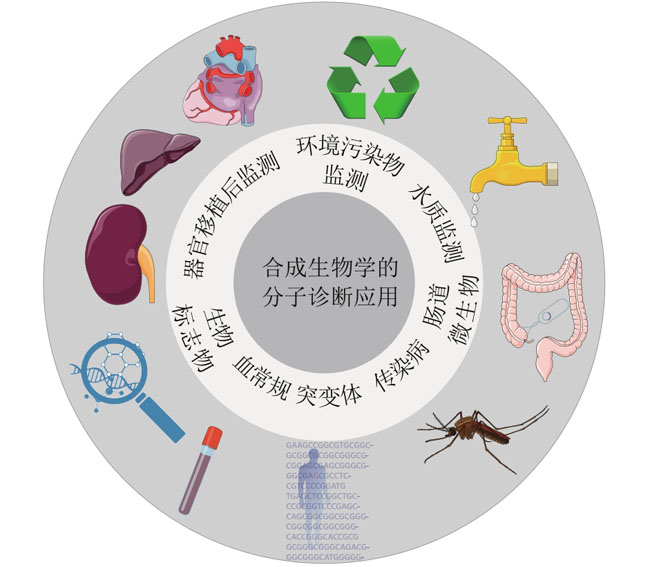
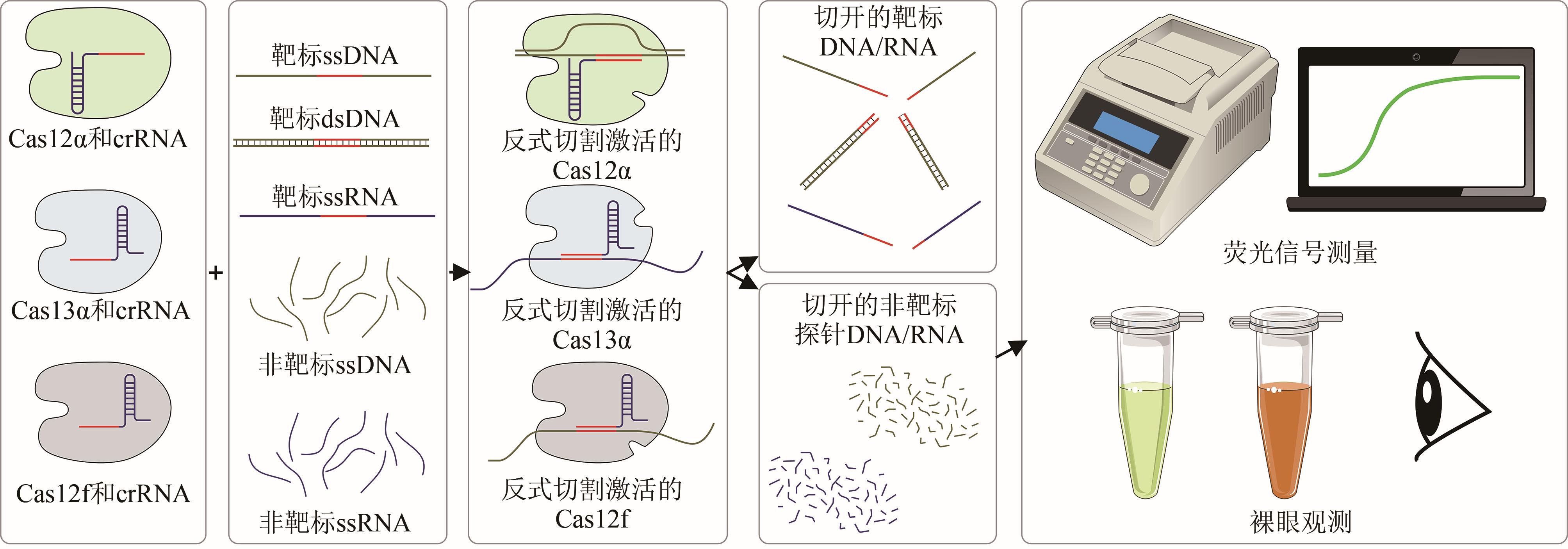
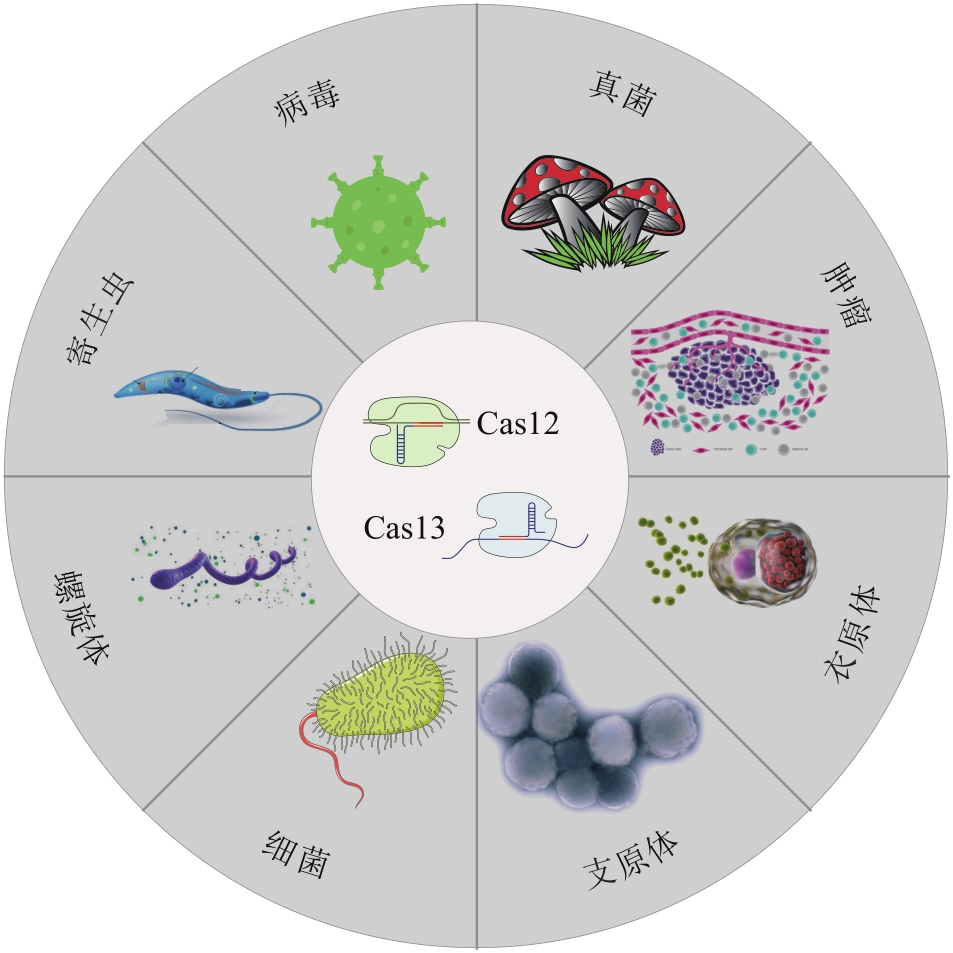
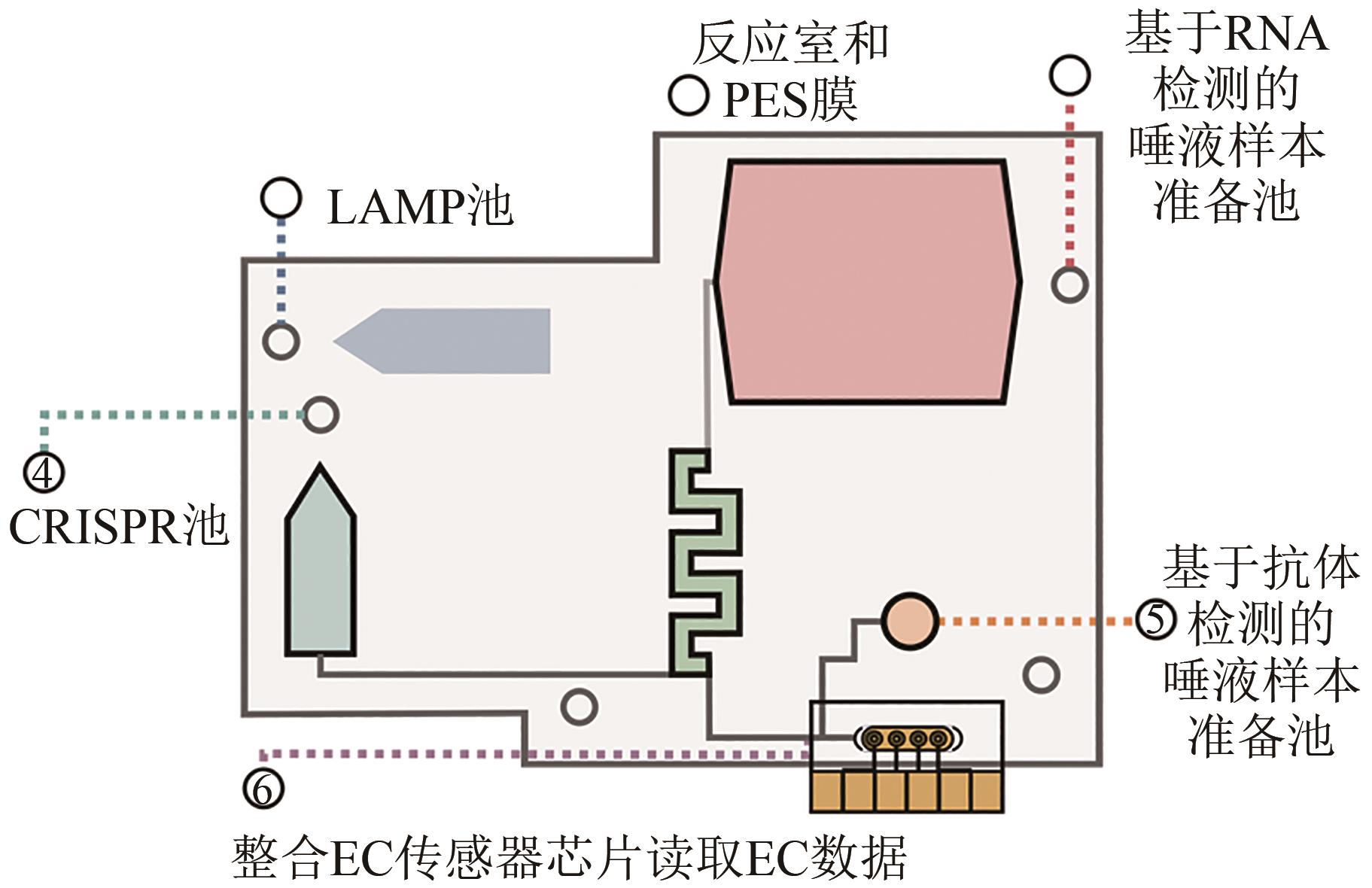
Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (2): 318-332.DOI: 10.12211/2096-8280.2022-061
• Invited Review • Previous Articles Next Articles
Synthetic biology for next-generation genetic diagnostics
LV Hailong1,2,3, WANG Jian2, LV Hao4, WANG Jin2,5, XU Yong3, GU Dayong3
- 1.Guangdong Key Laboratory for Biomedical Measurements and Ultrasound Imaging. School of Biomedical Engineering,School of Medicine,Shenzhen University,Shenzhen 518060,Guangdong,China
2.Shenzhen Institute of Translational Medicine,Health Science Center,The First Affiliated Hospital of Shenzhen University,Shenzhen Second People's Hospital,Shenzhen 518035,Guangdong,China
3.Department of Laboratory Medicine,The First Affiliated Hospital of Shenzhen University,Shenzhen Second People's Hospital,Shenzhen 518035,Guangdong,China
4.Department of Neurosurgery,Shenzhen Key Laboratory of Neurosurgery,the Shenzhen Second People’s Hospital,First Affiliated Hospital of Shenzhen University,Shenzhen 518037,Guangdong,China
5.Shanghai Tolo Biotechnology Co. ,Ltd. ,Shanghai 200233,China
-
Received:2022-11-02Revised:2022-12-17Online:2023-04-27Published:2023-04-30 -
Contact:GU Dayong
合成生物学在下一代基因诊断技术中的应用进展
吕海龙1,2,3, 王建2, 吕浩4, 王金2,5, 徐勇3, 顾大勇3
- 1.深圳大学医学部生物医学工程学院,广东省生物医学信息检测与超声成像重点实验室,广东 深圳 518060
2.深圳市第二人民医院,深圳市转化医学研究院,广东 深圳 518035
3.深圳市第二人民医院检验科,深圳市医学检验分子诊断重点实验室,广东 深圳 518035
4.深圳市第二人民医院神经外科,深圳市神经外科重点实验室,广东 深圳 518037
5.上海吐露港生物科技有限公司,上海 200233
-
通讯作者:顾大勇 -
作者简介:吕海龙 (1988—),男,博士,博士后。研究方向为合成生物学及CRISPR分子诊断技术开发。E-mail: lvhailong8828@163.com顾大勇 (1972—),男,博士,主任技师,博士生导师、博士后合作导师。研究方向为生物芯片、生物传感技术。E-mail:wanhood@163.com -
基金资助:国家重点研发计划(2018YFA0903700);深圳市医疗卫生三名工程(SZSM202011017)
CLC Number:
Cite this article
LV Hailong, WANG Jian, LV Hao, WANG Jin, XU Yong, GU Dayong. Synthetic biology for next-generation genetic diagnostics[J]. Synthetic Biology Journal, 2023, 4(2): 318-332.
吕海龙, 王建, 吕浩, 王金, 徐勇, 顾大勇. 合成生物学在下一代基因诊断技术中的应用进展[J]. 合成生物学, 2023, 4(2): 318-332.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-061
| 技术 类型 | 方法 | 原理 | 应用对象 | 效果 | 优点 | 参考文献 |
|---|---|---|---|---|---|---|
| 环境污染领域的检测分析 | 基因 修饰 | 大肠杆菌基因修饰合成可感知Cu2+并产生核黄素的孔蛋白,利用核黄素产量计算MFC生物传感器的最大电压,进而实现水中Cu2+的原位检测 | 水中Cu2+的原位检测 | 结果与现有的比色法、FAAS、ICP-OES等分析方法相一致 | 一种快速、经济的分析替代方案 | [ |
| 环境污染领域的检测分析 | 双色荧光报告基因 | 在一个基因结构中使用双色荧光报告基因来检测生物可利用的镉,双色荧光的产生与镉的暴露浓度成正比 | 环境水样中的Cd(Ⅱ) | 一种新型双传感、全细胞生物传感器的首次开发报告 | 可同时检测镉及其毒性效应 | [ |
| 环境污染领域的检测分析 | PCR,IFAT | PCR检测贾第虫、弓形虫和隐孢子虫属的DNA,免疫荧光抗体试验(IFAT)检测贾第虫和贾第虫属和隐孢子虫属的DNA的存在 | 饮用水中的原生生物 | 分析得出哥伦比亚的饮用水中致病性原生生物样本的流行率很高 | 两种检测方法相互验证 | [ |
| 环境污染领域的检测分析 | 逆转录巢式PCR | 开发RT-PCR引物集,优化甲型和乙型肝炎病毒(HAV和HEV)检测的标准模板 | 地下水中HAV和HEV | 开发的巢式RT-PCR有望通过监测HAV和HEV来评估水的安全性 | 可区分假阳性,提高了检测效率 | [ |
| 环境污染领域的检测分析 | qPCR | 采用HPLC-MS/MS测定抗生素,采用实时定量PCR定量,调查抗生素及其相应的ARGs的发生和分布 | 抗生素耐药性基因(ARGs) | 人为活动及暴雨天气对抗生素污染和ARGs水平均有显著影响 | 增加对污水污染与ARG之间联系的了解 | [ |
| 肠道微生物的检测分析 | NO感应开关 | 设计NO响应生物传感器,在大肠杆菌Nisle 1917和迷你Sim Cells中对电路进行了表征和优化 | NO | 优化的基因电路NO检测限有所提高,基于Sim Cells的NO生物传感器可以用作合成生物学的安全传感器底盘 | 特征良好的硝酸传感系统有助于细菌治疗和合成生物学的快速发展 | [ |
| 肠道微生物的检测分析 | 无细胞开关传感器,RT-qPCR | 在基于纸张的无细胞反应中使用RNA开关传感器进行按需和简单的微生物组样本分析 | 肠道微生物群和宿主生物标志物 | 可以用于快速、廉价地检测毒素mRNA来诊断艰难梭菌感染 | 快速、廉价,比标准的基于DNA的qPCR诊断更灵敏 | [ |
| 传染病 诊断 | 酶报告 基因 | 设计一种专门检测肠道中霍乱弧菌的群体感应信号乳杆菌菌株,并触发一种容易在粪便样本中检测到的酶报告基因的表达 | 霍乱弧菌 | 使用含有天然和工程乳杆菌菌株的发酵食品进行预防性饮食干预,可能会阻碍霍乱的进展 | 可提供快速保护,以抵御霍乱等快速进展的感染 | [ |
| 传染病 诊断 | 新型合成纳米体 | 通过筛选酵母表面显示的合成纳米体序列库,开发可破坏Spike和ACE2之间相互作用的纳米体 | SARS-CoV-2病毒 | 纳米体mNb6-tri对Spike和SARS-CoV-2的感染具有中和作用 | 气溶胶介导的纳米体中和剂可以直接传递到气道上皮细胞 | [ |
| 器官移植后检测 | mRNA 疫苗 | 设计一种编码ZIKV结构基因的mRNA疫苗,编码prM-E基因产生病毒样颗粒,导致中和抗体滴度高,保护ZIKV感染并赋予杀菌免疫 | ZIKV热病毒 | 改良的mRNA疫苗可以预防ZIKV疾病 | 可以降低个体对随后暴露于DENV的敏感性风险 | [ |
| 器官移植后检测 | 酶抑制剂 | 巨细胞病毒(CMV) | 用于人类巨细胞病毒感染的治疗和预防 | 与单独使用免疫抑制剂相比有更好的耐受性 | [ | |
| 遗传病 诊断 | 基因治疗 | 以基因矫正的热休克蛋白及其后代作为细胞载体,将分子传递到循环和组织中 | 自体造血干/祖细胞(HSPCs) | 有可能治愈由血液系统发育功能改变引起的单基因遗传性疾病 | 具有更安全的整合谱 | [ |
| 遗传病 诊断 | 下一代基因测序(NGS) | 通过针对所有鱼鳞病相关基因的多基因面板测试,使用NGS进行分子分析 | 中国人遗传学鱼鳞病 | 证明遗传性鱼鳞病是一组不同的角化症,扩大了中国人遗传性鱼鳞病的突变谱和临床表型 | NGS技术极大提高了遗传性鱼鳞病的诊断效率 | [ |
| 肿瘤基因诊断 | 工程免疫细胞 | 利用免疫细胞、核酸和细菌作为底盘的癌症基因电路疗法 | 肿瘤及癌症 | 合成生物学可创造出更有效的适应性疗法,使其能够特异性靶向癌细胞,同时保留健康细胞 | 可按需进行肿瘤及癌症的免疫治疗 | [ |
| 基因突变的检测 | 原核生物核糖核酸调节剂 | 从头设计一种在体内和无细胞转录-翻译反应中提供超特异性RNA检测能力的原核核糖体调节因子 | 核糖体调节 因子 | 开发用于与癌症、耐药性和遗传疾病相关的一系列突变的核糖体调节因子 | 实用,强大的分子探针 | [ |
Table 1 Applications of synthetic biology in gene diagnosis
| 技术 类型 | 方法 | 原理 | 应用对象 | 效果 | 优点 | 参考文献 |
|---|---|---|---|---|---|---|
| 环境污染领域的检测分析 | 基因 修饰 | 大肠杆菌基因修饰合成可感知Cu2+并产生核黄素的孔蛋白,利用核黄素产量计算MFC生物传感器的最大电压,进而实现水中Cu2+的原位检测 | 水中Cu2+的原位检测 | 结果与现有的比色法、FAAS、ICP-OES等分析方法相一致 | 一种快速、经济的分析替代方案 | [ |
| 环境污染领域的检测分析 | 双色荧光报告基因 | 在一个基因结构中使用双色荧光报告基因来检测生物可利用的镉,双色荧光的产生与镉的暴露浓度成正比 | 环境水样中的Cd(Ⅱ) | 一种新型双传感、全细胞生物传感器的首次开发报告 | 可同时检测镉及其毒性效应 | [ |
| 环境污染领域的检测分析 | PCR,IFAT | PCR检测贾第虫、弓形虫和隐孢子虫属的DNA,免疫荧光抗体试验(IFAT)检测贾第虫和贾第虫属和隐孢子虫属的DNA的存在 | 饮用水中的原生生物 | 分析得出哥伦比亚的饮用水中致病性原生生物样本的流行率很高 | 两种检测方法相互验证 | [ |
| 环境污染领域的检测分析 | 逆转录巢式PCR | 开发RT-PCR引物集,优化甲型和乙型肝炎病毒(HAV和HEV)检测的标准模板 | 地下水中HAV和HEV | 开发的巢式RT-PCR有望通过监测HAV和HEV来评估水的安全性 | 可区分假阳性,提高了检测效率 | [ |
| 环境污染领域的检测分析 | qPCR | 采用HPLC-MS/MS测定抗生素,采用实时定量PCR定量,调查抗生素及其相应的ARGs的发生和分布 | 抗生素耐药性基因(ARGs) | 人为活动及暴雨天气对抗生素污染和ARGs水平均有显著影响 | 增加对污水污染与ARG之间联系的了解 | [ |
| 肠道微生物的检测分析 | NO感应开关 | 设计NO响应生物传感器,在大肠杆菌Nisle 1917和迷你Sim Cells中对电路进行了表征和优化 | NO | 优化的基因电路NO检测限有所提高,基于Sim Cells的NO生物传感器可以用作合成生物学的安全传感器底盘 | 特征良好的硝酸传感系统有助于细菌治疗和合成生物学的快速发展 | [ |
| 肠道微生物的检测分析 | 无细胞开关传感器,RT-qPCR | 在基于纸张的无细胞反应中使用RNA开关传感器进行按需和简单的微生物组样本分析 | 肠道微生物群和宿主生物标志物 | 可以用于快速、廉价地检测毒素mRNA来诊断艰难梭菌感染 | 快速、廉价,比标准的基于DNA的qPCR诊断更灵敏 | [ |
| 传染病 诊断 | 酶报告 基因 | 设计一种专门检测肠道中霍乱弧菌的群体感应信号乳杆菌菌株,并触发一种容易在粪便样本中检测到的酶报告基因的表达 | 霍乱弧菌 | 使用含有天然和工程乳杆菌菌株的发酵食品进行预防性饮食干预,可能会阻碍霍乱的进展 | 可提供快速保护,以抵御霍乱等快速进展的感染 | [ |
| 传染病 诊断 | 新型合成纳米体 | 通过筛选酵母表面显示的合成纳米体序列库,开发可破坏Spike和ACE2之间相互作用的纳米体 | SARS-CoV-2病毒 | 纳米体mNb6-tri对Spike和SARS-CoV-2的感染具有中和作用 | 气溶胶介导的纳米体中和剂可以直接传递到气道上皮细胞 | [ |
| 器官移植后检测 | mRNA 疫苗 | 设计一种编码ZIKV结构基因的mRNA疫苗,编码prM-E基因产生病毒样颗粒,导致中和抗体滴度高,保护ZIKV感染并赋予杀菌免疫 | ZIKV热病毒 | 改良的mRNA疫苗可以预防ZIKV疾病 | 可以降低个体对随后暴露于DENV的敏感性风险 | [ |
| 器官移植后检测 | 酶抑制剂 | 巨细胞病毒(CMV) | 用于人类巨细胞病毒感染的治疗和预防 | 与单独使用免疫抑制剂相比有更好的耐受性 | [ | |
| 遗传病 诊断 | 基因治疗 | 以基因矫正的热休克蛋白及其后代作为细胞载体,将分子传递到循环和组织中 | 自体造血干/祖细胞(HSPCs) | 有可能治愈由血液系统发育功能改变引起的单基因遗传性疾病 | 具有更安全的整合谱 | [ |
| 遗传病 诊断 | 下一代基因测序(NGS) | 通过针对所有鱼鳞病相关基因的多基因面板测试,使用NGS进行分子分析 | 中国人遗传学鱼鳞病 | 证明遗传性鱼鳞病是一组不同的角化症,扩大了中国人遗传性鱼鳞病的突变谱和临床表型 | NGS技术极大提高了遗传性鱼鳞病的诊断效率 | [ |
| 肿瘤基因诊断 | 工程免疫细胞 | 利用免疫细胞、核酸和细菌作为底盘的癌症基因电路疗法 | 肿瘤及癌症 | 合成生物学可创造出更有效的适应性疗法,使其能够特异性靶向癌细胞,同时保留健康细胞 | 可按需进行肿瘤及癌症的免疫治疗 | [ |
| 基因突变的检测 | 原核生物核糖核酸调节剂 | 从头设计一种在体内和无细胞转录-翻译反应中提供超特异性RNA检测能力的原核核糖体调节因子 | 核糖体调节 因子 | 开发用于与癌症、耐药性和遗传疾病相关的一系列突变的核糖体调节因子 | 实用,强大的分子探针 | [ |
| 1 | TAN X, LETENDRE J H, COLLINS J J, et al. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics[J]. Cell, 2021, 184(4): 881-898. |
| 2 | ZHOU T Y, LI R, ZHANG S T, et al. A copper-specific microbial fuel cell biosensor based on riboflavin biosynthesis of engineered Escherichia coli [J]. Biotechnology and Bioengineering, 2021, 118(1): 210-222. |
| 3 | HUI C Y, GUO Y, WU J, et al. Detection of bioavailable cadmium by double-color fluorescence based on a dual-sensing bioreporter system[J]. Frontiers in Microbiology, 2021, 12: 696195. |
| 4 | TRIVIÑO-VALENCIA J, LORA F, ZULUAGA J D, et al. Detection by PCR of pathogenic protozoa in raw and drinkable water samples in Colombia[J]. Parasitology Research, 2016, 115(5): 1789-1797. |
| 5 | PINTO-DUARTE V A, HÉRNANDEZ-ARANGO N M, MARIN-GALLEGO B J, et al. Detection of Giardia duodenalis and Toxoplasma gondii in soil and water samples in the Quindío River Basin, Colombia[J]. Food and Waterborne Parasitology, 2022, 28: e00175. |
| 6 | BAE K S, LEE S W, LEE J Y, et al. Development of diagnostic systems for wide range and highly sensitive detection of two waterborne hepatitis viruses from groundwater using the conventional reverse transcription nested PCR assay[J]. Journal of Virological Methods, 2022, 299: 114344. |
| 7 | JIA J, GUAN Y J, CHENG M Q, et al. Occurrence and distribution of antibiotics and antibiotic resistance genes in Ba River, China[J]. Science of the Total Environment, 2018, 642: 1136-1144. |
| 8 | AHMED W, GYAWALI P, HAMILTON K A, et al. Antibiotic resistance and sewage-associated marker genes in untreated sewage and a river characterized during baseflow and stormflow[J]. Frontiers in Microbiology, 2021, 12: 632850. |
| 9 | CHEN X J, WANG B J, THOMPSON I P, et al. Rational design and characterization of nitric oxide biosensors in E. coli Nissle 1917 and mini SimCells[J]. ACS Synthetic Biology, 2021, 10(10): 2566-2578. |
| 10 | TAKAHASHI M K, TAN X, DY A J, et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers[J]. Nature Communications, 2018, 9: 3347. |
| 11 | TAKAHASHI M K, TAN X, DY A J. Cell-free paper-based analysis of gut microbiota and host biomarkers[J]. Methods in Molecular Biology, 2022, 2433: 351-374. |
| 12 | MAO N, CUBILLOS-RUIZ A, CAMERON D E, et al. Probiotic strains detect and suppress cholera in mice[J]. Science Translational Medicine, 2018, 10(445): eaao2586. |
| 13 | SCHOOF M, FAUST B, SAUNDERS R A, et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike[J]. Science, 2020, 370(6523): 1473-1479. |
| 14 | RICHNER J M, HIMANSU S, DOWD K A, et al. Modified mRNA vaccines protect against Zika virus infection[J]. Cell, 2017, 168(6): 1114-1125.e10. |
| 15 | MEESING A, RAZONABLE R R. New developments in the management of cytomegalovirus infection after transplantation[J]. Drugs, 2018, 78(11): 1085-1103. |
| 16 | TUCCI F, SCARAMUZZA S, AIUTI A, et al. Update on clinical ex vivo hematopoietic stem cell gene therapy for inherited monogenic diseases[J]. Molecular Therapy, 2021, 29(2): 489-504. |
| 17 | CHENG R H, LIANG J Y, LI Y, et al. Next-generation sequencing through multi-gene panel testing for diagnosis of hereditary ichthyosis in Chinese[J]. Clinical Genetics, 2020, 97(5): 770-778. |
| 18 | WU M R, JUSIAK B, LU T K. Engineering advanced cancer therapies with synthetic biology[J]. Nature Reviews Cancer, 2019, 19(4): 187-195. |
| 19 | SHIMABUKURO-VORNHAGEN A, BÖLL B, SCHELLONGOWSKI P, et al. Critical care management of chimeric antigen receptor T-cell therapy recipients[J]. CA: a Cancer Journal for Clinicians, 2022, 72(1): 78-93. |
| 20 | DIMITRI A, HERBST F, FRAIETTA J A. Engineering the next-generation of CAR T-cells with CRISPR-Cas9 gene editing[J]. Molecular Cancer, 2022, 21(1): 78. |
| 21 | HONG F, MA D, WU K Y, et al. Precise and programmable detection of mutations using ultraspecific riboregulators[J]. Cell, 2020, 180(5): 1018-1032.e16. |
| 22 | RYLOTT E L, BRUCE N C. How synthetic biology can help bioremediation[J]. Current Opinion in Chemical Biology, 2020, 58: 86-95. |
| 23 | PARUCH L. Molecular diagnostic tools applied for assessing microbial water quality[J]. International Journal of Environmental Research and Public Health, 2022, 19(9): 5128. |
| 24 | YAASHIKAA P R, DEVI M K, KUMAR P S. Engineering microbes for enhancing the degradation of environmental pollutants: a detailed review on synthetic biology[J]. Environmental Research, 2022, 214(Pt 1): 113868. |
| 25 | THAVARAJAH W, VEROSLOFF M S, JUNG J K, et al. A primer on emerging field-deployable synthetic biology tools for global water quality monitoring[J]. Npj Clean Water, 2020, 3: 18. |
| 26 | DOU J, BENNETT M R. Synthetic biology and the gut microbiome[J]. Biotechnology Journal, 2018, 13(5): 1700159. |
| 27 | RAMACHANDRAN G, BIKARD D. Editing the microbiome the CRISPR way[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2019, 374(1772): 20180103. |
| 28 | MIMEE M, TUCKER A C, VOIGT C A, et al. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota[J]. Cell Systems, 2015, 1(1): 62-71. |
| 29 | SINGH R P, SHADAN A, MA Y. Biotechnological applications of probiotics: a multifarious weapon to disease and metabolic abnormality[J]. Probiotics and Antimicrobial Proteins, 2022, 14(6): 1184-1210. |
| 30 | MILLS H, ACQUAH R, TANG N, et al. The use of bacteria in cancer treatment: a review from the perspective of cellular microbiology[J]. Emergency Medicine International, 2022, 2022: 8127137. |
| 31 | CRESCI G A M, LAMPE J W, GIBSON G. Targeted approaches for in situ gut microbiome manipulation[J]. JPEN Journal of Parenteral and Enteral Nutrition, 2020, 44(4): 581-588. |
| 32 | BOBER J R, BEISEL C L, NAIR N U. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications[J]. Annual Review of Biomedical Engineering, 2018, 20: 277-300. |
| 33 | CAÑEZ C, SELLE K, GOH Y J, et al. Outcomes and characterization of chromosomal self-targeting by native CRISPR-Cas systems in Streptococcus thermophilus [J]. FEMS Microbiology Letters, 2019, 366(9): fnz105. |
| 34 | KUMAR P, SINHA R, SHUKLA P. Artificial intelligence and synthetic biology approaches for human gut microbiome[J]. Critical Reviews in Food Science and Nutrition, 2022, 62(8): 2103-2121. |
| 35 | VOORHEES P J, CRUZ-TERAN C, EDELSTEIN J, et al. Challenges & opportunities for phage-based in situ microbiome engineering in the gut[J]. Journal of Controlled Release, 2020, 326: 106-119. |
| 36 | BARANI M, RAHDAR A, SARGAZI S, et al. Nanotechnology for inflammatory bowel disease management: detection, imaging and treatment[J]. Sensing and Bio-Sensing Research, 2021, 32: 100417. |
| 37 | NJUE F, CHIH S. When to intervene for donor-specific antibody after heart transplantation[J]. Current Opinion in Organ Transplantation, 2019, 24(3): 271-278. |
| 38 | SU J A, BAXTER-LOWE L A, KANTOR P F, et al. The clinical impact of donor-specific antibodies on antibody-mediated rejection and long-term prognosis after heart transplantation[J]. Current Opinion in Organ Transplantation, 2019, 24(3): 245-251. |
| 39 | ROTH T L, MARSON A. Genetic disease and therapy[J]. Annual Review of Pathology, 2021, 16: 145-166. |
| 40 | BHARATHKUMAR N, SUNIL A, MEERA P, et al. CRISPR/Cas-based modifications for therapeutic applications: a review[J]. Molecular Biotechnology, 2022, 64(4): 355-372. |
| 41 | LIN H F, LI G, PENG X W, et al. The use of CRISPR/Cas9 as a tool to study human infectious viruses[J]. Frontiers in Cellular and Infection Microbiology, 2021, 11: 590989. |
| 42 | SHARMA G, SHARMA A R, BHATTACHARYA M, et al. CRISPR-Cas9: a preclinical and clinical perspective for the treatment of human diseases[J]. Molecular Therapy, 2021, 29(2): 571-586. |
| 43 | KANG K, SONG Y, KIM I, et al. Therapeutic applications of the CRISPR-Cas system[J]. Bioengineering, 2022, 9(9): 477. |
| 44 | PICKAR-OLIVER A, GERSBACH C A. The next generation of CRISPR-Cas technologies and applications[J]. Nature Reviews Molecular Cell Biology, 2019, 20(8): 490-507. |
| 45 | WANG X S, XIONG E H, TIAN T, et al. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay[J]. ACS Nano, 2020, 14(2): 2497-2508. |
| 46 | KLEINSTIVER B P, SOUSA A A, WALTON R T, et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing[J]. Nature Biotechnology, 2019, 37(3): 276-282. |
| 47 | CHEN J S, MA E B, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity[J]. Science, 2018, 360(6387): 436-439. |
| 48 | LI S Y, CHENG Q X, WANG J M, et al. CRISPR-Cas12a-assisted nucleic acid detection[J]. Cell Discovery, 2018, 4: 20. |
| 49 | LV H L, WANG J, ZHANG J, et al. Definition of CRISPR Cas12a trans-cleavage units to facilitate CRISPR diagnostics[J]. Frontiers in Microbiology, 2021, 12: 766464. |
| 50 | LI Y, LI S Y, WANG J, et al. CRISPR/Cas systems towards next-generation biosensing[J]. Trends in Biotechnology, 2019, 37(7): 730-743. |
| 51 | KHAN A, OSTAKU J, ARAS E, et al. Combating infectious diseases with synthetic biology[J]. ACS Synthetic Biology, 2022, 11(2): 528-537. |
| 52 | HUANG Z, TIAN D, LIU Y, et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis[J]. Biosensors & Bioelectronics, 2020, 164: 112316. |
| 53 | BROTO M, KAMINSKI M M, ADRIANUS C, et al. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs[J]. Nature Nanotechnology, 2022, 17(10): 1120-1126. |
| 54 | ZOU Y P, MASON M G, BOTELLA J R. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics[J]. PLoS One, 2020, 15(6): e0235216. |
| 55 | MUSTAFA M I, MAKHAWI A M. SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases[J]. Journal of Clinical Microbiology, 2021, 59(3): e00745-e00720. |
| 56 | LEE R A, PUIG H, NGUYEN P Q, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(41): 25722-25731. |
| 57 | MANGHWAR H, LINDSEY K, ZHANG X L, et al. CRISPR/Cas system: recent advances and future prospects for genome editing[J]. Trends in Plant Science, 2019, 24(12): 1102-1125. |
| 58 | KIM J, CAMPBELL A S, DE ÁVILA B E F, et al. Wearable biosensors for healthcare monitoring[J]. Nature Biotechnology, 2019, 37(4): 389-406. |
| 59 | NGUYEN P Q, SOENKSEN L R, DONGHIA N M, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection[J]. Nature Biotechnology, 2021, 39(11): 1366-1374. |
| 60 | RHEA K A, MCDONALD N D, COLE S D, et al. Variability in cell-free expression reactions can impact qualitative genetic circuit characterization[J]. Synthetic Biology, 2022, 7(1): ysac011. |
| 61 | SADAT MOUSAVI P, SMITH S J, CHEN J B, et al. A multiplexed, electrochemical interface for gene-circuit-based sensors[J]. Nature Chemistry, 2020, 12(1): 48-55. |
| 62 | LAI W, XIONG X W, WANG F, et al. Nonlinear regulation of enzyme-free DNA circuitry with ultrasensitive switches[J]. ACS Synthetic Biology, 2019, 8(9): 2106-2112. |
| 63 | KIM H, SKINNER D J, GLASS D S, et al. 4-Bit adhesion logic enables universal multicellular interface patterning[J]. Nature, 2022, 608(7922): 324-329. |
| 64 | NAJJAR D, RAINBOW J, SHARMA TIMILSINA S, et al. A lab-on-a-chip for the concurrent electrochemical detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva and plasma[J]. Nature Biomedical Engineering, 2022, 6(8): 968-978. |
| 65 | BHADRA S, POTHUKUCHY A, SHROFF R, et al. Cellular reagents for diagnostics and synthetic biology[J]. PLoS One, 2018, 13(8): e0201681. |
| 66 | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 67 | OOIJEVAAR R E, TERVEER E M, VERSPAGET H W, et al. Clinical application and potential of fecal microbiota transplantation [J]. Annual Review of Medicine, 2019, 70: 335-351. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems [J]. Synthetic Biology Journal, 2025, 6(1): 105-117. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | CHEN Yingying, LIU Yang, SHI Junjie, MA Junying, JU Jianhua. CRISPR/Cas systems and their applications in gene editing with filamentous fungi [J]. Synthetic Biology Journal, 2024, 5(3): 672-693. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||