Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (2): 212-225.DOI: 10.12211/2096-8280.2020-006
• Invited Review • Previous Articles Next Articles
Compatibility between synthetic pathway and chassis cells from the viewpoint of post-translational modifications
YOU Di1, YE Bangce1,2
- 1.Laboratory of Biosystems and Microanalysis,State Key Laboratory of Bioreactor Engineering,East China University of Science and Technology,Shanghai 200237,China
2.Institute of Engineering Biology and Health,Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals,College of Pharmaceutical Sciences,Zhejiang University of Technology,Hangzhou 310014,Zhejiang,China
-
Received:2020-02-28Revised:2020-04-08Online:2020-08-04Published:2020-04-30 -
Contact:YE Bangce
从翻译后修饰角度解析人工合成途径与底盘细胞的适配性
尤迪1, 叶邦策1,2
- 1.华东理工大学微分析与生物系统工程实验室,生物反应器工程国家重点实验室,上海 200237
2.浙江工业大学工程生物学与健康研究中心,长三角绿色制药协同创新中心,药学院,杭州 浙江 310014
-
通讯作者:叶邦策 -
作者简介:尤迪(1989—),女,博士,副研究员,研究方向为微生物代谢调控与翻译后修饰研究。E-mail:030111115@mail.ecust.edu.cn
叶邦策(1967—),男,博士,教授,研究方向为合成生物学及智能生物系统设计、生物分析及生物传感研究。E-mail:bcye@ecust.edu.cn -
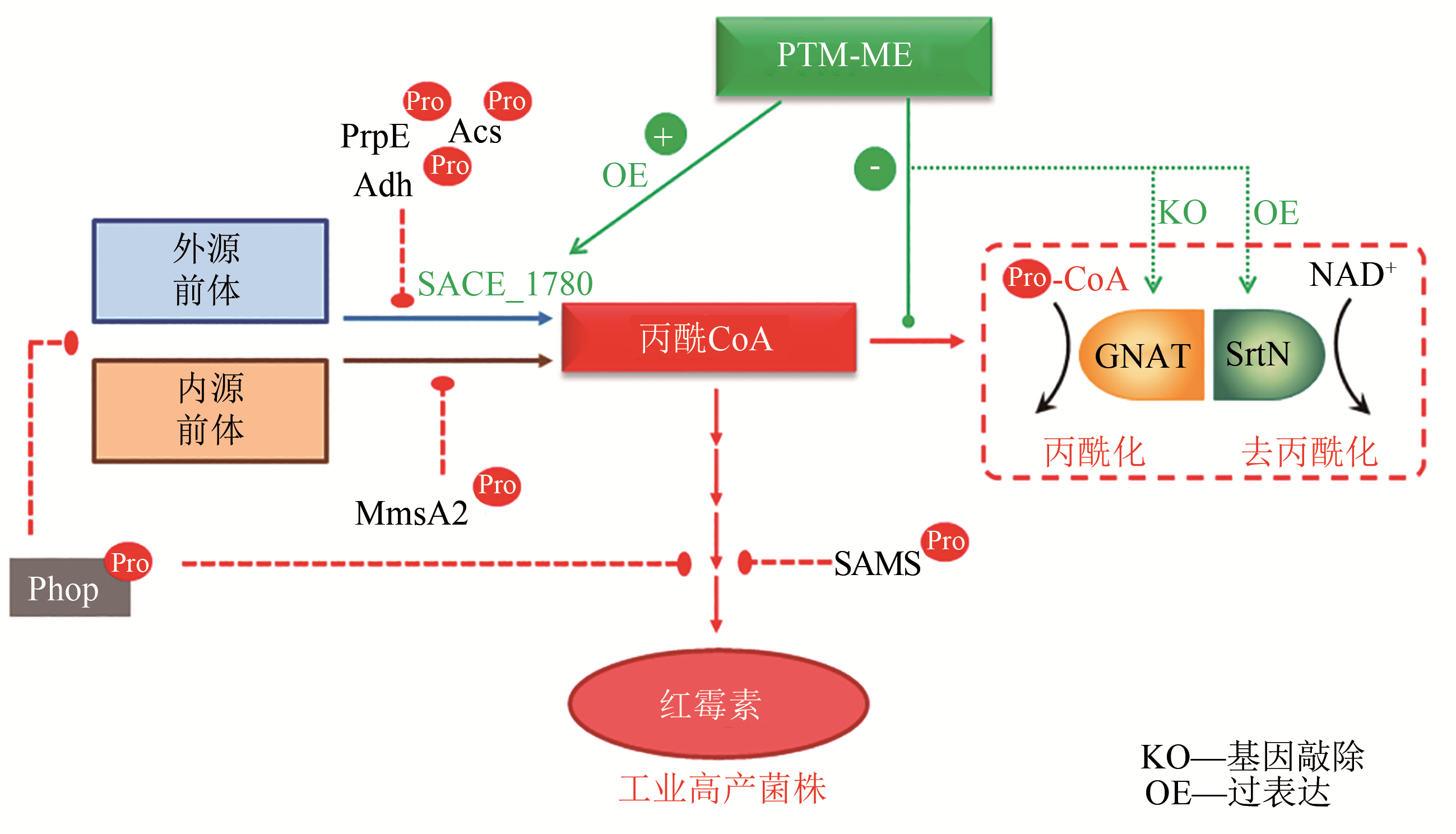
基金资助:国家自然科学基金(31730004);国家重点研发计划“合成生物学”重点专项(2018YFA0900404)
CLC Number:
Cite this article
YOU Di, YE Bangce. Compatibility between synthetic pathway and chassis cells from the viewpoint of post-translational modifications[J]. Synthetic Biology Journal, 2020, 1(2): 212-225.
尤迪, 叶邦策. 从翻译后修饰角度解析人工合成途径与底盘细胞的适配性[J]. 合成生物学, 2020, 1(2): 212-225.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-006
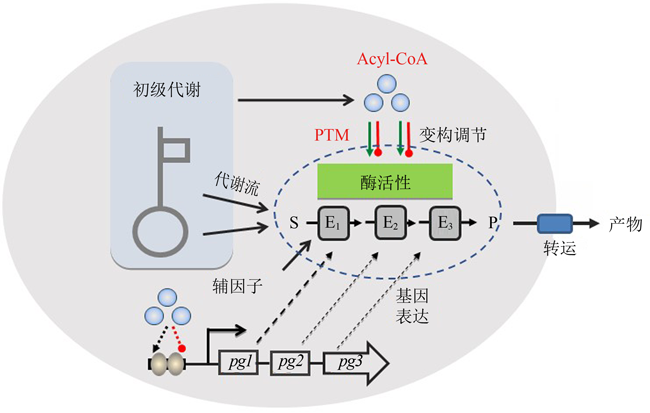
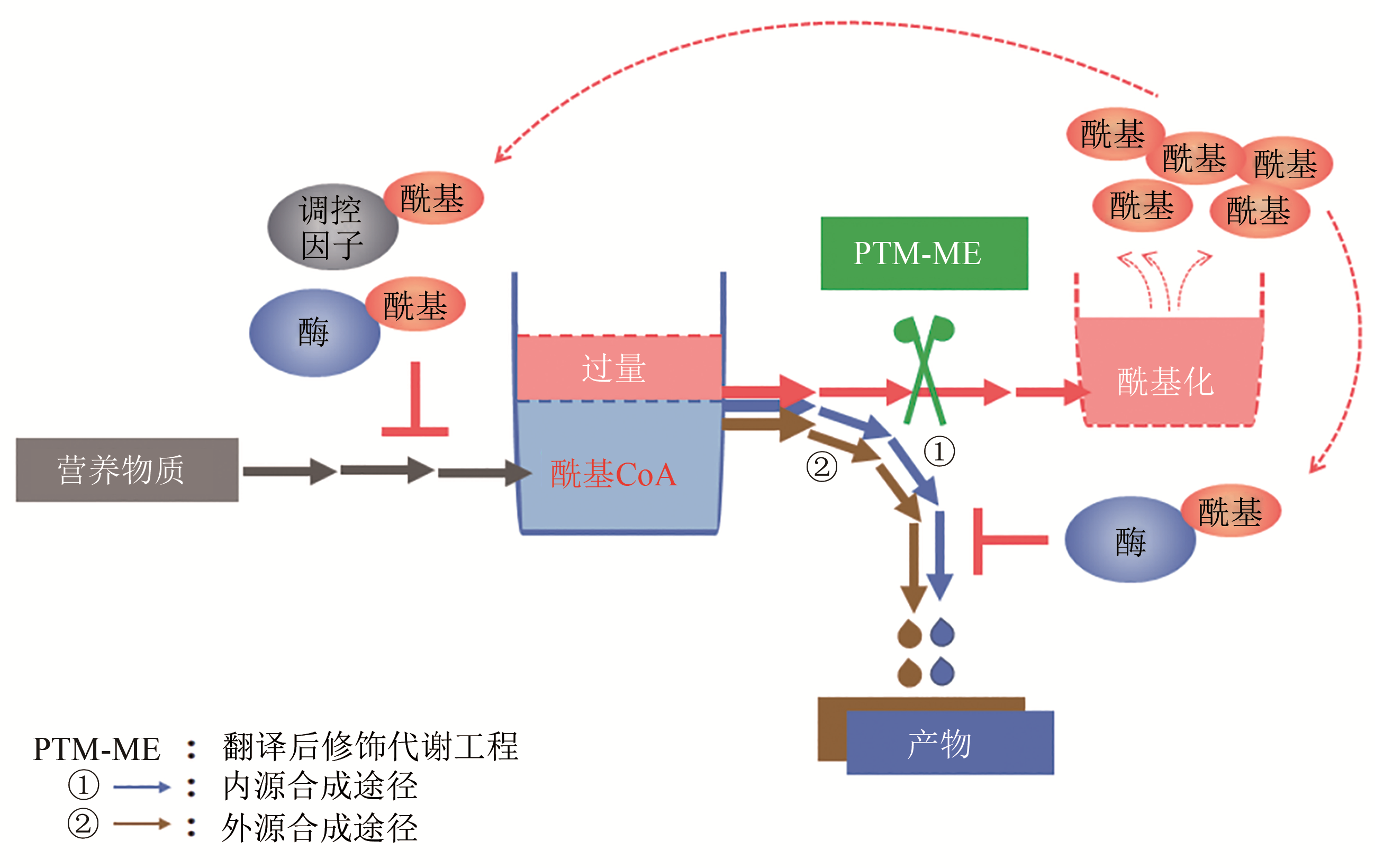
Fig. 1 Effect of chassis cells on artificial biosynthesis pathways (gene expression regulation, precursor and cofactor supply, enzyme activity regulation, product transport, etc.)
| 1 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes [J]. Nature Review Microbiology,2019, 17(12):725-741. |
| 2 | ELOUNDOU-MBEBI J M, KUKEN A, OMRANIAN N, et al. A network property necessary for concentration robustness [J]. Nature Communications,2016, 7:13255. |
| 3 | MAIA P, ROCHA M, ROCHA I. In silico constraint-based strain optimization methods: the quest for optimal cell factories [J]. Microbiology and Molecular Biology Reviews, 2016, 80(1):45-67. |
| 4 | FUHRER T, ZAMPIERI M, SEVIN D C, et al. Genomewide landscape of gene-metabolome associations in Escherichia coli [J]. Molecular Systems Biology, 2017, 13(1):907. |
| 5 | SHAO Y, LU N, WU Z, et al. Creating a functional single-chromosome yeast [J]. Nature,2018, 560(7718):331-335. |
| 6 | SHAO Y, LU N, XUE X, et al. Creating functional chromosome fusions in yeast with CRISPR-Cas9 [J]. Nature Protocols, 2019, 14(8):2521-2545. |
| 7 | FEIST A M, HERRGARD M J, THIELE I, et al. Reconstruction of biochemical networks in microorganisms [J]. Nature Review Microbiology,2009, 7(2):129-143. |
| 8 | FENDT S M, BUESCHER J M, RUDROFF F, et al. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity [J]. Molecular Systems Biology,2010, 6:356. |
| 9 | HEINEMANN M, SAUER U. Systems biology of microbial metabolism [J]. Current Opinion in Microbiology,2010, 13(3):337-343. |
| 10 | LINK H, KOCHANOWSKI K, SAUER U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo [J]. Nature Biotechnology,2013, 31(4):357-361. |
| 11 | ORTH J D, CONRAD T M, NA J, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism [J]. Molecular Systems Biology,2011, 7:535. |
| 12 | WANG Q, ZHANG Y, YANG C, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux [J]. Science, 2010, 327(5968):1004-1007. |
| 13 | XU Y, YOU D, YAO L L, et al. Phosphate regulator PhoP directly and indirectly controls transcription of the erythromycin biosynthesis genes in Saccharopolyspora erythraea [J]. Microbial Cell Factories, 2019, 18(1):206. |
| 14 | LIAO C H, XU Y, RIGALI S, et al. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea [J]. Applied Microbiology and Biotechnology, 2015, 99(23):10215-10224. |
| 15 | RIGALI S, TITGEMEYER F, BARENDS S, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by streptomyces [J]. EMBO Reports,2008, 9(7):670-675. |
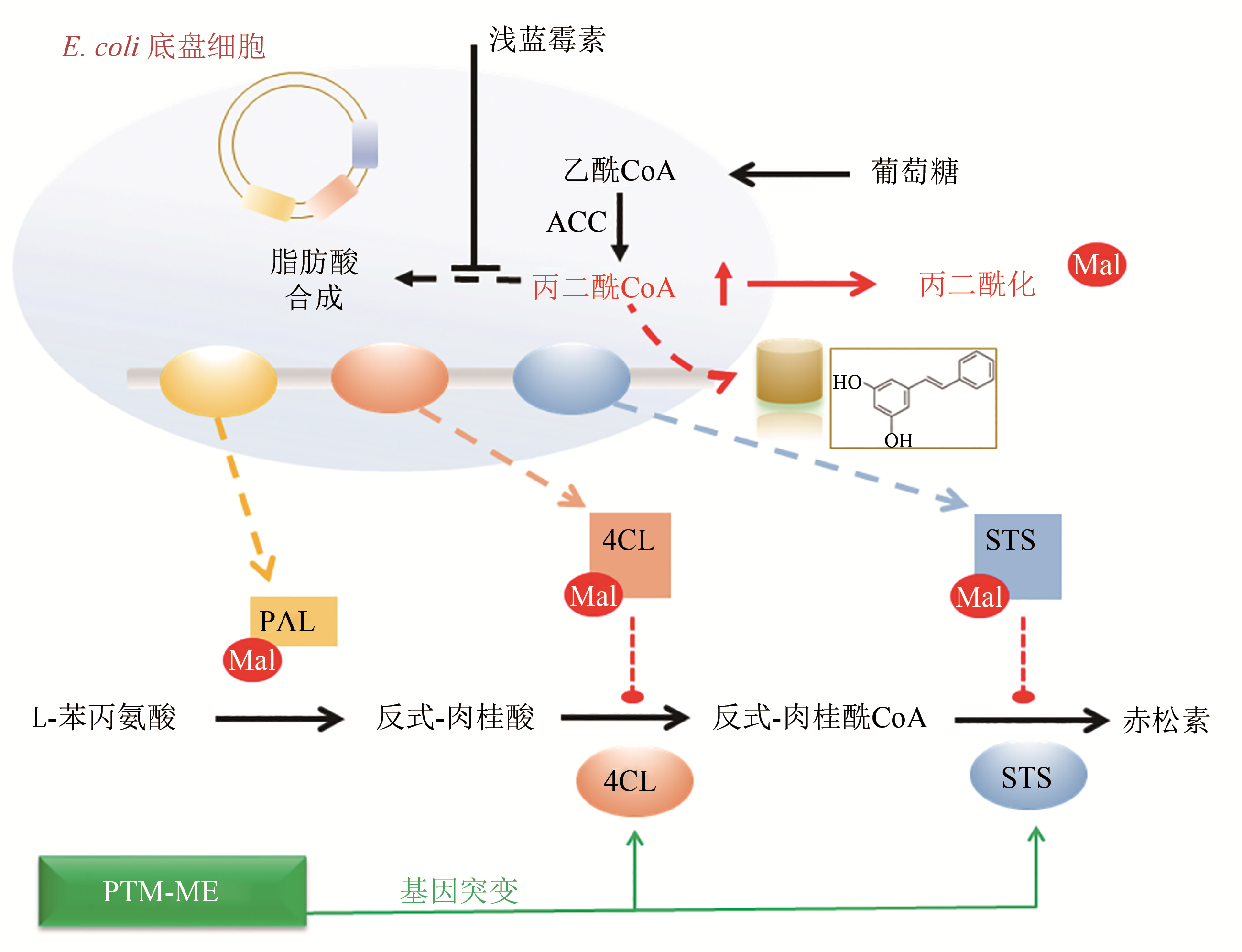
| 16 | XU J Y, XU Y, CHU X H, et al. Protein acylation affects the artificial biosynthetic pathway for pinosylvin production in engineered E. coli [J]. ACS Chemical Biology,2018, 13(5):1200-1208. |
| 17 | YOU D, WANG M M, YIN B C, et al. Precursor supply for erythromycin biosynthesis: engineering of propionate assimilation pathway based on propionylation modification [J]. ACS Synthetic Biology,2019, 8(2):371-380. |
| 18 | BILYK O, LUZHETSKYY A. Metabolic engineering of natural product biosynthesis in Actinobacteria [J]. Current Opinion in Biotechnology,2016, 42:98-107. |
| 19 | JUNG W S, KIM E, YOO Y J, et al. Characterization and engineering of the ethylmalonyl-CoA pathway towards the improved heterologous production of polyketides in Streptomyces venezuelae [J]. Advances in Applied Microbiology,2014, 98:3701-3713. |
| 20 | LI Y, SMOLKE C D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast [J]. Nature Communications, 2016, 7:12137. |
| 21 | MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production [J]. Nature,2016, 537(7622):694-697. |
| 22 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin [J]. Nature,2013, 496(7446):528-532. |
| 23 | THODEY K, GALANIE S, SMOLKE C D. A microbial biomanufacturing platform for natural and semisynthetic opioids [J]. Nature Chemical Biology,2014, 10(10):837-844. |
| 24 | BECKER J, LANGE A, FABARIUS J, et al. Top value platform chemicals: bio-based production of organic acids [J]. Current Opinion in Biotechnology,2015, 36:168-175. |
| 25 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: the future of chemical production [J]. Science,2017, 355(6320):AAG0804. |
| 26 | DELLOMONACO C, CLOMBURG J M, MILLER E N, et al. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals [J]. Nature,2011, 476(7360):355-359. |
| 27 | DUGAR D, STEPHANOPOULOS G. Relative potential of biosynthetic pathways for biofuels and bio-based products [J]. Nature Biotechnology, 2011, 29(12):1074-1078. |
| 28 | CHUNG H, YANG J E, HA J Y, et al. Bio-based production of monomers and polymers by metabolically engineered microorganisms [J]. Current Opinion in Biotechnology, 2015, 36:73-84. |
| 29 | CLOMBURG J M, BLANKSCHIEN M D, VICK J E, et al. Integrated engineering of beta-oxidation reversal and omega-oxidation pathways for the synthesis of medium chain omega-functionalized carboxylic acids [J]. Metabolic Engineering, 2015, 28:202-212. |
| 30 | KIND S, NEUBAUER S, BECKER J, et al. From zero to hero-production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum [J]. Metabolic Engineering. 2014, 25:113-123. |
| 31 | ALLFREY V G, FAULKNER R, MIRSKY A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis [J]. PNAS, 1964, 51:786-794. |
| 32 | ALLFREY V G, MIRSKY A E. Structural modifications of histones and their possible role in the regulation of RNA synthesis [J]. Science, 1964, 144(3618):559. |
| 33 | LUO J, LI M, TANG Y, et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo [J]. PNAS, 2004, 101(8):2259-2264. |
| 34 | STARAI V J, CELIC I, COLE R N, et al. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine [J]. Science, 2002, 298:2390-2392. |
| 35 | CHOUDHARY C, WEINERT B T, NISHIDA Y, et al. The growing landscape of lysine acetylation links metabolism and cell signalling [J]. Nature Reviews: Molecular Cell Biology, 2014, 15(8):536-550. |
| 36 | SABARI B R, ZHANG D, ALLIS C D, et al. Metabolic regulation of gene expression through histone acylations [J]. Nature Reviews: Molecular Cell Biology, 2017, 18(2):90-101. |
| 37 | WAGNER G R, PAYNE R M. Widespread and enzyme-independent nepsilon-acetylation and nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix [J]. Journal of Biological Chemistry, 2013, 288(40):29036-29045. |
| 38 | WEINERT B T, LESMANTAVICIUS V, WAGNER S A, et al. Acetylphosphate is a critical determinant of lysine acetylation in E. coli . [J]. Molecular Cell, 2013, 51:265-272. |
| 39 | GARRITY J G J, HAWSE W, WOLBERGER C, et al. N-lysine propionylation controls the activity of propionyl-CoA synthetase [J]. Journal of Biological Chemistry, 2007, 282(41):30239-30245. |
| 40 | CHEN Y, SPRUNG R, TANG Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones [J]. Molecular & Cellular Proteomics, 2007, 6(5):812-819. |
| 41 | ZHANG Z, TAN M, XIE Z, et al. Identification of lysine succinylation as a new post-translational modification [J]. Nature Chemical Biology, 2011, 7(1):58-63. |
| 42 | PENG C, LU Z, XIE Z, et al. The first identification of lysine malonylation substrates and its regulatory enzyme [J]. Molecular & Cellular Proteomics, 2011, 10(12):M111 012658. |
| 43 | TAN M, PENG C, ANDERSON K A, et al. Lysine glutarylation is a protein posttranslational modification regulated by Sirt5 [J]. Cell Metabolism, 2014, 19(4):605-617. |
| 44 | TAN M, LUO H, LEE S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification [J]. Cell, 2011, 146(6):1016-1028. |
| 45 | XIE Z, ZHANG D, CHUNG D, et al. Metabolic regulation of gene expression by histone lysine beta-hydroxybutyrylation [J]. Molecular Cell, 2016, 62(2):194-206. |
| 46 | HUANG H, ZHANG D, WANG Y, et al. Lysine benzoylation is a histone mark regulated by Sirt2 [J]. Nature Communication, 2018, 9(1):3374. |
| 47 | ZHANG D, TANG Z, HUANG H, et al. Metabolic regulation of gene expression by histone lactylation [J]. Nature, 2019, 574(7779):575-580. |
| 48 | KUHN M L, ZEMAITAITIS B, HU L I, et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation [J]. PLoS One, 2014, 9(4):E94816. |
| 49 | SCHILLING B, CHRISTENSEN D, DAVIS R, et al. Protein acetylation dynamics in response to carbon overflow in Escherichia coli [J]. Molecular Microbiology, 2015, 98(5):847-863. |
| 50 | COLAK G, XIE Z, ZHU A Y, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli [J]. Molecular & Cellular Proteomics, 2013, 12(12):3509-3520. |
| 51 | QIAN L, NIE L, CHEN M, et al. Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism [J]. Journal of Proteome Research, 2016, 15(6):2060-2071. |
| 52 | SUN M, XU J, WU Z, et al. Characterization of protein lysine propionylation in Escherichia coli: global profiling, dynamic change, and enzymatic regulation [J]. Journal of Proteome Research, 2016, 15(12):4696-4708. |
| 53 | CHEN Y, HUANG M, WANG Z, et al. Controlling the feed rate of glucose and propanol for the enhancement of erythromycin production and exploration of propanol metabolism fate by quantitative metabolic flux analysis [J]. Bioprocess Biosystem Engneering, 2013, 36(10):1445-1453. |
| 54 | EL-ENSHASY H A, MOHAMED N A, FARID M A, et al. Improvement of erythromycin production by Saccharopolyspora erythraea in molasses based medium through cultivation medium optimization [J]. Bioresource Technology, 2008, 99(10):4263-4268. |
| 55 | XU Z, LIU Y, YE B C. PccD regulates branched-chain amino acid degradation and exerts a negative effect on erythromycin production in Saccharopolyspora erythraea [J]. Applied and Environmental Microbiology, 2018. DOI: 10.1128/AEM.00049-18 . |
| 56 | RYU Y G, BUTLER M J, CHATER K F, et al. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor [J]. Applied and Environmental Microbiology, 2006, 72(11):7132-7139. |
| 57 | LIM C G, FOWLER Z L, HUELLER T, et al. High-yield resveratrol production in engineered Escherichia coli [J]. Applied and Environmental Microbiology,2011, 77(10):3451-3460. |
| 58 | SUMMEREN-WESENHAGEN P V VAN, MARIENHAGEN J. Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin [J]. Applied and Environmental Microbiology, 2015, 81(3):840-849. |
| 59 | SHIBA Y, PARADISE E M, KIRBY J, et al. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids [J]. Metabolic Engineering, 2007, 9(2):160-168. |
| 60 | MEADOWA A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production [J]. Nature, 2016, 537(7622):694-697. |
| 61 | SCHADEWEG V, BOLES E. N-butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA [J]. Biotechnol. Biofuels, 2016, 9:44. |
| 62 | DE MEY M, DE MAESENEIRE S, SOETAERT W, et al. Minimizing acetate formation in E.coli fermentations [J]. Journal of Industrial Microbiology and Biotechnology, 2007, 34(11):689-700. |
| 63 | DITTRICH C R, BENNETT G N, SAN K Y. Characterization of the acetate-producing pathways in Escherichia coli [J]. Biotechnology Progress, 2005, 21(4):1062-1067. |
| 64 | BAUER K A, BEN-BASSAT A, DAWSON M, et al. Improved expression of human interleukin-2 in high-cell-density fermentor cultures of Escherichia coli K-12 by a phosphotransacetylase mutant [J]. Applied and Environmental Microbiology, 1990, 56(5):1296-1302. |
| 65 | HAHM D H, PAN J, RHEE J S. Characterization and evaluation of a pta (phosphotransacetylase) negative mutant of Escherichia coli HB101 as production host of foreign lipase [J]. Applied Microbiology and Biotechnology, 1994, 42(1):100-107. |
| 66 | LIN H, CASTRO N M, BENNETT G N, et al. Acetyl-CoA synthetase overexpression in Escherichia coli demonstrates more efficient acetate assimilation and lower acetate accumulation: a potential tool in metabolic engineering [J]. Applied Microbiology and Biotechnology, 2006, 71(6):870-874. |
| 67 | VADALI R V, HORTON C E, RUDOLPH F B, et al. Production of isoamyl acetate in ackA-pta and/or ldh mutants of Escherichia coli with overexpression of yeast AFT2 [J]. Applied Microbiology and Biotechnology, 2004, 63(6):698-704. |
| 68 | DITTRICH C R, VADALI R V, BENNETT G N, et al. Redistribution of metabolic fluxes in the central aerobic metabolic pathway of E. coli mutant strains with deletion of the ackA-pta and poxb pathways for the synthesis of isoamyl acetate [J]. Biotechnology Progress, 2005, 21(2):627-631. |
| 69 | CENTENO-LEIJA S, HUERTA-BERISTAIN G, GILES-GOMEZ M, et al. Improving poly-3-hydroxybutyrate production in Escherichia coli by combining the increase in the NADPH pool and acetyl-CoA availability [J]. Antonie van Leeuwenhoek, 2014, 105(4):687-696. |
| 70 | CHANG D E, SHIN S, RHEE J S, et al. Acetate metabolism in a pta mutant of Escherichia coli W3110: Importance of maintaining acetyl coenzyme a flux for growth and survival [J]. Journal of Bacteriology, 1999, 181(21):6656-6663. |
| 71 | MIYAKE M, MIYAMOTO C, SCHNACKENBERG J, et al. Phosphotransacetylase as a key factor in biological production of polyhydroxybutyrate [J]. Applied Biochemistry and Biotechnology, 2000, 84-86:1039-1044. |
| 72 | MCCLEARY W R, STOCK J B, NINFA A J. Is acetyl phosphate a global signal in Escherichia coli? [J]. Journal of Bacteriology, 1993, 175(10):2793-2798. |
| 73 | KIM J Y, CHA H J. Down-regulation of acetate pathway through antisense strategy in Escherichia coli: improved foreign protein production [J]. Biotechnology and Bioengineering, 2003, 83(7):841-853. |
| 74 | ZHOU S, IVERSON A G, GRAYBURN W S. Engineering a native homoethanol pathway in Escherichia coli B for ethanol production [J]. Biotechnology Letters, 2008, 30(2):335-342. |
| 75 | ATSUMI S, CANN A F, CONNOR M R, et al. Metabolic engineering of Escherichia coli for 1-butanol production [J]. Metabolic Engineering, 2008, 10(6):305-311. |
| 76 | BAEK J M, MAZUMDAR S, LEE S W, et al. Butyrate production in engineered Escherichia coli with synthetic scaffolds [J]. Biotechnology and Bioengineering, 2013, 110(10):2790-2794. |
| 77 | CHRISTENSEN D G, XIE X, BASISTY N, et al. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions [J]. Frontiers in Microbiology, 2019. DOI: 10.3389/fmicb. 2019. 01604 . |
| 78 | VANDRISSE C M, ESCALANTE-SEMERENA J C. Protein acetylation in bacteria [J]. Annual Review of Microbiology, 2019, 73:111-132. |
| 79 | MARTIN J F, LIRAS P. The balance metabolism safety net: Integration of stress signals by interacting transcriptional factors in streptomyces and related actinobacteria [J]. Frontiers in Microbiology, 2020. DOI: 10.3389/fmicb. 2019.03120 . |
| 80 | WEI W, LIU T, LI X, et al. Lysine acetylation regulates the function of the global anaerobic transcription factor FnrL in Rhodobacter sphaeroides [J]. Molecular Microbiology, 2017, 104(2):278-293. |
| 81 | QIN R, SANG Y, REN J, et al. The bacterial two-hybrid system uncovers the involvement of acetylation in regulating of Lrp activity in Salmonella typhimurium [J]. Frontiers in Microbiology, 2016, 7:1864. |
| 82 | REN J, SANG Y, TAN Y, et al. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence [J]. PLoS Pathogens, 2016, 12(3):E1005458. |
| 83 | SANG Y, REN J, NI J, et al. Protein acetylation is involved in Salmonella enterica serovar typhimurium virulence [J]. The Journal of Infectious Diseases, 2016, 213(11):1836-1845. |
| 84 | AMIN R, FRANZ-WACHTEL M, TIFFERT Y, et al. Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145 [J]. Frontiers in Molecular Biosciences, 2016, 3:38. |
| 85 | XU Y, LI Y X, YE B C. Lysine propionylation modulates the transcriptional activity of phosphate regulator PhoP in Saccharopolyspora erythraea [J]. Molecular Microbiology, 2018, 110(4):648-661. |
| 86 | LIAO C H, YAO L L, YE B C. Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP [J]. Molecular Microbiology, 2014, 94(5):1065-1084. |
| 87 | YOU D, ZHANG B Q, YE B C. GntR family regulator DasR controls acetate assimilation by directly repressing the acsa gene in Saccharopolyspora erythraea [J]. Journal of Bacteriology, 2018. DOI: 10.1128/JB.00685-17 |
| 88 | CHAN C H, GARRITY J, CROSBY H A, et al. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate [J]. Molecular Microbiology, 2011, 80:168-183. |
| 89 | CASTANO-CEREZO S, BERNAL V, BLANCO-CATALA J, et al. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli [J]. Molecular Microbiology, 2011, 82(5):1110-1128. |
| 90 | XU H, HEGDE S S, BLANCHARD J S. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP [J]. Biochemistry, 2011, 50(26):5883-5892. |
| 91 | CROSBY H A, HEINIGER E K, HARWOOD C S, et al. Reversible N-lysine acetylation regulates the activity of acyl-CoA synthetases involved in anaerobic benzoate catabolism in Rhodopseudomonas palustris [J]. Molecular Microbiology, 2010, 76(4):874-888. |
| 92 | GARDNER J G, GRUNDY F J, HENKIN T M, et al. Control of acetyl-coenzyme a synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis [J]. Journal of Bacteriology, 2006, 188:5460-5468. |
| 93 | LIU X X, LIU W B, YE B C. Regulation of a protein acetyltransferase in Myxococcus xanthus by the coenzyme NADP [J]. Journal of Bacteriology, 2015, 198(4):623-632. |
| 94 | YOU D, WANG M M, YE B C. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels [J]. Molecular Microbiology, 2017, 103(5):845-859. |
| 95 | YOU D, YAO L L, HUANG D, et al. Acetyl coenzyme A synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-type N-acetyltransferase (GNAT) domain in Saccharopolyspora erythraea [J]. Journal of Bacteriology, 2014, 196(17):3169-3178. |
| 96 | XU J Y, YOU D, LENG P Q, et al. Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine [J]. Journal of Biological Chemistry, 2014, 289:27034-27045. |
| 97 | LIU X X, SHEN M J, LIU W B, et al. Glnr-mediated regulation of short-chain fatty acid assimilation in Mycobacterium smegmatis [J]. Frontiers in Microbiology, 2018, 9:1311. |
| 98 | STARAI V J, TAKAHASHI H, BOEKE J D, et al. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires Sir2 protein function in Salmonella enterica and Saccharomyces cerevisiae [J]. Genetics, 2003, 163:545-555. |
| 99 | YU B J, KIM J A, MOON J H, et al. The diversity of lysine-acetylated proteins in Escherichia coli [J]. Journal of Microbiology and Biotechnology, 2008, 18(9):1529-1536. |
| 100 | XU J Y, XU Z, ZHOU Y, et al. Lysine malonylome may affect the central metabolism and erythromycin biosynthesis pathway in Saccharopolyspora erythraea [J]. Journal of Proteome Research, 2016, 15(5):1685-1701. |
| 101 | XU J Y, XU Y, XU Z, et al. Protein acylation is a general regulatory mechanism in biosynthetic pathway of acyl-CoA-derived natural products [J]. Cell Chemical Biology, 2018, 25:1-12. |
| [1] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [2] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||