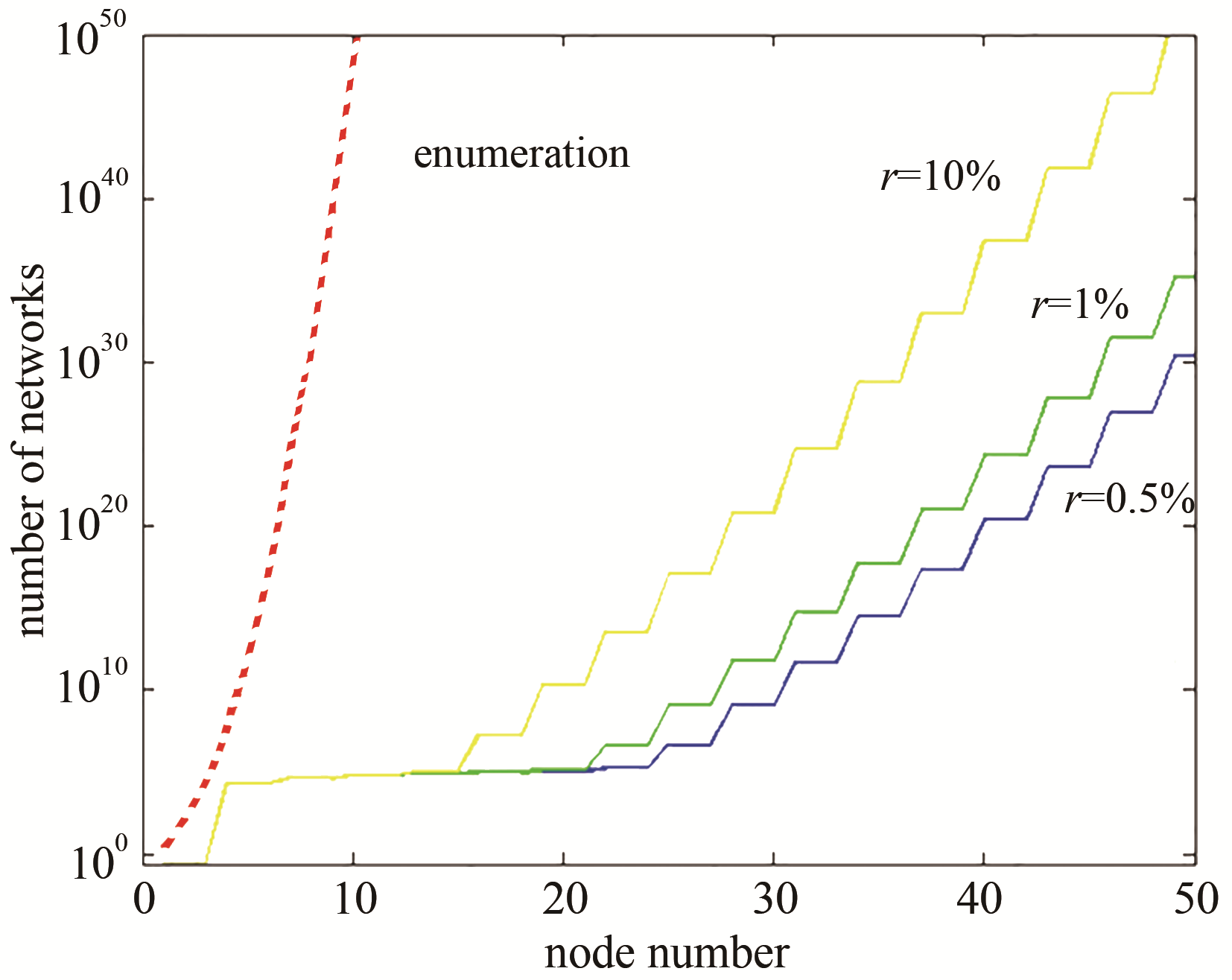
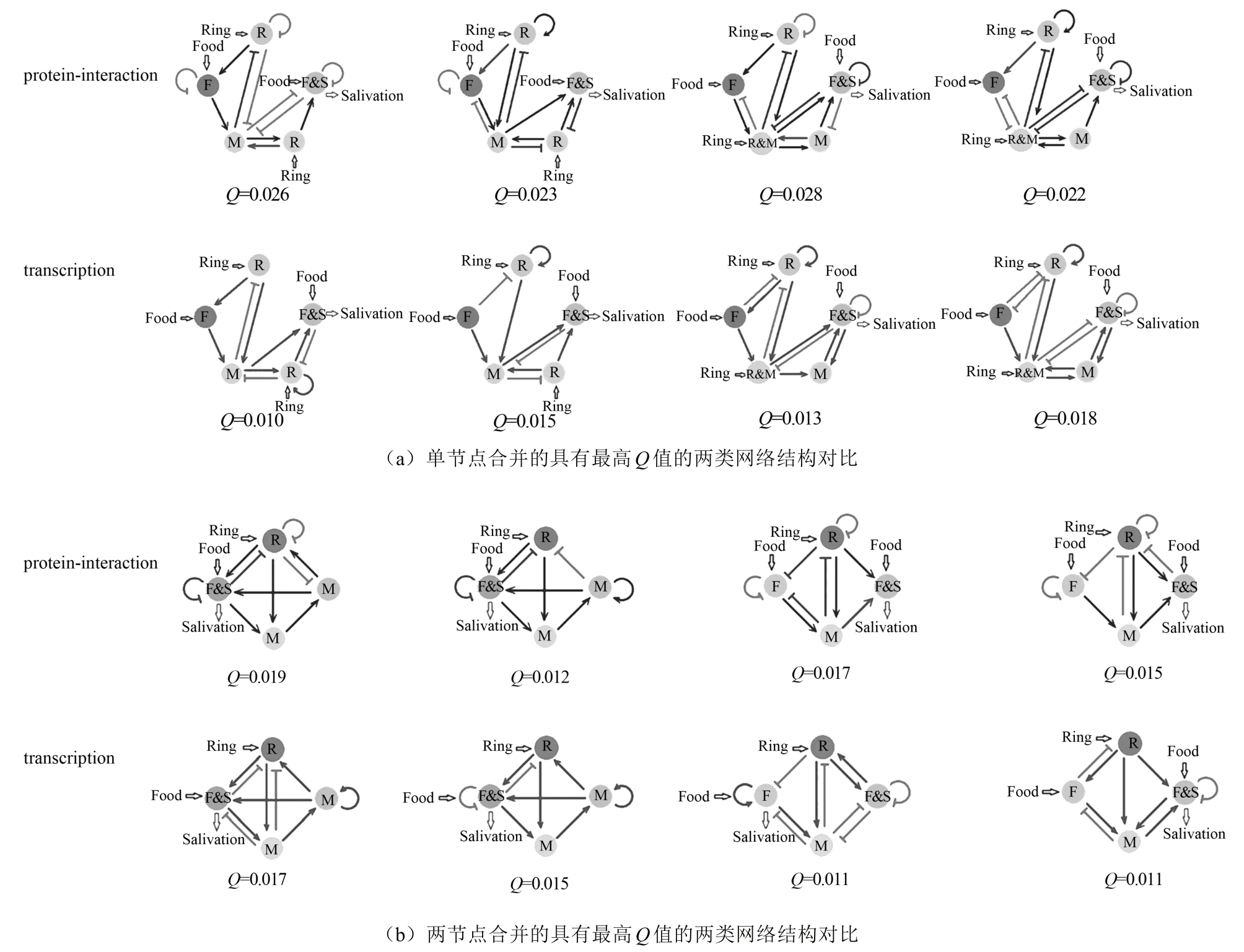
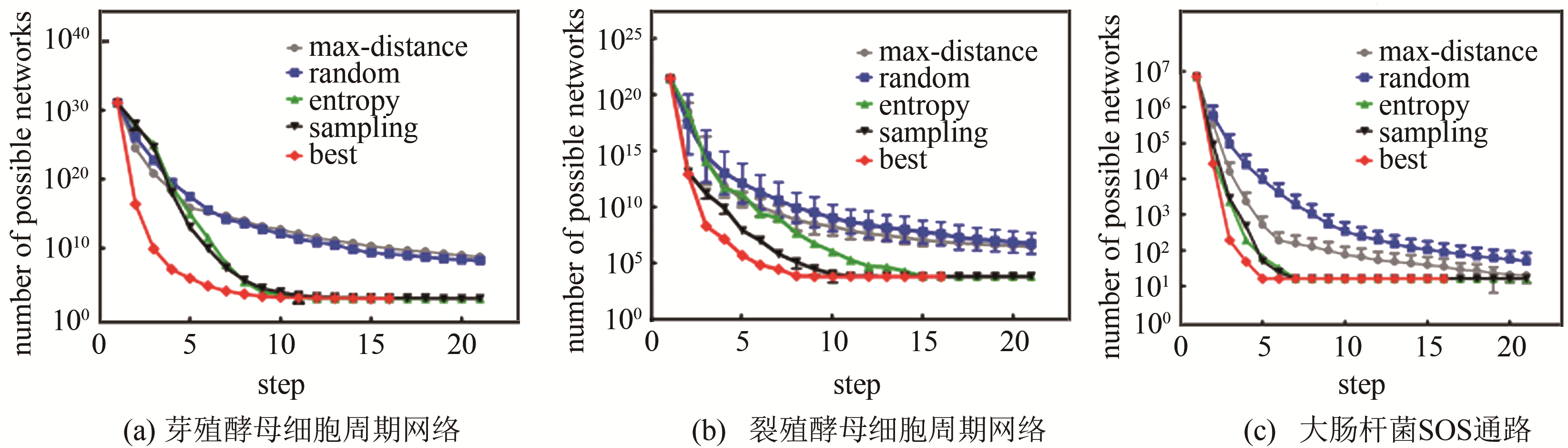
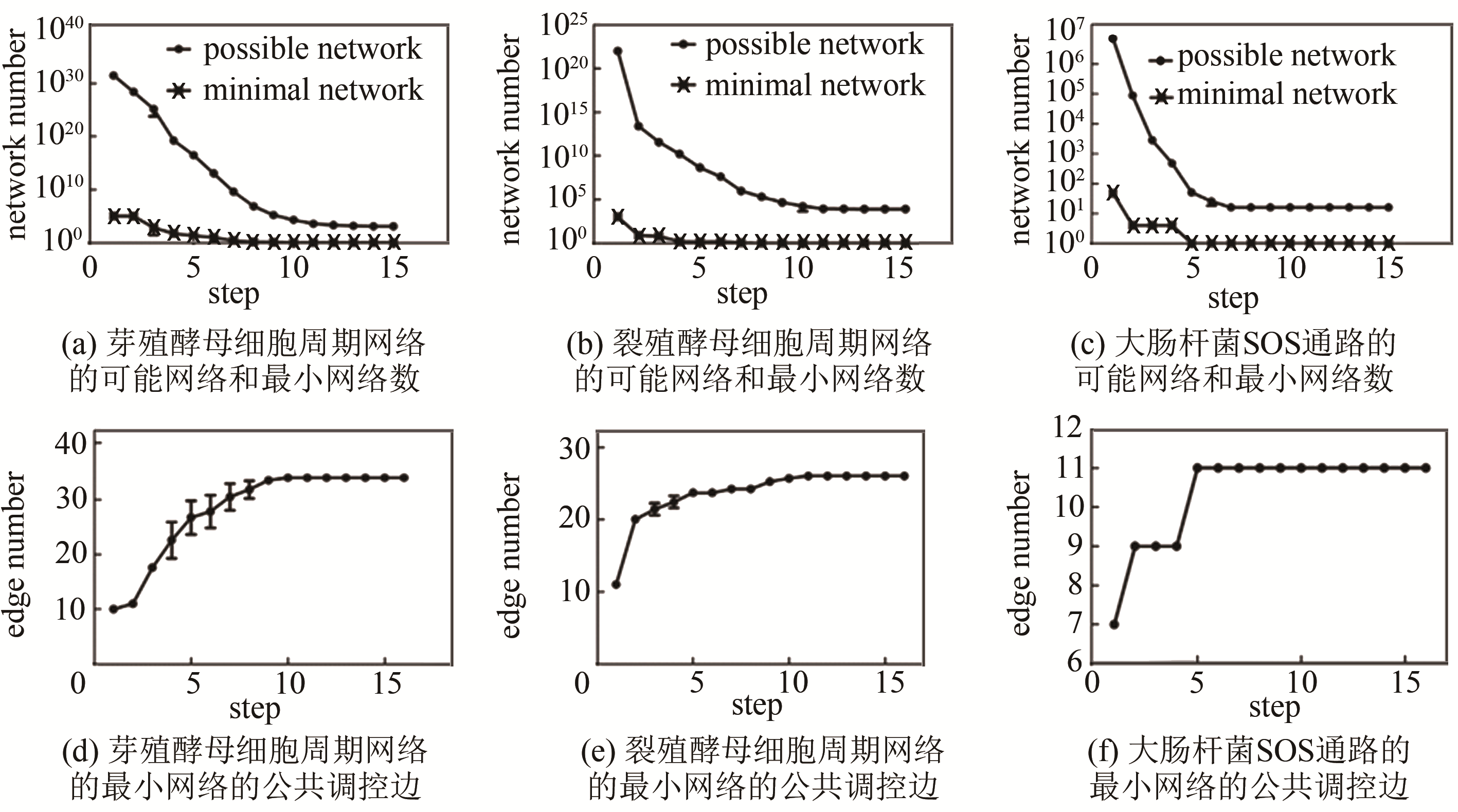
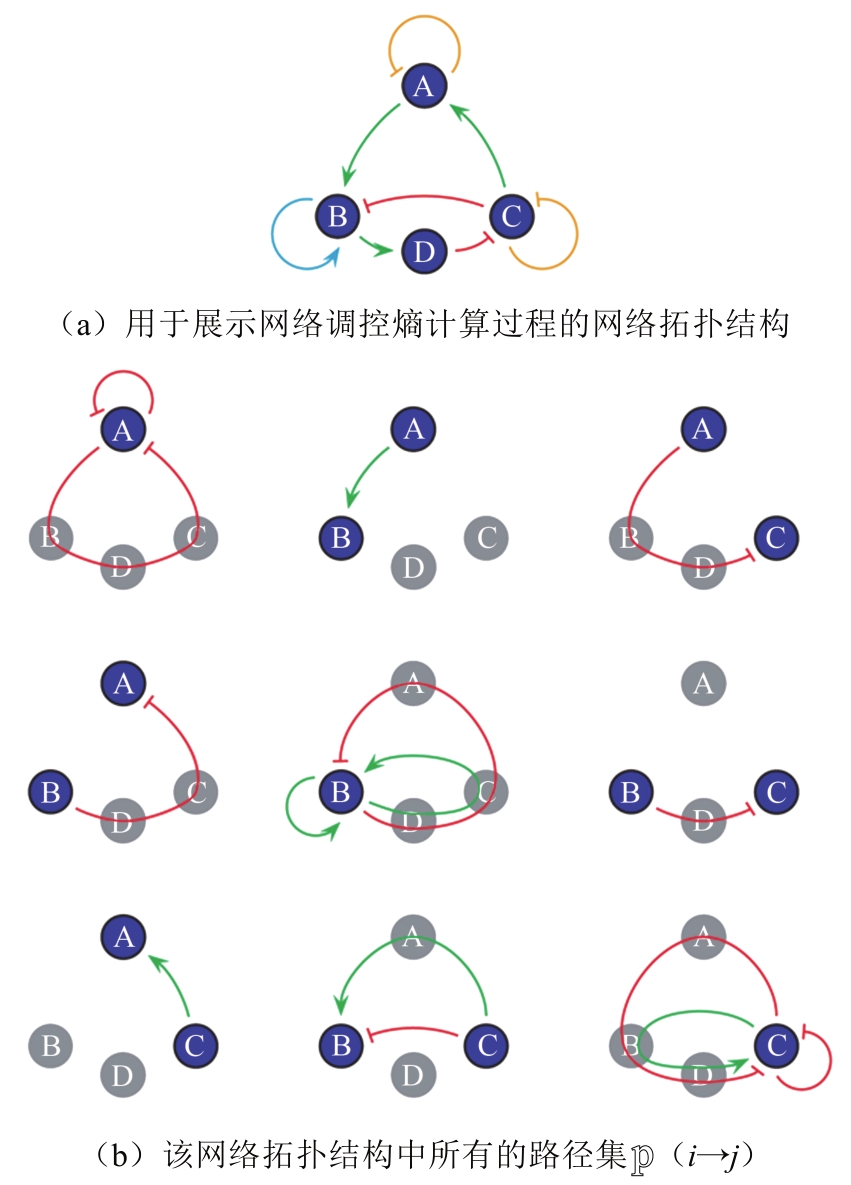
图 5 传统网络穷举与子网络合并的计算复杂度对比[47](虚线表示传统网络穷举方法的计算复杂度随网络节点数的变化,余下折线对应子网络合并方法的计算复杂度的变化)
Fig. 5 The complexity comparison between traditional enumeration and sub-networks combination[47]
| Time | SK | Cdc2 | Ste9 | Rum1 | Slp1 | Cdc2* | Wee1 | Cdc25 | PP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 5 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 6 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| 7 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| 8 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| 9 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
表 1 裂殖酵母细胞周期功能的动力学轨迹[55]
Tab. 1 Biological pathway of fission yeast cell cycle network [55]
| Time | SK | Cdc2 | Ste9 | Rum1 | Slp1 | Cdc2* | Wee1 | Cdc25 | PP |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 5 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| 6 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| 7 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| 8 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| 9 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
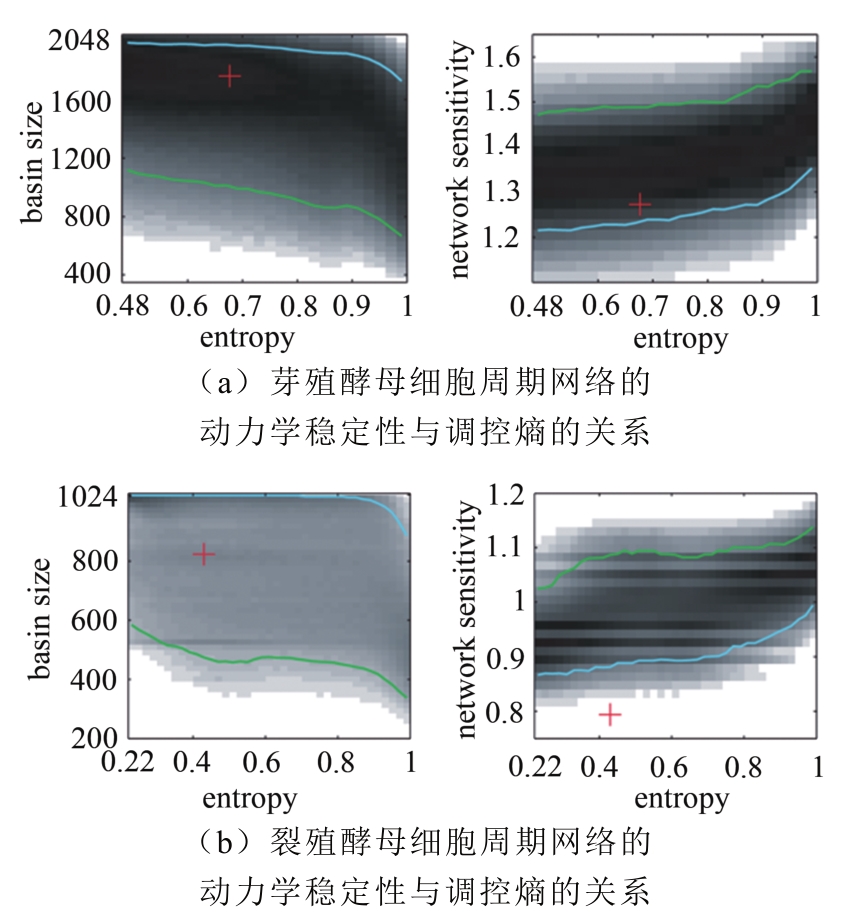
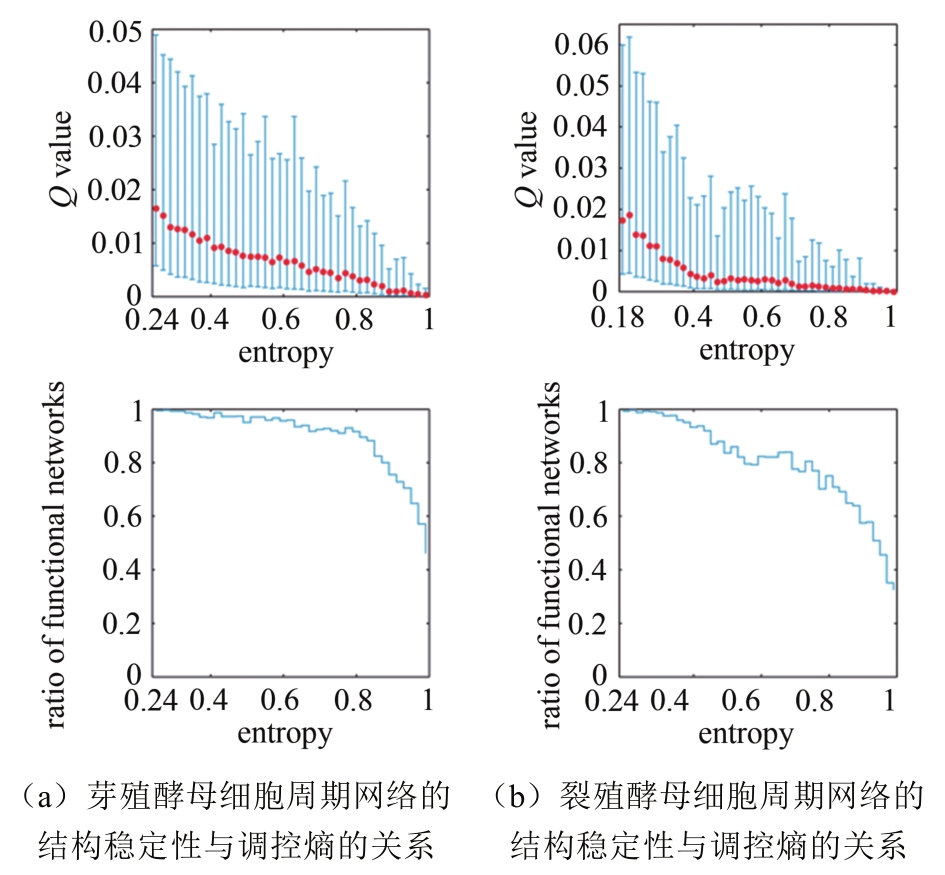
图 11 网络调控熵与动力学稳定性的关系[56](蓝色和绿色线分别表示稳定性最高的前5%和后5%的随机网络对应的动力学稳定性,红十字表示相应的生物网络的动力学稳定性和调控熵)
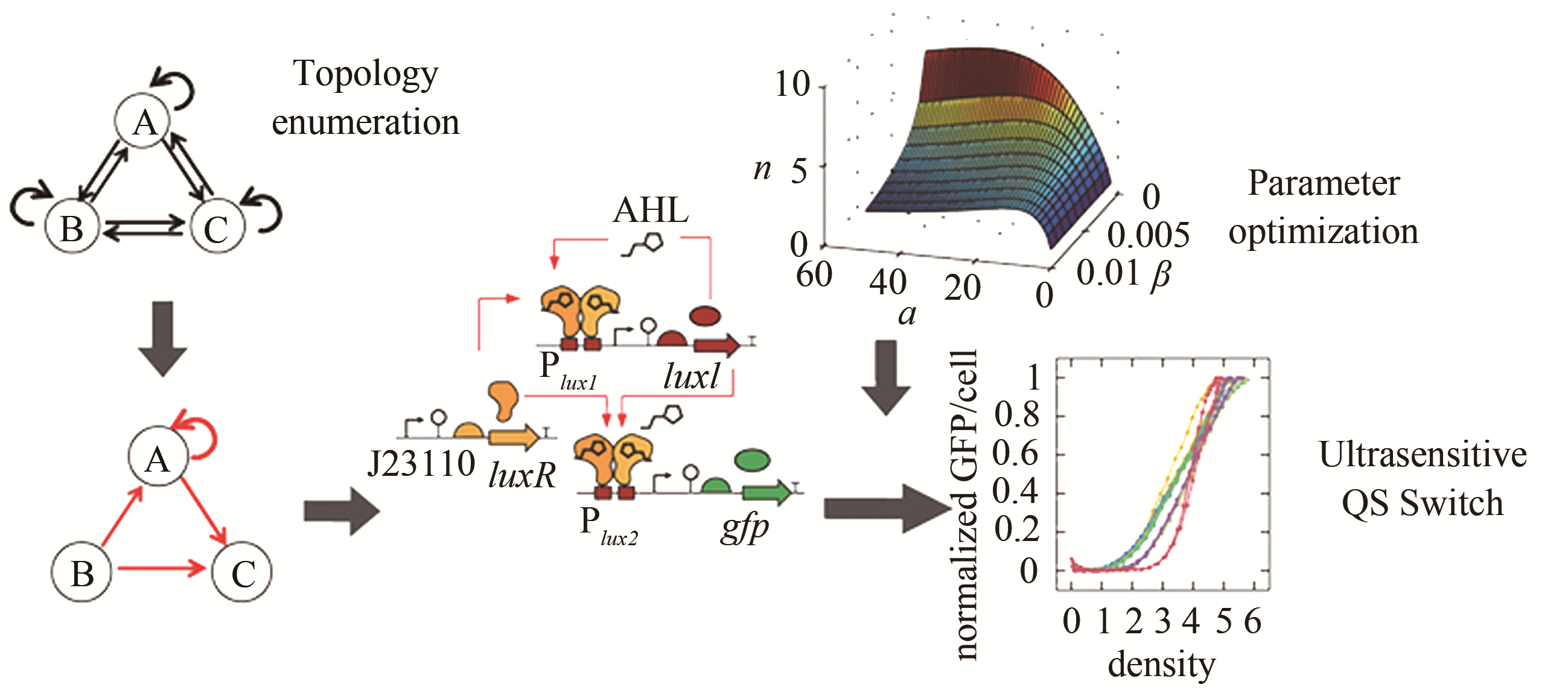
Fig. 11 The relationship of network regulation entropy and dynamic stability [56]
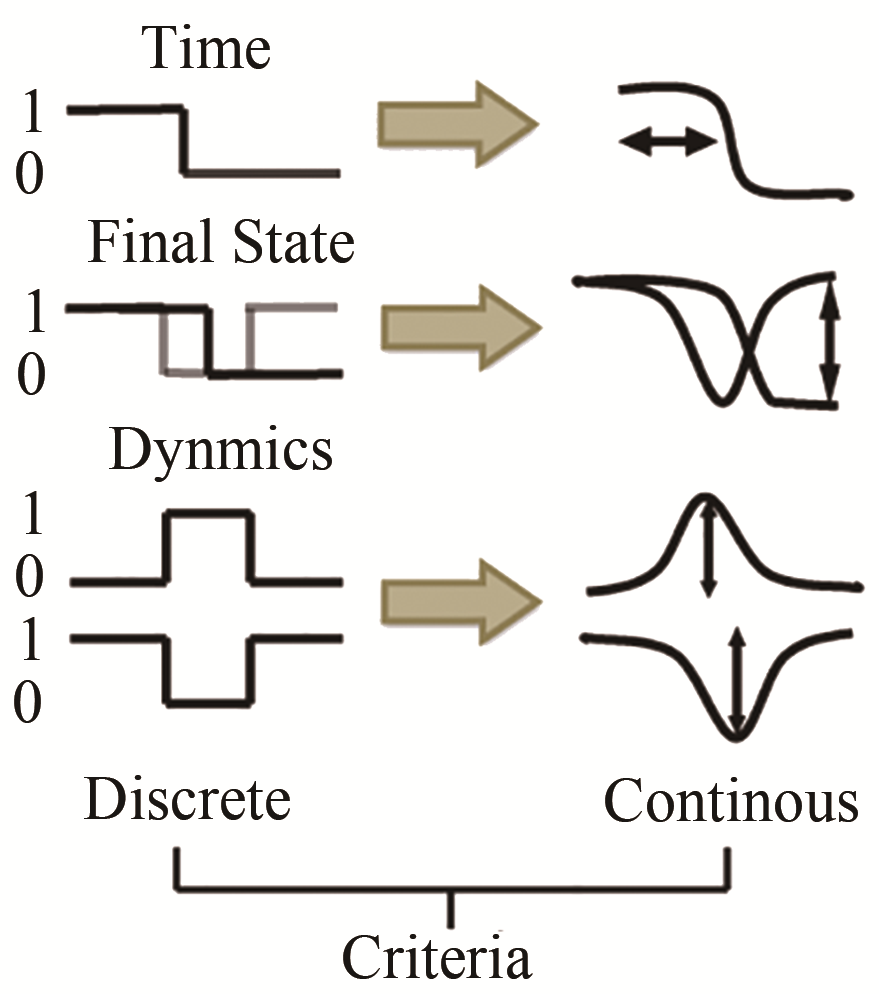
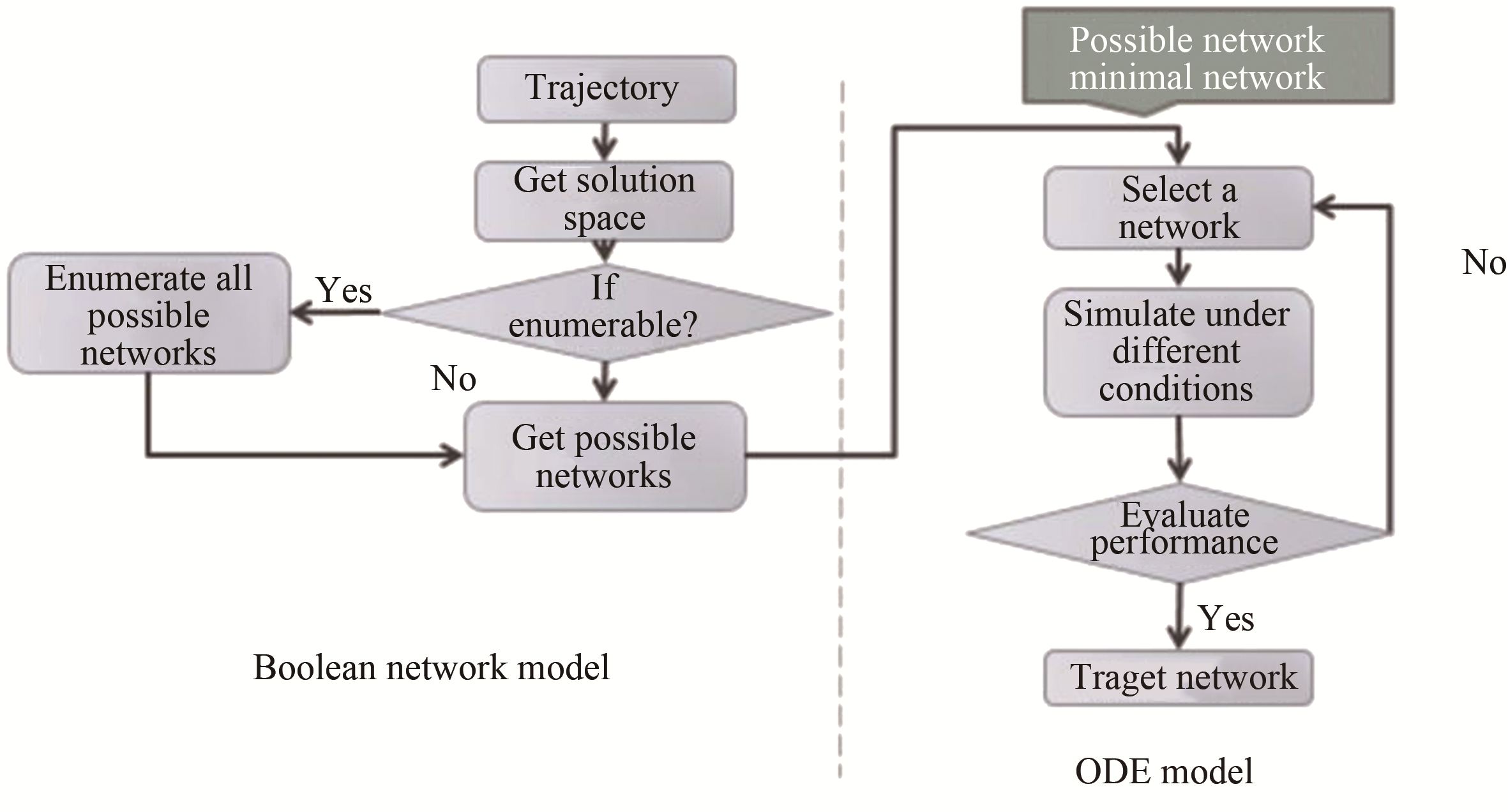
图 15 在布尔网络模型和连续模型下的三个限制条件[58][第一个限制条件是ssDNA的浓度在结束计算时应下调;第二个是系统在经历响应反应后应回到初态(ssDNA除外);第三个是节点在两种模型下的动力学行为特征应该保持一致]
Fig. 15 The three criteria in the Boolean network model and continuous model[58]
| 1 | IDEKER T, THORSSON V, RANISH J A, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network[J]. Science, 2001, 292(5518): 929-934. |
| 2 | BARABÁSI A, CRANDALL R E. Linked: the new science of networks[J]. American Journal of Physics, 2002, 71(4): 243-270. |
| 3 | WOLKENHAUER O. Systems biology: the reincarnation of systems theory applied in biology?[J]. Briefings in Bioinformatics, 2001, 2(3): 258-270. |
| 4 | BORNHOLDT S. Less is more in modeling large genetic networks[J]. Science, 2005, 310(5747): 449-451. |
| 5 | FRIEDMAN N. Inferring cellular networks using probabilistic graphical models[J]. Science, 2004, 303(5659): 799-805. |
| 6 | SAEZ‐RODRIGUEZ J, ALEXOPOULOS L G, EPPERLEIN J, et al. Discrete logic modelling as a means to link protein signaling networks with functional analysis of mammalian signal transduction[J]. Molecular Systems Biology, 2009, 5(1): e331. |
| 7 | BASSO K, MARGOLIN A A, STOLOVITZKY G, et al. Reverse engineering of regulatory networks in human B cells[J]. Nature Genetics, 2005, 37(4): 382-390. |
| 8 | ZOPPOLI P, MORGANELLA S, CECCARELLI M. TimeDelay-ARACNE: reverse engineering of gene networks from time-course data by an information theoretic approach[J]. BMC Bioinformatics, 2010, 11(1): 154-169. |
| 9 | YEUNG M K S, TEGNÉR J, COLLINS J J. Reverse engineering gene networks using singular value decomposition and robust regression[J]. PNAS, 2002, 99(9): 6163-6168. |
| 10 | TEGNER J, YEUNG M K S, HASTY J, et al. Reverse engineering gene networks: integrating genetic perturbations with dynamical modeling[J]. PNAS, 2003, 100(10): 5944-5949. |
| 11 | BONGARD J, LIPSON H. Automated reverse engineering of nonlinear dynamical systems[J]. PNAS, 2007, 104(24): 9943-9948. |
| 12 | D’HAESELEER P, LIANG S, SOMOGYI R. Genetic network inference: from co-expression clustering to reverse engineering[J]. Bioinformatics, 2000, 16(8): 707-726. |
| 13 | GARDNER T S, FAITH J J. Reverse-engineering transcription control networks [J]. Physics of Life Reviews, 2005, 2(1): 65-88. |
| 14 | VILLAVERDE A F, BANGA J R. Reverse engineering and identification in systems biology: strategies, perspectives and challenges[J]. Journal of the Royal Society Interface, 2014, 11(91): 20130505. |
| 15 | SLUSARCZYK A L, LIN A, WEISS R. Foundations for the design and implementation of synthetic genetic circuits[J]. Nature Reviews Genetics, 2012, 13(6): 406-420. |
| 16 | CANTON B, LABNO A, ENDY D. Refinement and standardization of synthetic biological parts and devices[J]. Nature Biotechnology, 2008, 26(7): 787-793. |
| 17 | ENDY D. Foundations for engineering biology[J]. Nature, 2005, 438(7067): 449-453. |
| 18 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 19 | GARDNER T S, CANTOR C R, Collins J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 20 | BASHOR C J, HORWITZ A A, PEISAJOVICH S G, et al. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems[J]. Annual Review of Biophysics, 2010, 39: 515-537. |
| 21 | MAYO A E, SETTY Y, SHAVIT S, et al. Plasticity of the cis-regulatory input function of a gene[J]. PLoS Biology, 2006, 4(4):555-561. |
| 22 | ANDERSON J C, VOIGT C A, ARKIN A P. Environmental signal integration by a modular AND gate[J]. Molecular Systems Biology, 2007, 3(1): e133. |
| 23 | TAMSIR A, TABOR J J, VOIGT C A. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’[J]. Nature, 2011, 469(7329): 212-215. |
| 24 | MOON T S, LOU C, TAMSIR A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491(7423): 249-253. |
| 25 | LOU C, LIU X, NI M, et al. Synthesizing a novel genetic sequential logic circuit: a push‐on push‐off switch[J]. Molecular Systems Biology, 2010, 6(1): e350. |
| 26 | WANG B, KITNEY R I, JOLY N, et al. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology[J]. Nature Communications, 2011, 2(1): 1-9. |
| 27 | BALAGADDÉ F K, SONG H, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem[J]. Molecular Systems Biology, 2008, 4(1): e187. |
| 28 | MARTIN V J J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids[J]. Nature Biotechnology, 2003, 21(7): 796-802. |
| 29 | RO D K, PARADISE E M, OUELLET M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast[J]. Nature, 2006, 440(7086): 940-943. |
| 30 | ZHANG H, LIN M, SHI H, et al. Programming a Pavlovian-like conditioning circuit in Escherichia coli [J]. Nature Communications, 2014, 5(1): 1-10. |
| 31 | MATSUURA S, ONO H, KAWASAKI S, et al. Synthetic RNA-based logic computation in mammalian cells[J]. Nature Communications, 2018, 9(1): 1-8. |
| 32 | LIU W, CREMER J, LI D, et al. An evolutionarily stable strategy to colonize spatially extended habitats[J]. Nature, 2019, 575(7784): 664-668. |
| 33 | BASHOR C J, PATEL N, CHOUBEY S, et al. Complex signal processing in synthetic gene circuits using cooperative regulatory assemblies[J]. Science, 2019, 364(6440): 593-597. |
| 34 | SHAO Y, LU N, WU Z, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 35 | MCKAY M D, BECKMAN R J, Conover W J. A comparison of three methods for selecting values of input variables in the analysis of output from a computer code[J]. Technometrics, 2000, 42(1): 55-61. |
| 36 | MA W, TRUSINA A, El-SAMAD H, et al. Defining network topologies that can achieve biochemical adaptation[J]. Cell, 2009, 138(4): 760-773. |
| 37 | MA W, LAI L, OUYANG Q, et al. Robustness and modular design of the Drosophila segment polarity network[J]. Molecular Systems Biology, 2006, 2(1): e70. |
| 38 | YAN L, OUYANG Q, WANG H. Dose-response aligned circuits in signaling systems[J]. PLoS One, 2012, 7(4): e34727. |
| 39 | CHAU A H, WALTER J M, GERARDIN J, et al. Designing synthetic regulatory networks capable of self-organizing cell polarization[J]. Cell, 2012, 151(2): 320-332. |
| 40 | ZHENG M M, SHAO B, OUYANG Q. Identifying network topologies that can generate turing pattern[J]. Journal of Theoretical Biology, 2016, 408: 88-96. |
| 41 | WU L, OUYANG Q, WANG H. Robust network topologies for generating oscillations with temperature-independent periods[J]. PLoS One, 2017, 12(2): e0171263. |
| 42 | GERARDIN J, REDDY N R, LIM W A. The design principles of biochemical timers: circuits that discriminate between transient and sustained stimulation[J]. Cell Systems, 2019, 9(3): 297-308. |
| 43 | GUET C C, ELOWITZ M B, HSING W, et al. Combinatorial synthesis of genetic networks[J]. Science, 2002, 296(5572): 1466-1470. |
| 44 | MANGAN S, ALON U. Structure and function of the feed-forward loop network motif[J]. PNAS, 2003, 100(21): 11980-11985. |
| 45 | WHITAKER W R, DAVIS S A, ARKIN A P, et al. Engineering robust control of two-component system phosphotransfer using modular scaffolds[J]. PNAS, 2012, 109(4): 18090-18095. |
| 46 | 习静怡. 生物网络逆向工程的子网络合并法以及生物系统中的量子热力学[D]. 北京:北京大学, 2017. |
| XI J Y. Sub-network combination in biological network reverse engineering and quantum thermodynamics in biological systems[D]. Beijing: Peking University, 2017. | |
| 47 | XI J Y, OUYANG Q. Using sub-network combinations to scale up an enumeration method for determining the network structures of biological functions[J]. PLoS One, 2016, 11(12): e0168214. |
| 48 | WU L, WANG H, OUYANG Q. Constructing network topologies for multiple signal-encoding functions[J]. BMC Systems Biology, 2019, 13(1): 6. |
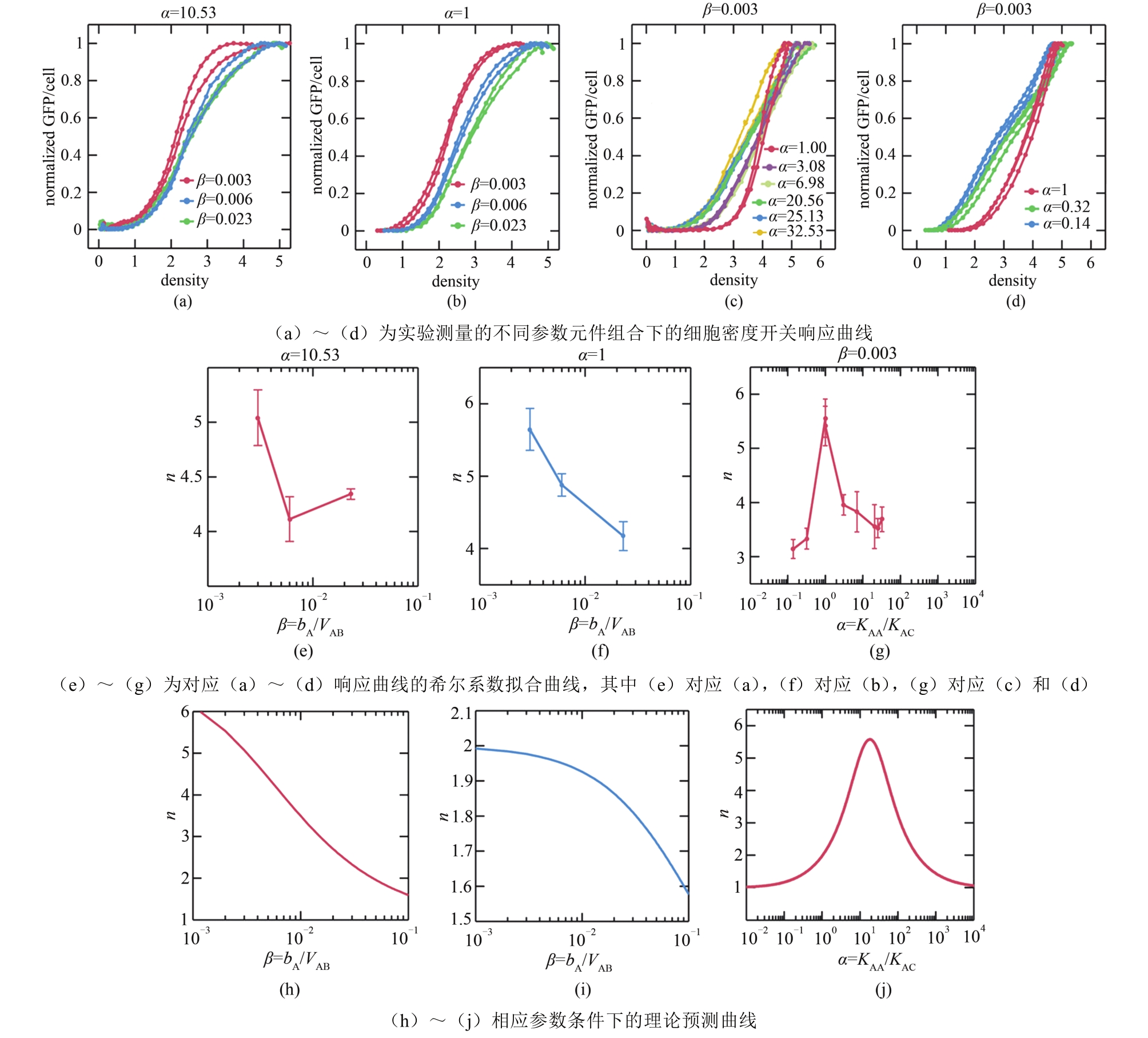
| 49 | ZENG W, DU P, LOU Q, et al. Rational design of an ultrasensitive quorum-sensing switch[J]. ACS Synthetic Biology, 2017, 6(8): 1445-1452. |
| 50 | 曾维倩.基于逆向工程的超敏细胞密度开关的理性设计[D]. 北京: 北京大学, 2017. |
| ZENG W Q. Rational design of an ultrasensitive cell density switch based on reverse engineering[D]. Beijing: Peking University, 2017. | |
| 51 | LI F, LONG T, LU Y, et al. The yeast cell-cycle network is robustly designed[J]. PNAS, 2004, 101(14): 4781-4786. |
| 52 | WANG G, DU C, CHEN H, et al. Process-based network decomposition reveals backbone motif structure[J]. PNAS, 2010, 107(23): 10478-10483. |
| 53 | 张晓萌.布尔网络动力学研究及其反向工程[D]. 北京: 北京大学, 2011. |
| ZHANG X M. Dynamics and reverse engineering of Boolean network[D]. Beijing: Peking University, 2011. | |
| 54 | ZHANG X, SHAO B, WU Y, et al. A reverse engineering approach to optimize experiments for the construction of biological regulatory networks[J]. PLoS One, 2013, 8(9): e75931. |
| 55 | SHAO B, WU J, TIAN B, et al. Minimum network constraint on reverse engineering to develop biological regulatory networks[J]. Journal of Theoretical Biology, 2015, 380: 9-15. |
| 56 | WU Y, ZHANG X, YU J, et al. Identification of a topological characteristic responsible for the biological robustness of regulatory networks[J]. PLoS Computational Biology, 2009, 5(7): e1000442. |
| 57 | 邵斌.生物网络逆向工程及其应用[D]. 北京: 北京大学, 2016. |
| SHAO B. Biological network reverse engineering and its applications[D]. Beijing: Peking University, 2016. | |
| 58 | SHAO B, LIU X, ZHANG D, et al. From Boolean network model to continuous model helps in design of functional circuits[J]. PLoS One, 2015, 10(6):e0128630. |
| 59 | 张浩千.细菌转录调控线路的精准制造[D]. 北京: 北京大学, 2016. |
| ZHANG H Q. High-precision manufacturing of transcriptional regulatory circuits in bacteria [D]. Beijing: Peking University, 2016. | |
| 60 | ZONG Y Q, ZHANG H M, LYU C, et al. Insulated transcriptional elements enable precise design of genetic circuits[J]. Nature Communications, 2017, 8(1): 1-13. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||